Abstract
The relative probabilities that different pairs of chromosomal loci will collide with one another in vegetatively growing diploid yeast cells have been assessed using a genetic assay for Cre/loxP site-specific recombination. Recombination rates have been determined for 18 different pairs of loxP sites representing diverse pairs of positions within the genome. Overall, relative collision probabilities vary over an eightfold range. Within this range, a hierarchy comprising three levels of organization can be discerned. First, collisions between loci on nonhomologous chromosomes are governed by nonspecific centromere clustering. Second, a sequence is closer to allelic or nearby sequences on its homolog than to sequences on nonhomologous chromosomes, an effect most simply attributed to homolog pairing. Third, a sequence can be closer to other sequences nearby on the same chromosome than to sequences on other chromosomes. These findings provide a framework for assessing the role of chromosome disposition in cellular processes such as DNA repair and gene expression. Also the possibility is raised that genome-wide coalignment of homologs is not the fundamental raison d’etre of the somatic pairing process. We suggest instead that pairing may exist to promote juxtaposition of homologous regions within irregular genome complements.
Keywords: Saccharomyces cerevisiae, site-specific recombination, chromosome, nucleus, homolog pairing, DNA repair
Chromosome organization, disposition, and behavior have been investigated by many experimental approaches. A handful of studies have used site-specific recombination reactions to probe chromosome status in living cells. In bacteria, the phage λ Int reaction has been used to assess in vivo supercoiling levels (Bliska and Cozzarelli 1987); the bacteriophage P1 Cre/loxP recombination reaction has been used to assess effective DNA concentration (Hildebrandt and Cozzarelli 1995); and the γ–δ resolvase reaction has been used to probe for domainal organization of chromosomes (e.g., Staczek and Higgins 1998). Also, in Drosophila, the yeast Flp/frt recombination reaction has been used to assess the effect of chromosomal inversions on pairing of allelic regions lying distal to the rearrangement breakpoints (Golic and Golic 1996b) and to compare intrachromosomal versus interchromosomal recombination rates (Golic and Golic 1996a).
The current study exploits Cre/loxP site-specific recombination to examine chromosome status in vegetatively growing diploid budding yeast cells. A genetic assay for Cre/loxP recombination in yeast has been developed and used to determine in vivo recombination rates for 18 different pairs of loxP inserts representing various pairs of positions within the genome. The goal of this study was to assess the relative probabilities with which different regions of the genome collide with one another. Cre/loxP recombination seemed to be an appropriate tool for such analysis because the amount of product formed depends on the square of the loxP concentration in vitro and retains its dependence on loxP concentration in vivo in Escherichia coli (Hildebrandt and Cozzarelli 1995). Thus, the relative levels of recombination for pairs of loxP sites inserted at various positions in the yeast genome should reflect the relative probabilities of collision for the corresponding pairs of loci.
The array of recombination rates among various pairs of loxP inserts in the yeast genome could provide several types of information. Suitable comparisons among particular recombination rates should identify specific features that govern interlocus collisions. Thus, for example, if homolog pairing significantly constrains interchromosomal collisions, the recombination rates between allelic loci on homologs should be greater than those for loci on nonhomologous chromosomes. Furthermore, if more than one feature is found to affect recombination rates, their relative importance should be reflected by quantitative differences in the corresponding rates. Finally, the total range of values observed among different pairs of loci should give an indication of the extent to which interlocus collisions are constrained by various features within the nucleus.
In budding yeast, several candidates for features that might constrain relative chromosomal positions are known from cytological studies. Centromeric regions are clustered, and the cluster occurs at the nuclear periphery, two features which together comprise the so-called Rabl orientation (Rabl 1885). This configuration appears to be present essentially throughout the cell cycle (Jin et al. 1998). Also, telomeres tend to form multiple clusters and to occur at or near the nuclear periphery (e.g., Gotta et al. 1996); whether interstitial regions behave similarly, as in Drosophila (Marshall et al. 1996), is unknown. Also, in diploid yeast cells, homologs are directly paired, coaligned along their lengths via multiple interstitial interactions that occur roughly once per 70 kb (Burgess et al. 1999).
Overall, a complete data set should provide a baseline for assessing whether any endogenous process involving contact between two regions of the genome, for example, recombinational repair of DNA damage, shows a similar sensitivity to chromosome disposition.
The findings presented below suggest that interchromosomal collisions in diploid yeast cells are governed by at least three different features.
Results
The assay
Cre recombinase promotes crossing over between a pair of 34-bp loxP sites (Hoess et al. 1982; Abremski et al. 1983; Fig. 1A). To determine Cre/loxP recombination rates in vivo by a genetic assay, two differentially marked loxP sites were constructed (Fig. 1B). In one construct, a loxP site is immediately downstream of a strong promoter (pGPD1–loxP); in the other construct, loxP is immediately upstream of a promoterless ura3 gene (loxP–ura3). Cre-promoted recombination between the two sites creates a pGPD1–loxP–ura3 fusion that confers a Ura+ phenotype. The frequency of Ura+ prototrophs in a growing yeast culture should reflect the rate of Cre/loxP recombination between the two marked loci.
Figure 1.
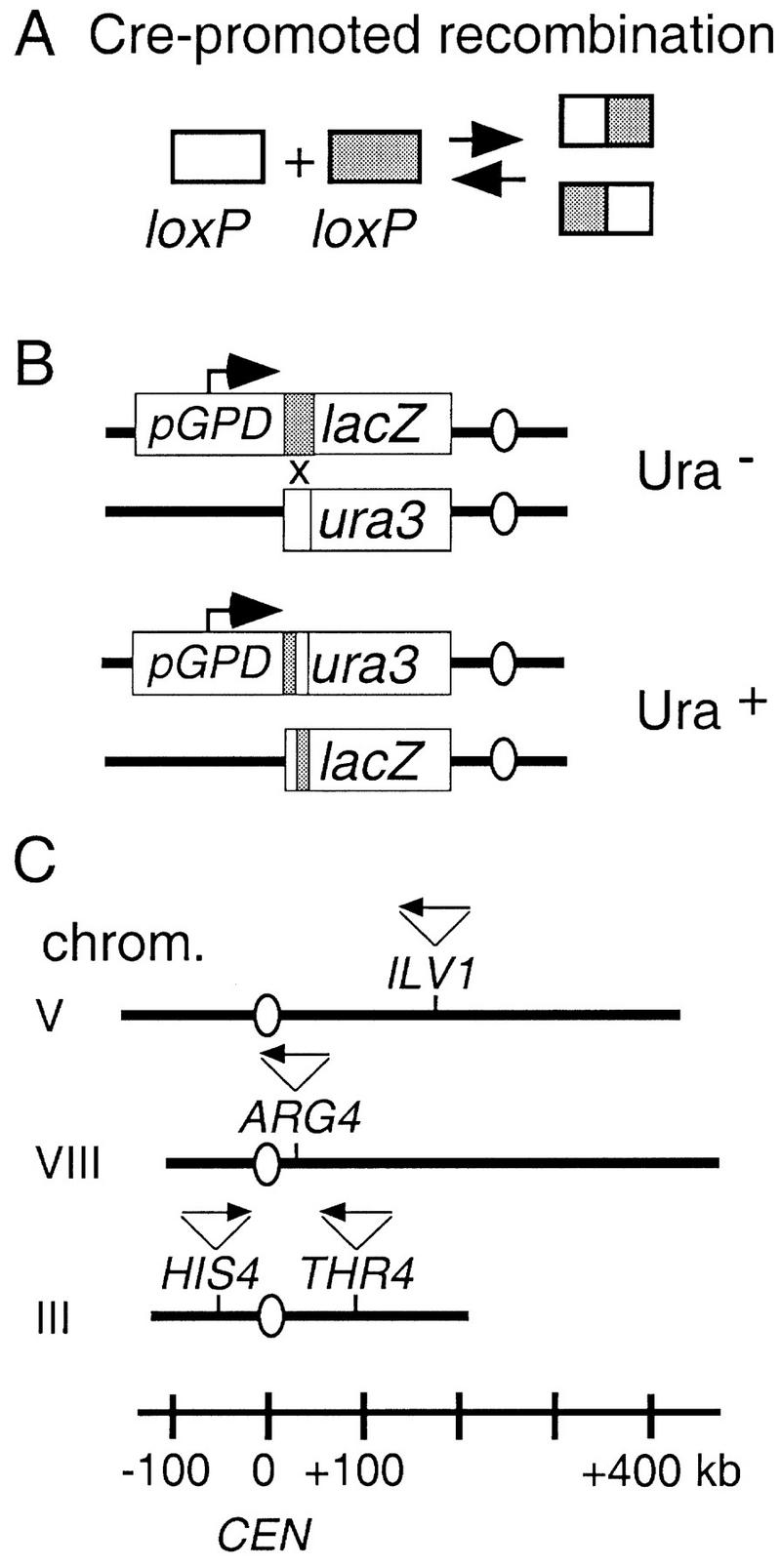
Cre/loxP recombination assay. (A) Cre-mediated recombination occurs between two 34-bp loxP sites (shown in white and gray boxes). (B) Cre/loxP recombination creates a pGPD1–loxP–ura3 fusion and a Ura+ phenotype. (C) Position and orientation of loxP sites integrated into the yeast genome for this study. The loxP sites are oriented so that recombination between sites located on two chromosomes results in the reciprocal exchange of chromosome arms. For loxP sites located on the same chromosome (e.g., at THR4 and HIS4) recombination results in an inversion of the region between the two loci. The position of the centromere is represented by an open circle.
Each version of the loxP construct was integrated at each of four loci (HIS4, ARG4, ILV1, and THR4; Fig. 1C) and diploid strains were constructed that contain one loxP site of each type (below). None of the Cre-promoted recombination events in these strains will generate acentric or dicentric chromosomes. Cre recombinase is expressed from a GAL1 promoter provided on an autonomous plasmid.
Formation of Ura+ prototrophs is dependent on Cre recombinase: The presence of an uninduced Cre gene increases the level of prototrophs by three to four orders of magnitude (Table 1; cf. first four columns). Induction of Cre expression by exposure of cells to galactose prior to plating increases the level an additional three orders of magnitude (Table 1; cf. fourth and sixth columns). Formation of Ura+ prototrophs also strongly depends on the presence of one loxP site of each type: Elimination of either insert reduces the level of Ura+ prototrophs by several orders of magnitude, essentially to background levels (Table 1, cf. line 1 with lines 3 and 4). A small Cre-dependent increase in prototroph formation can be observed under conditions of induced Cre expression for the loxP–ura3 construct, but not the pGPD1–loxP construct. This increase probably reflects recombination between the marked insert and endogenous pseudo-loxP sites located downstream of promoter regions, as seen in other studies (Sauer 1992). Finally, for the two representative reactions shown in Table 1, the relative levels of recombination products detected after Cre induction by physical analysis, that is, as PCR products or as diagnostic restriction fragments, are comparable with those detected genetically (data not shown), suggesting that variations in the frequencies of Ura+ cells reflect differences in the rates of recombination rather than in the viability or recovery of cells containing different types of recombinant chromosomes (see also below).
Table 1.
Dependence of Ura+ prototroph formation on Cre protein and two loxP inserts
| Recombination ratea
|
Recombinant frequencyb
|
||||||
|---|---|---|---|---|---|---|---|
| Sites of loxP inserts
|
−Gal | −Gal | +Gal | ||||
|
pGPD–loxP
|
loxP–ura3
|
−Cre
|
+Cre
|
−Cre
|
+Cre
|
−Cre
|
+Cre
|
| 1. ARG4 | ARG4 | 2.3 × 10−8 | 1.7 × 10−5 | 6 × 10−8 | 3.8 × 10−4 | 3 × 10−7 | 1.2 × 10−1 |
| 2. ARG4 | ILV1 | 2.7 × 10−8 | 0.8 × 10−5 | 0.8 × 10−8 | 0.9 × 10−4 | 5 × 10−7 | 0.4 × 10−1 |
| 3. None | ARG4 | <1 × 10−7 | 1.8 × 10−6 | ||||
| 4. ARG4 | none | <1 × 10−7 | <1 × 10−7 | ||||
Each value represents the average of three independent cultures assayed in parallel; s.d. ± 20%.
Determined by fluctuation test, which gives the rate of recombination per cell generation (Materials and Methods).
Determined by plating of cultures on selective and nonselective media with or without an immediately preceding 2-hr exposure to galactose; galactose was absent after plating (Materials and Methods). Recombinant frequency is the number of recombinants per viable colony forming unit.
Data set
Eighteen different diploid strains were analyzed, each carrying one loxP construct of each type, and together representing all possible pairwise combinations of inserts (Fig. 2A). In four of these strains, the two loxP sites are at allelic positions on homologs; the corresponding interactions are referred to as allelic. In 10 strains, the two loxP sites are present on two nonhomologous chromosomes; the corresponding interactions are referred to as nonhomolog. In the four remaining strains, the two loxP inserts are present on homologs but at nonallelic positions lying ∼150 kb apart, at HIS4 and THR4 on Chromosome III. In two of these strains, the two constructs were present on homologs; in the other two they were present on the same chromosome. These two situations are referred to, respectively, as nearby interhomolog (III-trans) and intrachromosomal (III-cis).
Figure 2.
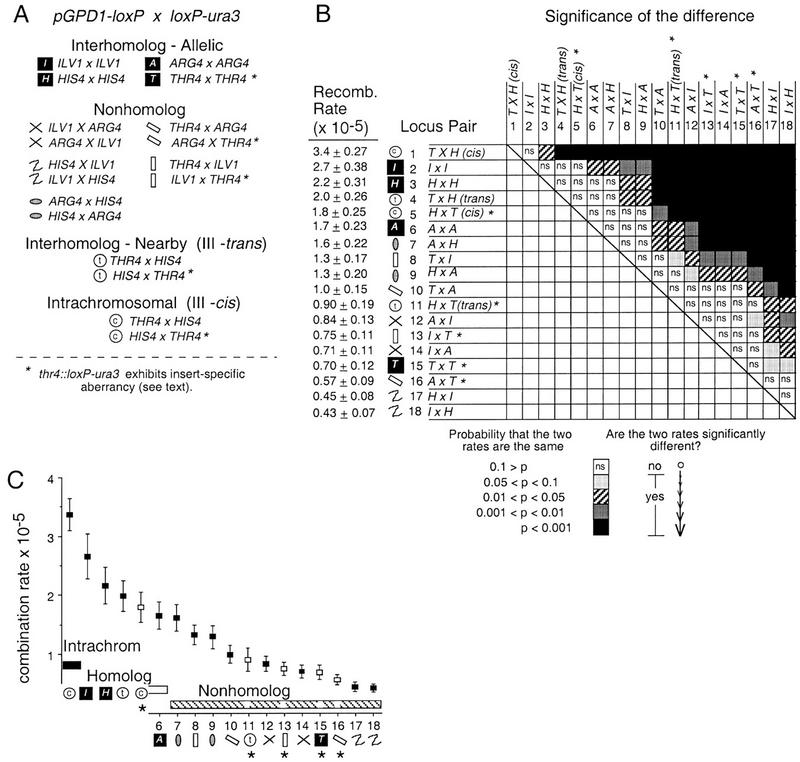
Recombination rates among 18 pairs of loci containing loxP site constructs. (A) Four different types of interactions were measured by Cre/loxP recombination: (1) interhomolog-allelic: loxP sites located at allelic positions on homologous chromosomes, (2) nonhomolog: loxP sites located on nonhomologous chromosomes, (3) interhomolog-nearby: loxP sites at nonallelic positions on homologous chromosomes, and (4) intrachromosomal: loxP sites on the same physical chromosome. Reciprocal pairs of loxP tester constructs are denoted by the same symbol. (B) For each strain, a recombination rate (± s.d.) was determined by fluctuation analysis and is reported in descending rank order at left. Loci are abbreviated by the first letter of the locus name. The type of loxP site insertion is always represented as pGPD1–loxP–lacZ × loxP–ura3. The probability of any two measured rates being different from one another was determined by a two-tailed z-test. Results of this analysis are indicated in the matrix by the appropriate shading corresponding to the calculated p value (ns, not significant). (C) Plot of recombination rate values for all 18 strains (identified by number in B) in descending rank order from left to right. The type of chromosome interaction being measured is indicated by the same symbols as in A and B. Strains containing the thr4::loxP–ura3 construct are marked with an asterisk and are not included within the groupings as indicated by lack of shading (see text for details).
Recombination rates were measured in all 18 strains by fluctuation analysis. Data are presented for cells grown and assayed under conditions in which Cre recombinase expression was not induced. Each single fluctuation test provides the rate of recombination per cell generation, and its corresponding variance (and hence its standard deviation), all of which were calculated by the method of the median as described by Lea and Colson (1948). The rates observed for the 18 strains, plus evaluation of the statistical significance of the differences between all possible pairs of rates, are presented in Figure 2B. For convenience, the data have been arrayed in rank order, from the greatest to the lowest rate, and the strains have been numbered correspondingly from 1 to 18. For easier visualization, the recombination rates (with their standard deviations) are also presented graphically in rank order (Fig. 2C). It can be noted that fluctuation tests are extremely precise. The standard deviation for any single test is only 10% to 15% of the measured rate (Fig. 2B); the rates obtained from independent fluctuation tests of any single strain vary by <15% of the mean value (data not shown).
Analogous fluctuation analysis was not possible under conditions of constitutive Cre expression because cell growth and viability are compromised. When examined by a pulse-induction protocol, however, the 18 strains exhibited patterns of relative recombinant levels analogous to those observed by fluctuation analysis under noninducing conditions (see Table 1; data not shown).
Evaluation of insert-specific effects
The 14 strains carrying nonallelic inserts represent 7 different locus combinations, each represented by a pair of reciprocally related strains that differ with respect to which loxP construct is at which locus (Fig. 2A). In the absence of complications, the two members of each reciprocal pair should exhibit the same recombination rate. For all combinations of inserts at ILV1, ARG4, and HIS4, reciprocally related pairs of strains exhibit indistinguishable recombination rates (Table 2, I), implying an absence of insert-specific effects for the six loxP inserts represented by this group. In contrast, for recombination between THR4 and each of the other three loci, reciprocity is not observed. Instead, for each case involving THR4, the strain carrying thr4::loxP–ura3 exhibits half the recombination rate of its reciprocal partner. There are four different interactions of this type, two nonhomolog cases and the two different arrangements of HIS4 and THR4 (cis and trans) (Table 2, II). These data suggest that at least one of the two thr4 inserts is aberrant.
Table 2.
Comparison of reciprocal interaction rates
| Interactions compareda
|
Recombination rates (×10−5)b
|
Likelihood that rates differ (confidence level, %)b
|
|---|---|---|
| I. Not involving THR4 | ||
| H × I; I × H | 0.45 vs. 0.43 | <90 |
| A × I; I × A | 0.84 vs. 0.71 | <90 |
| A × H; H × A | 1.6 vs. 1.3 | <90 |
| II. Involving THR4 | ||
| T × A; A × T | 1.0 vs. 0.57 | >99 |
| T × I; I × T | 1.3 vs. 0.75 | >95 |
| T × H (cis); H × T (cis) | 3.4 vs. 1.8 | >99.9 |
| T × H (trans); H × T (trans) | 2.0 vs. 0.9 | >99.9 |
aAll loci are represented as pGPD–loxP × loxP–ura3.
From Fig. 2; Confidence level = [(1 − P value) × 100%].
Two types of aberrancies are possible: the thr4:: pGPD1–loxP insert could exhibit an aberrantly high recombination rate or the thr4::loxP–ura3 insert could exhibit an aberrantly low rate. Additional data support the latter possibility. Allelic recombination rates for HIS4, ARG4, and ILV1 differ relatively little from one another and are as high or greater than any other rates (Table 3, I; further discussion below). In contrast, the allelic recombination rate for THR4 is substantially lower (2.4- to 3.8-fold) than any of the other allelic rates (Table 3, II), which suggests that at least one of the thr4 inserts is exerting a depressive effect on recombination rate. Thus, all data are explained if thr4::loxP–ura3 confers a construct-specific depression of recombination rate irrespective of its recombinational partner. We conclude that thr4::loxP–ura3 has an insert-specific aberrancy, whereas the other seven inserts lack any such peculiarity. These findings suggest that, overall, locus-specific effects are the exception rather than the rule and that, when they do occur, they can be sensitively detected. The atypical behavior of the thr4::loxP–ura3 construct could reflect, for example, reduced Cre binding or a reduced probability that collision with another loxP site is effective in generating a recombinant.
Table 3.
Comparison of allelic interaction rates
| Allelic interactions compared
|
Recombination rates (×10−5)a
|
Likelihood that rates differ (confidence level, %)a
|
|---|---|---|
| I. Not involving THR4 | ||
| H × H; I × I | 2.2 vs. 2.7 | <90 |
| A × A; I × I | 1.7 vs. 2.7 | >95 |
| A × A; H × H | 1.7 vs. 2.2 | <90 |
| II. Involving THR4 | ||
| T × T; A × A | 0.7 vs. 1.7 | >99.9 |
| T × T; I × I | 0.7 vs. 2.7 | >99.9 |
| T × T; H × H | 0.7 vs. 2.2 | >99.9 |
From Fig. 2; Confidence level = [(1 − P value) × 100%].
In light of these findings, it would be legitimate to exclude recombination rates for the five strains carrying thr4::loxP–ura3 from consideration during analysis of the data. However, most of the patterns described below are sufficiently robust that they are apparent and statistically significant even if the known-to-be-aberrant cases are included (Discussion).
Recombination rates vary over a relatively small range
Recombination rates for loxP pairs located on different chromosomes, whether homologs or nonhomologs, vary over a total range of about sixfold (Fig. 2B,C; strain 2 vs. strain 18) or, when the intrachromosomal rate is included (strain 1), about eightfold. Whereas this total range of values is relatively small, the differences in recombination rate for most pairs of strains (101/153) are significantly different from one another as judged by a two-tailed z-test, P < 0.05 (Fig. 2B).
The intrachromosomal recombination rate between HIS4 and THR4 is greater than the corresponding interhomolog recombination rate
The intrachromosomal rate for thr4::pGPD1–loxP by his4::loxP–ura3 is the greatest of all the rates observed, significantly so for all but the next greatest rate (Fig. 2B). The rate of recombination between THR4 and HIS4 is nearly twice as high when the corresponding loxP inserts are present on the same homolog (in cis) as when they are present on two homologs (in trans). This can be observed both for strains carrying thr4::pGPD1–loxP and his4::loxP–ura3 inserts (strain 1 vs. strain 4), and for the reciprocally related pair of strains carrying his4::pGPD1–loxP and thr4::loxP–ura3 (strain 5 vs. strain 11); in the latter case, the aberrant behavior of the thr4::loxP–ura3 insert apparently affects cis and trans situations similarly.
Allelic interhomolog recombination rates are greater than nonhomolog recombination rates
When recombination rates are arrayed in rank order, a consistent pattern emerges: All interhomolog interaction rates for allelic loci are greater than all nonhomolog interaction rates (omitting thr4::loxP–ura3 strains; Fig. 2C).
This pattern is confirmed by statistical evaluation. First, the mean rate for all allelic pairs is twice the mean rate for all nonhomolog pairs, irrespective of whether the aberrant construct is included or not, with a confidence level of 99.9% in the latter case (Table 4). Second, every allelic interaction rate is greater than any rate for a nonhomolog interaction involving the same locus and all of these differences are statistically significant except in some of the cases involving both HIS4 and ARG4 (which are accounted for below). This feature can be seen in both the total data set (Fig. 3A) and by considering loci two at a time (Fig. 3B).
Table 4.
Comparison of mean rates for all allelic homolog interactions and nonhomolog interactions
| Comparison type
|
Mean ratea (×10−5)
|
Likelihood that mean rates differ (confidence level, %)b
|
|---|---|---|
| I. All pairs excluding those containing thr4::loxP–ura3c | ||
| Allelic | 2.2 ± 0.5 | |
| Nonhomolog | 0.95 ± 0.50 | 99.9 |
| II. All pairs including those containing thr4::loxP–ura3d | ||
| Allelic | 1.8 ± 0.9 | |
| Nonhomolog | 0.90 ± 0.4 | 95–99 |
Calculated from individual values in Fig. 2.
Determined by two-tailed t-test. Confidence level = [(1 − P value) × 100%].
Allele pairs: H × H; A × A; I × I. Nonhomolog pairs: H × A; A × H; A × I; I × A; H × I; I × H; T × H; T × I; T × A.
Allelic pairs: H × H; A × A; I × I; T × T. Nonhomolog pairs: H × A; A × H; A × I; I × A; H × I; I × H; T × H; H × T; T × I; I × T; T × A; A × T.
Figure 3.
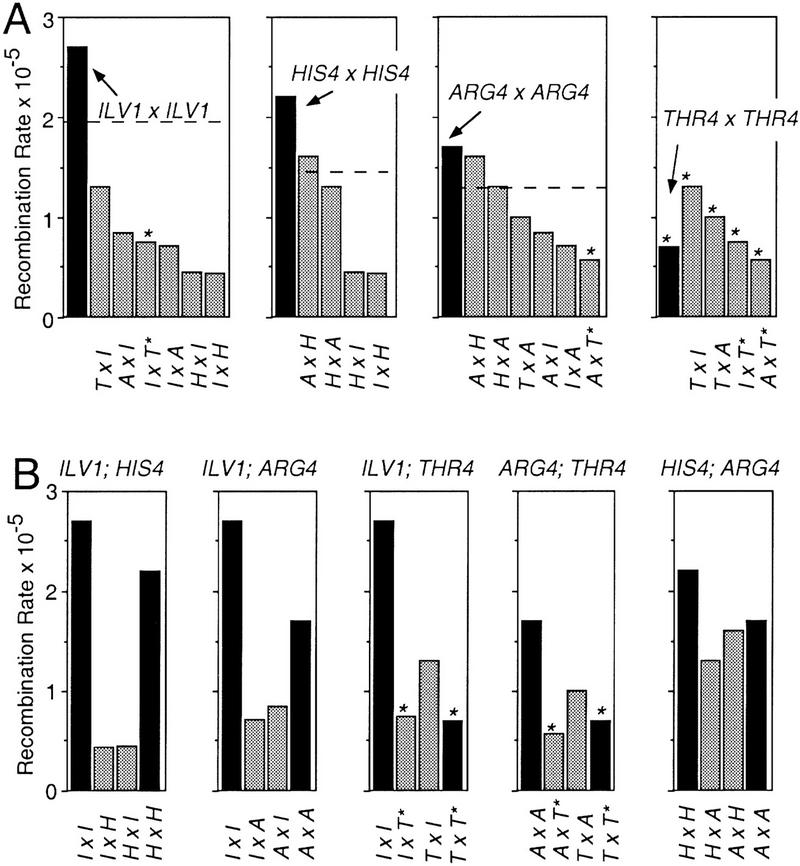
Allelic rates versus related nonhomolog rates. (A) Each interhomolog-allelic recombination rate (black bars) is shown separately with all related nonhomolog recombination rates (gray bars). The broken line in each panel represents a value equal to two standard deviations from the mean interhomolog recombination rate (95% confidence level). (Asterisks) Strains containing the thr4::loxP–ura3 construct, which behaves aberrantly (see text for details). (B) Interhomolog-allelic rates for any two loci are shown with the two related reciprocal nonhomolog rates as in A.
Interestingly, the recombination rates for the three allelic interactions (excluding THR4) are very similar, ranging from 1.7 ×10−5 to 2.7 × 10−5. Among these rates, only the two extreme values, for ARG4 and ILV1, are signficantly different (Table 3, I).
Recombination rates for loci on nonhomologous chromosomes vary directly with differential locus-to-centromere distance
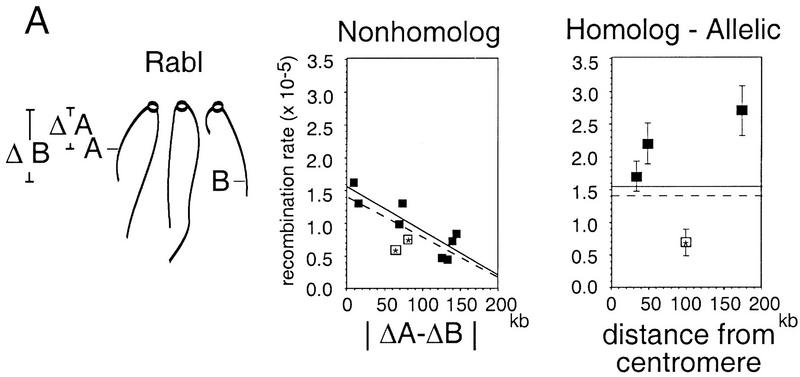
Centromere clustering should have the effect of colocalizing all loci located at similar distances from their centromeres, particularly so in the Rabl configuration in which centromeres are clustered at one edge of the nucleus (Fig. 4A). Such an effect should be reflected in the rates of recombination between loxP sites located on nonhomologous chromosomes, in which pairing and other possible homolog-specific determinants are absent. Rates of recombination for pairs of loci on nonhomologs should be greater when the two loci are located at similar distances from their centromeres and lower when their locus-to-centromere distances are more disparate. Such a relationship is seen among the pairs of loci examined in this study, with or without inclusion of the aberrant thr4::loxP–ura3 construct; the data are best fit by a linear curve (Fig. 4A, middle; see Fig. 2B for significance of individual rate differences).
Figure 4.
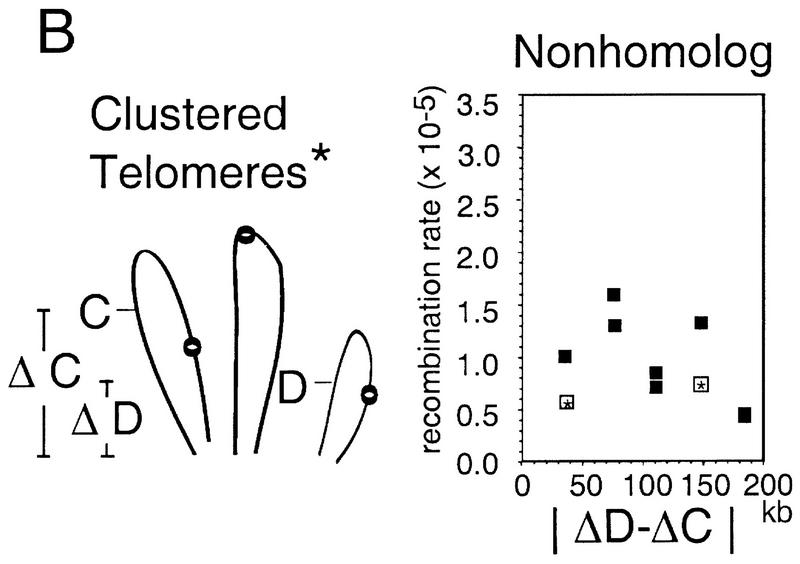
Relative contributions of the Rabl orientation and homolog pairing to Cre/loxP recombination rates. (A) The Rabl configuration of chromosomes with centromeres marked by open circles is shown. The distance in kb each loxP site is from its respective centromere is denoted by ΔA and ΔB (left). Each nonhomolog recombination rate is plotted against the absolute difference in the distance the two sites are from their respective centromeres (|ΔA − ΔB|; middle). The correlation between the relative position of loxP sites in the Rabl and the observed recombination rate is significant (broken line, R2 = 0.59, P < 0.01). For eight points, not including the thr4::loxP–ura3 construct (open boxes with asterisks), this significance is even greater (solid line, R2 = 0.77, P < 0.01). The y intercepts of the fitted lines from the left-hand plot (where |ΔA − ΔB| = 0) represent minimum rates expected for two allelic loci by virtue of their relative chromosome position in the Rabl orientation. Significant differences from these values are observed for the HIS4 × HIS4, ILV1 × ILV1, and THR4 × THR4 pairs (z-test, P < 0.05). (B) No contribution of telomere clustering on Cre/loxP recombination rates between nonhomologous chromosomes is detected. (Asterisk) One of many possible arrangements of telomeres within the nucleus that was tested. For this particular arrangement, each nonhomolog rate is plotted against the absolute difference in distance each site is from the nearest telomere (|ΔD − ΔC|). No significant effect on recombination rate was found for loxP sites positioned in this particular arrangement (R2 = 0.19, P > 0.1) or in other related variations (not shown).
This approach can also be used analogously to determine whether recombination rates are significantly affected by telomere clustering. No evidence of such an effect can be seen (Fig. 4B).
It is worth noting that none of the loci examined in this study is farther than ∼200 kb from its respective centromere; thus it is possible that the effects of centromere clustering would be less for more centromere-distal loci. Similarly, none of the loci examined are located near the ends of long chromosomes (and thus far from their centromeres); thus, effects of telomere clustering might be detected by examination of more telomere–proximal loci.
Recombination rates for loci on homologs are greater than expected from centromere clustering
Because allelic loci on homologs necessarily lie at the same distance from their respective centromeres, nonspecific centromere clustering could in principle account for their greater rates of recombination as compared with nonallelic loci. This possibility can be evaluated by comparing homolog recombination rates with the recombination rate predicted for nonhomologous loci located at the same distance from their respective contromeres. The latter value is given by extrapolation of the relationship between locus-to-centromere distance and recombination rate to zero difference, which yields a predicted Cre/loxP recombination rate of 1.6 × 10−5 ± 0.1 × 10−5. Because this extrapolation is based on analysis of relatively centromere–proximal locus pairs, it is likely to be a maximum estimate of clustering influence (above). Even so, the rates of recombination for loci on homologs, both allelic and nearby, are all greater than this value (excluding thr4::loxP–ura3 strains), significantly so in all but one case (1.7 × 10−5 to 2.7 × 10−5; Fig. 4A, right). Thus, the greater recombination rates for allelic or nearby loci on homologs, as compared with nonallelic loci, appear to reflect the existence of one or more homolog-specific constraints (Discussion).
Relative contributions of homolog-specific effects and centromere clustering to colocalization of homologs
Homolog-specific effects and centromere clustering can each contribute to the colocalization of allelic loci. We wondered, however, whether homolog-specific effects might predominate such that centromere clustering is effectively irrelevant for allelic loci. That is, if it were possible to subtract out any effect of the Rabl, would the relative proximity of allelic loci decrease or remain unchanged? The available data seem to favor the latter possibility. Overall, allelic interhomolog rates tend to be more uniform than rates among loci on nonhomologous chromosomes (ranges of 1.6-fold and 2.6-fold, respectively), which could reflect an overriding homolog-specific effect. Also, if centromere clustering played a significant role for allelic loci, it should result in greater recombination rates for allelic near-centromere loci than for allelic centromere–distal loci. No such effect is apparent, however, interhomolog recombination at the ILV1 locus, located nearly 200 kb from its centromere, is the greatest among those examined, whereas interhomolog recombination at the ARG4 locus, located ∼25 kb from its centromere, is least among those examined (excluding THR4) (Fig. 4A, right).
As a consequence of the above patterns, recombination rates for loci located on nonhomologous chromosomes, but at similar distances from their centromeres, approach the general rate for allelic interhomolog interactions. This effect presumably accounts for the fact that allelic rates at HIS4 and ARG4 are only slightly greater than the corresponding nonhomolog rates (above) because HIS4 and ARG4 exhibit the smallest locus-to-centromere distance among the various pairs of loci studied (Δ10 kb, Fig. 4A).
Interhomolog recombination rates are the same for two nearby pericentric loci as for allelic loci
HIS4 and THR4 both map to Chromosome III and are separated by a total of ∼150 kb (Fig. 1D). Recombination between thr4::pGPD1–loxP and his4::lox–ura3, in a strain in which the two constructs are located on homologs, occurs at a rate of 2.0 × 10−5 (Fig. 2B,C). This value is greater than the rates for any of the nonhomolog interactions examined (Fig. 2B) or the mean of such interactions (Table 5). This value is also statistically indistinguishable from the allelic interhomolog recombination rate at HIS4 or at any other locus (excluding THR4) (Table 5). Thus, the rate of recombination between two loci on homologs can be greater than that for nonhomolog rates, even if those two loci are at somewhat different positions on their respective chromosomes.
Table 5.
Rate for nearby loci on homologs as compared to allelic homolog and nonhomolog rates
| Interactions compared
|
Recombination rate(s) (×10−5)a
|
Likelihood that rate differs from THR4 × HIS4 (confidence level, %)a
|
|---|---|---|
| THR4 × HIS4 (trans) | 2.0 | |
| I. Allelic pairs: | ||
| I × I; H × H; A × A | 2.7; 2.2; 1.7 | <90; <90; <90 |
| II. Nonhomolog pairs | ||
| T × A; T × I | 1.0; 1.3 | >99.9; >95 |
| A × I; I × A | 0.84; 0.71 | >95; >99 |
| H × I; I × H | 0.45; 0.43 | >99.5; > 99.9 |
| A × H; H × A | 1.6; 1.3 | <90; >95 |
From Fig. 2; Confidence level = [(1 − P value) × 100%].
HIS4 and THR4 are somewhat special in that they happen to lie on either side of the chromosome III centromere (Fig. 1C). The Rabl configuration should tend to give each individual chromosome a U shape, which could, in turn, tend to specifically promote colocalization of pericentrically disposed loci. It remains to be determined whether or not such an effect contributes significantly to the HIS4 × THR4 interhomolog recombination rate. Conversely, systematic analysis of how intrachromosomal recombination rates vary with inter–locus distance along a chromosome arm might reveal whether chromosome stiffness ever becomes a limiting factor for recombinational interactions.
Most detected Cre-loxP recombination events occur prior to DNA replication
Ura+ prototrophs could arise from a Cre-loxP recombination event between unreplicated chromosomes (G1) or after the loxP regions have been replicated (G2; we will ignore the presumably minor contribution of events that might occur during S phase between one replicated loxP and one unreplicated loxP). It was of interest to know the relative contributions of events in the two situations and, in particular, whether strains carrying different types of loxP partners were similar or different in this regard.
If a recombination event occurs in telophase or G1, the ensuing round of DNA replication, chromosome segregation, and cell division will yield two identical Ura+ cells, each containing a pair of reciprocally related recombinant chromosomes. If recombination occurs after DNA replication, ensuing events will yield only one Ura+ cell; moreover, according to whether the two recombinant chromosomes segregate to the same or different daughter cells, the Ura+ daughter will have either the balanced G1 chromosome configuration or an unbalanced configuration comprising one recombinant and one nonrecombinant chromosome. The relative frequencies of the balanced and unbalanced configurations among Ura+ prototrophs was examined in strains carrying either allelic or nonhomolog loxP pairs by PCR analysis. Products diagnostic for each nonrecombinant or recombinant loxP containing construct (nonrecombinant loxP–ura3 and pGPD1–loxP–lacZ or recombinant pGPD–loxP–ura3 and loxP–lacZ) could be detected independent of the sites of the inserts with suitable pairs of primers homologous to sequences within the constructs themselves.
In three strains containing loxP sites at allelic-homolog positions (ILV1 × ILV1; ARG4 × ARG4, and HIS4 × HIS4), among independently arising Ura+ prototrophs, the fractions containing the unbalanced configuration were 1/29, 0/19, and 1/30, respectively. Among these 78 Ura+ prototrophs, 74 represented 37 prereplication events, each of which yielded two recombinant cells, whereas 4 represented 4 postreplication events, each of which yielded a single recombinant cell (Materials and Methods). Thus, most Ura+ prototrophs, 74/78, represent prereplication events, whereas only 4/78 represent postreplication events. Furthermore, among recombination events that are represented by Ura+ prototrophs, 37/41 (90%) occurred between unreplicated chromosomes, whereas 4/41 (10%) occurred between replicated chromosomes.
Very similar results were observed in five nonhomolog strains: In ARG4 × HIS4, HIS4 × ARG4, ARG4 × ILV1, ILV1 × ARG4, and ILV1 × HIS4, the unbalanced configuration was observed in 2/20, 0/20, 3/26, 0/30, and 3/29 independent Ura+ colonies, respectively. Thus, unbalanced recombinant constructs were recovered for each of the three nonhomolog locus pairs represented (ARG4 and HIS4, 2/40; ARG4 and ILV1, 3/56; ILV1 and HIS4, 3/29). Correspondingly, 109/125 Ura+ prototrophs represent prereplication events, whereas 16/125 represent postreplication events. Furthermore, 55/71 (77%) events represented by Ura+ prototrophs arose between unreplicated chromosomes, whereas 16/69 (23%) arose between replicated chromosomes.
In strains that examine nonhomolog recombinational interactions, the occurrence of the unbalanced configuration of loxP constructs is accompanied by an unbalanced chromosome complement. In contrast, in strains that examine allelic-homolog interactions, no chromosome inbalance occurs. The fact that similar fractions of pre- and postreplication events are recovered in both situations suggests that cells with unbalanced chromosome complements are recovered efficiently as Ura+ prototrophs. In accord with this suggestion, Ura+ strains expected to carry unbalanced chromosome complements exhibited no apparent reduction in colony size.
Because pre- and postreplication events contribute differently to the final pool of Ura+ recombinants (above), differences in recombinant frequencies could, in principle, reflect systematic differences in the relative frequencies of the two types of events. This does not appear to be the case. The five strains in which pairs of loxP sites are located on nonhomologs include two in which the loxP sites are located at very similar distances from their respective centromeres, HIS4 × ARG4 and ARG4 × HIS4, and three in which the loxP sites lie at very disparate locus-to-centromere distances. The same fraction of unbalanced Ura+ events was observed in both cases. Similarly, pre- and postreplication events occur in similar or identical proportions in both allelic-homolog and nonhomolog strain sets (2/78 and 8/125 are not statistically different from one another by z-test, P > 0.1). Thus, variations in the G1/G2 ratio appear not to affect the data that point to centromere clustering and homolog-specific effects as important determinants of recombination rate.
Altogether, the 203 Ura+ recombinant progeny examined with respect to loxP configuration represented a total of 112 recombination events. Among these, 82% (92/112) occurred between unreplicated loxP sites, whereas 18% (20/112) occurred between replicated loxP sites.
Discussion
Relative rates of Cre-promoted recombination for different loxP pairs reflect relative in vivo collision probabilities
The number of Cre-promoted recombination events that occur per unit time (the recombination rate) should be determined by three factors: (1) the rate of collision between two loxP sites of different types, (2) the fraction of cases in which the two colliding loxP sites will have appropriate complements of Cre recombinase, and (3) the probability that a collision between two appropriately Cre-bound loxP sites will be effective (will be followed by the next step in the reaction).
The current study reports significant differences in recombination rates among strains that are identical except that they carry different pairs of loxP sites. Three lines of evidence suggest that these differences result primarily from differences in the rates of collision between loxP partners. First, relative recombination rates vary systematically according to patterns expected from known constraints on the spatial organization of chromosomes, as discussed below. Second, differences that reflect variations in Cre binding or occurrence of a protein–DNA geometry suitable for effective interaction would reflect local peculiarities of the different insert constructs; reciprocally related pairs of loxP inserts, however, which differ with regard to the local context of the loxP construct, usually exhibit very similar recombination rates, suggesting that local effects are the exception rather than the rule. Third, if variations in Cre-binding affinity among different sites were responsible for the observed rate differences, a substantial increase in Cre protein concentration should reduce or eliminate such differences; the patterns of rate differences are not affected, however, by pulse induction of Cre protein with a concomitant 1000-fold increase in the level of recombinants.
Chromosome collisions in vivo are governed by three levels of organization
The rate of inter-locus collision in vivo, as inferred from relative rates of Cre/loxP recombination for different loxP pairs, appears to be constrained in at least three different ways. First, collisions occur more frequently between nearby segments on the same chromosome than between the genetically identical segments when they are present on homologs. Second, for loci on nonhomologous chromosomes, collisions are more frequent if the interacting segments are located at equivalent distances from their respective centromeres, and increasingly less frequent as the disparity in locus-to-centromere distance increases. This is the pattern predicted from the presence of centromere clustering in the Rabl orientation (Jin et al. 1998). Third, collisions between allelic loci on homologs are more frequent than collisions between loci on nonhomologous chromosomes, apparently over and above any differences predicted from centromere clustering alone. This is the constraint predicted from the presence of direct homolog pairing (Burgess et al. 1999). Alternatively, such a constraint could reflect a nuclear addressing process in which homologs tend to be colocalized in the absence of direct contacts; such processes have been documented in Drosophila (for review, see Marshall et al. 1997) but are not yet reported for yeast. For addressing to be a general effect for most chromosomal loci, however, addressing determinants would have to occur frequently along the chromosomes and/or the chromosomes would have to be relatively stiff and compact so that a few determinants would suffice.
These levels of spatial organization are presumably mechanistically independent of one another, but we favor the idea that centromere clustering effects are effectively irrelevant for homologs, whose relative positions are determined by homolog-specific effects (presumptively direct pairing), as discussed above.
Detected recombinational interactions have occurred primarily between nonreplicated loxP sites
Of the detected Cre/loxP recombination events, ∼80% occurred between nonreplicated chromosomes and ∼20% occurred between replicated chromosomes. Thus, for most events, recombination has occurred in a situation in which the nucleus contains two, and only two, loxP sites, one of each type.
In the strain used for this analysis, under conditions analogous to those used for fluctuation analysis, >70% of cells are unbudded, that is, in telophase/G1. Thus, recombination may be equally probable at most or all stages of the cell cycle, with the relative fractions of pre- and postreplication events reflecting the relative fractions of the cell cycle occupied by the corresponding stages. Alternatively, or in addition, postreplication events might occur but be inefficiently recovered, for example, because they generate mitotic chiasmata, which lead to aberrant chromosome segregation and/or cell cycle arrest (Chua and Jinks-Robertson 1991; Kleckner, 1996; Beumer et al. 1998) or because interactions between identical loxP sites on sister chromatids compete with interactions between pGPD1–loxP and loxP–ura3.
There does not seem to be any significant difference in the ratio of pre- and postreplication recombinants among eight different strains that represent the entire range of interchromosomal loxP partner configurations, suggesting that such differences do not contribute to differences between measured recombination rates. This conclusion is in accord with the fact that both the Rabl orientation and homolog pairing are known to be present both before and after DNA replication (Jin et al. 1998; Burgess et al. 1999).
Organizational constraints confer quantitatively modest effects
The recombination rates for the 18 loxP combinations examined in this analysis differ by approximately sixfold for interchromosomal interactions and approximately eightfold if the intrachromosomal interaction is included. If relative recombination rates accurately reflect relative probabilities of interlocus collision for the corresponding DNA segments, this result would suggest that spatial constraints, although cytologically prominent, have relatively modest quantitative effects on the probability with which two loci in the genome will collide with one another.
This conclusion can be framed in another way. The recombination rate for each particular loxP pair defines an effective volume, which is the volume that would produce the observed rate in a simple in vitro reaction between untethered sites. Because the recombination rate depends on the product of the concentrations of the two loxP sites, the effective volume varies inversely with the square root of the recombination rate. The observed eightfold range in recombination rates thus corresponds to a range of only ∼2.9-fold in relative effective volumes. The absolute effective volumes that correspond to these relative volumes remain to be determined.
Implications for other cellular processes that involve interaction of two DNA segments
A number of normal cellular processes involve the coming together of two DNA segments (e.g., DNA repair). Three such processes are recombinational repair, nonhomologous end joining, and single-strand annealing of two broken DNA ends that share local homology.
The current study provides a baseline for evaluating the nature of such processes. If the level of product formed by a particular process exhibits the same three types of systematic variation with the relative chromosomal positions of the interacting segments as observed for Cre/loxP site-specific recombination, the probability of obtaining the corresponding product is likely dependent on the rate at which the two interacting segments collide with one another. If no such relationship is observed, then either the rate of collision is irrelevant to the probability that a product will form, or, alternatively, the rate of collision is an important variable but its influence is masked by locus-specific variations in other aspects of the process.
In Saccharomyces cerevisiae, the very limited available data provides some insight into two types of repair processes. First, the level of spontaneous mitotic recombination is thought to reflect the occurrence of recombinational repair in response to spontaneous DNA damage (see Petes et al. 1991). Two studies have compared the frequency of such recombination either between sequences located at allelic positions on homologs or between a sequence present at its endogenous position with another copy of the sequence present elsewhere (ectopically). In one such comparison, the two rates were essentially the same (Lichten and Haber 1989); in another case, the allelic rate was severalfold greater than the ectopic rate (Jinks-Robertson and Petes 1986). In both cases, the ectopic position was located on a different chromosome from the endogenous position, but, in the first case, the two positions were at similar distances from their respective centromeres (Δ10 kb), whereas in the second case, the locus-to-centromere distances were very different (Δ370 kb). The first study also found that recombination between marker loci separated by ∼20 kb in cis, that is, within a chromatid or between sisters, occurred at 6–13 times the frequency recombination between the same pair of markers in trans (when present on homologs). Although limited, these data are consistent with dependence of recombinational repair frequency on inter-locus collision rate. A similar conclusion was drawn from a third study in which the frequency of recombination between a chromosomal locus and an homologous sequence increased when an increased number of copies of the partner sequence were provided on a multicopy plasmid (Melamed and Kupiec 1992).
Other studies show that mitotic recombinational repair events in G2 cells occur between sister chromatids in preference to between homologs (Fabre et al. 1984; Kadyk and Hartwell 1992). It would be interesting to know whether this bias reflects closer proximity of sister chromatids and/or the operation of specific intersister targeting factors.
Second, partner choice during single-strand annealing has also been examined with respect to the effects of chromosome disposition (Haber and Leung 1996). Following induction of double-strand breaks simultaneously at loci on two different chromosomes in a haploid strain, repair products can arise either by annealing of left and right ends from the same locus or from different loci. The two types of products arise at equivalent frequencies, regardless of whether the two loci were at the same or different distances from their respective centromeres. In the original study, these data were interpreted as evidence for absence of order within the yeast genome. This interpretation would assume that the probability of repair would be sensitive to the rate of collision between ends. In light of the current results, we suggest, instead, that genomic order is present, but irrelevant, to the probability of effective single-strand annealing.
In mouse, Jasin and colleagues (Richardson et al. 1998) find that the frequency of recombinational repair in one test situation is eightfold higher when the interacting regions are at allelic positions than when they are located on two different, nonhomologous chromosomes. In the latter case, the positions of the interacting segments differ in locus-to-centromere distance by ∼30%, the length of an average mouse chromosome (25 and ∼75 cM, respectively; Dietrich et al. 1996; Richardson et al. 1998). This corresponds approximately to the relationship between the HIS4 and ILV1 loci in yeast, which are separated by ∼20% of the length of an average yeast chromosome (130 and ∼75 kb, respectively; Cherry et al. 1997) and for which the nonhomolog Cre/loxP recombination rates differ from the allelic rates by approximately sixfold. These data are therefore consistent with a general similarity between recombinational repair in the two organisms.
In Drosophila, the frequencies of Flp-promoted site-specific recombination between frt sites exhibit variations similar to those observed here for Cre/loxP recombination in yeast. For sites located on the same chromosome, recombination frequencies vary over a range of 1000-fold as interlocus distances vary from 0.005 to 15 Mb; furthermore, for a pair of loci separated by ∼10% of the length of chromosome 2, the intrachromosomal recombination frequency was severalfold greater than the corresponding interhomolog frequency (Golic and Golic 1996b). In contrast, for recombination events promoted by P-element excision, although intrachromosomal events are more frequent than interhomolog events, intrachromosomal events exhibit no dependence on interlocus distance (Engels et al. 1994). Interestingly, these events likely occur by synthesis-dependent single-strand annealing (Nassif et al. 1994), perhaps again pointing to nondependence of this type of process on intersegment collision rate.
What is the evolutionary raison d’etre of somatic pairing?
For the pairs of loci examined thus far, homolog pairing confers a rate of Cre/loxP recombination that is no more than twofold higher than the rate predicted to be achieved by the Rabl orientation alone, independent of homolog-specific effects. It is usually assumed that somatic pairing of homologs exists to promote genome-wide colocalization of homologs, for example, for modulation of gene expression (for reviews, see Henikoff and Comai 1998; Wu and Morris 1999). But if that goal can be achieved nearly as effectively by the Rabl orientation, perhaps the ability of homologs to identify one another by direct contact is evolutionarily advantageous for other reasons, with genome-wide pairing occurring as one secondary consequence. We could suggest specifically that the homolog pairing process is maintained through evolution primarily because it provides a way of getting homologous regions together in cells containing irregular genome complements, that is, translocations, inversions, and/or extensive homeologies. In such cases, the Rabl orientation will actually tend to place homologous regions at disparate positions within the nucleus, and a direct pairing process would then serve as a counterforce, for example, to enhance recombinational repair (above). Also, homology-directed interactions that might be mechanistically related to homolog pairing (Campbell 1993; Kleckner and Weiner 1993; Wu and Morris 1999) are already known to occur between disparate regions in filamentous fungi and plants (Selker 1997; Bender 1998; Maloisel and Rossignol 1998). Other epigenetic phenomena provide additional possible cases, most notably paramutation in plants which involves trans interactions between homologous genomes that may be mediated by direct contacts (for review, see Hollick et al. 1997; Que and Jorgensen 1998).
It has also been suggested, however, that somatic pairing exists to facilitate eventual meiotic pairing (Stack and Brown 1969). Although it is clear that some, and possibly many, organisms carry out meiosis without any premeiotic pairing, the process might be maintained in nonmeiotic cells of a few organisms as an evolutionary reservoir, to be called on in cases in which the meiotic process is damaged or lost.
These several possibilities can be addressed further once mutants specifically defective in homolog pairing become available. Suitable application of the Cre/loxP assay should make possible the identification of yeast mutants that are defective either in mitotic or meiotic pairing or in other features that affect chromosome disposition in vivo.
Materials and methods
Yeast and media
All strains are derivatives of NKY3061 (MATa/MATα ura3–52/′′lys2–Δ202/′′trp1Δ63/′′leu2Δ1/′′his3Δ200/′′) made from isogenic S288C parents (Winston et al. 1995). Yeast media and genetic methods are described in Rose et al. (1990); synthetic complete medium (SC) and SC medium lacking specific amino acids (SC − aa) contained 2% glucose unless otherwise noted and were supplemented with additional threonine (160 mg/liter) and isoleucine (40 mg/liter).
Plasmids and strain construction
Both loxP constructs contain the 1.2-kb LEU2 gene from YEp13 (XhoI–SalI fragment; Rose et al. 1990) and the 66-bp loxP region from pBS43 (BamHI–BamHI fragment; Sauer 1987). The LEU2 gene is oriented in opposite directions in the two constructs. LEU2–loxP–ura3 contains a 1.1-kb ura3 fragment from yDpU (Pst1–SmaI fragment; Berben et al. 1991) and LEU2–pGPD1–loxP–lacZ contains the GPD1 promoter from PG1 (Schena et al. 1991), lacZ from LRD1 (BamHI–ScaI; West et al. 1984), and the PGK1 transcriptional terminator of PG1 (Schena et al. 1991). Restriction sites from original plasmids were not always maintained during cloning. Details are available on request. The LEU2–pGDP1–loxP–lacZ and the LEU2–loxP–ura3 constructs were inserted at the SacI site of ARG4, between XhoI–BglII of HIS4, and at the HpaI sites of ILV1 and THR4 on integrating plasmids. These alleles were introduced into the same loci of the yeast genome by one-step allele replacement into NKY3061 or its haploid derivatives (Rothstein 1991). Genomic integrations were confirmed by Southern analysis (Sambrook et al. 1989). Haploid strains were mated to make all possible combinations of diploid strains bearing one of each loxP-marked construct. Because slow growth conferred by the Ilv− auxotrophy confounds fluctuation analysis, the strain carrying two inserts at ILV1 also carries a spontaneous mutation to Ilv+. This mutation likely affects CHA1 expression (Pedersen et al. 1997). The suppressor mutation is not linked to ILV1 and does not affect Cre/loxP recombination rates as tested in ilv1/ILV1 heterozygotes (data not shown). The pGAL1–cre transcriptional fusion was derived from pBS49 (Sauer 1987) by replacement of URA3 with LYS2 from yDpK (Berben et al. 1991).
Recombination assay
The rate of recombination per cell generation in the absence of Cre induction was determined for each strain by fluctuation analysis, method of the median (Lea and Coulson 1948), for colonies arising after 3 days of growth on SC–LYS at 30°C. For each strain, 11 colonies were each suspended in 1 ml of water, sonicated in water at half power for 3 sec with a microtip, diluted appropriately in water, and plated onto SC–URA and SC–LYS media; Cre plasmid loss was negligible as determined by comparing plating efficiencies on SC–LYS with SC. Ura+ prototrophs were counted after 5 days at 30°C; colonies on SC–LYS were counted after 3-days growth at 30°C.
Recombination frequencies in the presence of Cre induction were determined by direct plating of cells after exposure to galactose for 2 hr. Briefly, individual colonies grown 3 days on SC–LYS were used to inoculate 5 ml of SC–LYS medium containing 2% raffinose instead of glucose. Cultures were incubated overnight at 30°C. The next day, cultures were diluted 1:8 in the same medium and incubated an additional 4 hr. Galactose was added to a 2% final concentration and the cultures were incubated an additional 2 hr. One milliliter of cells was pelleted, washed once in water, sonicated, and diluted appropriately before plating onto selective and nonselective medium for analysis as described above.
G1 vs. G2 analysis
PCR analysis of Ura+ prototrophs was performed on individual colonies with the following oligonucleotide primers to the loci noted below: (A) GPD1, 5′-ACTAATAAGTATATAAAGACGG-3′; (B) lacZ, 5′-CATTAATGAATCGGCCAACG-3′; (C) LEU2, 5′-ATGGCTCACGTAAGG-3′; and (D) ura3, 5′-CCTTGCATGACAATTCTGC-3′. Oligonucleotide pairs AB and CD give 300 and 800 bp nonrecombinant (parental) products, respectively. Oligonucleotide pairs AD and CB give 600 and 450 bp recombinant products, respectively. PCR products were resolved on a 1.5% agarose 1× TBE gel.
The percentages of Ura+ prototrophs arising by recombination between unreplicated and replicated chromosomes (see text) were calculated by assuming that (1) each unbalanced recombinant reflected a postreplication event, (2) a number of balanced recombinants equal to the number of unbalanced recombinants also reflected postreplication events, and (3) each postreplication event yields a single Ura+ recombinant progeny cell, whereas each prereplication event yields two Ura+ recombinant progeny cells. Thus, for the strains representing allelic-homolog interactions, among 78 independently arising Ura+ recombinants, 2 were unbalanced and reflected 2 postreplication recombination events; correspondingly, 2 of the 76 balanced recombinants also reflected (an additional) 2 postreplication events; the remaining 74 recombinants thus reflected 37 prereplication events; the total number of recombination events represented in the sample was therefore 4 + 37 = 41 and the percentages of prereplication and postreplication events among this sample were 37/41 = 90% and 4/41 = 10%, respectively. Percentages for nonhomolog strains were calculated analogously.
Acknowledgments
We thank Job Dekker, Neil Hunter, and Scott Keeney for comments on the manuscript and Howard Nash and Guido Guidotti for discussion. This work was supported by a grant to N.K. from the National Institutes of Health (RO1-GM44794). S.M.B. was supported by the Helen Hay Whitney Foundation and an Office of Naval Research Science Scholar Fellowship from the Mary Ingraham Bunting Institute of Radcliffe College.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kleckner@fas.harvard.edu; FAX (617) 495-0758.
References
- Ambremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: Evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- Bender JC. Cytosine methylation of repeated sequences in eukaryotes: The role of DNA pairing. Trends Biochem Sci. 1998;23:252–256. doi: 10.1016/s0968-0004(98)01225-0. [DOI] [PubMed] [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Beumer KJ, Pimpinelli S, Golic KG. Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics. 1998;150:173–188. doi: 10.1093/genetics/150.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: Existence and modulation. Genes & Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AM. Co-chairman’s remarks: Genetic recombination in the molecular era. Gene. 1993;135:147–151. doi: 10.1016/0378-1119(93)90059-c. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weg S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, Botstein D. Genetic and physical maps of Saccharomyces cerevisiae. Nature (Suppl.) 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- Chua P, Jinks-Roberson S. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics. 1991;129:359–369. doi: 10.1093/genetics/129.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain A, Dredge R, Daly MJ, Ingalls KA, O’Connor TJ, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Engels WR, Preston CR, Johnson-Schlitz DM. Long-range cis preference in DNA homology search over the length of a Drosophila chromosome. Science. 1994;263:1623–1625. doi: 10.1126/science.8128250. [DOI] [PubMed] [Google Scholar]
- Fabre F, Boulet A, Roman H. Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae. Mol & Gen Genet. 1984;195:139–143. doi: 10.1007/BF00332736. [DOI] [PubMed] [Google Scholar]
- Golic KG, Golic MM. Engineering the Drosophila genome: Chromosome rearrangements by design. Genetics. 1996a;144:1693–1711. doi: 10.1093/genetics/144.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic MM, Golic KG. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics. 1996b;143:385–400. doi: 10.1093/genetics/143.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Leung WY. Lack of chromosome territoriality in yeast: Promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Comai L. Trans-sensing effects: The ups and downs of being together. Cell. 1998;93:329–332. doi: 10.1016/s0092-8674(00)81161-7. [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Cozzarelli NR. Comparison of recombination in vitro and in E. coli cells: Measure of the effective concentration of DNA in vivo. Cell. 1995;81:331–340. doi: 10.1016/0092-8674(95)90386-0. [DOI] [PubMed] [Google Scholar]
- Hoess RH, Ziese M, Sternberg N. P1 site-specific recombination: Nucleotide sequence of the recombining sites. Proc Natl Acad Sci. 1982;79:3398–3462. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB, Dorweiler JE, Chandler VL. Paramutation and related allelic interactions. Trends Genet. 1997;13:302–308. doi: 10.1016/s0168-9525(97)01184-0. [DOI] [PubMed] [Google Scholar]
- Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Petes TD. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Meiosis: How could it work? Proc Natl Acad Sci. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the number of mutants in bacterial populations. J Genet. 1948;49:264–284. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Lichten M, Haber JE. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989;123:261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L, Rossignol JL. Suppression of crossing-over by DNA methylation in Ascobolus. Genes & Dev. 1998;12:1381–1389. doi: 10.1101/gad.12.9.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard D A, Sedat JW. Specific interactions of chromatin with the nuclear envelope: Positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Fung JC, Sedat JW. Deconstructing the nucleus: Global architecture from local interactions. Curr Opin Genet Dev. 1997;7:259–263. doi: 10.1016/s0959-437x(97)80136-0. [DOI] [PubMed] [Google Scholar]
- Melamed C, Kupiec M. Effect of donor copy number on the rate of gene conversion in the yeast Saccharomyces cerevisiae. Mol & Gen Genet. 1992;235:97–103. doi: 10.1007/BF00286186. [DOI] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JO, Rodriguez MA, Praetorius-Ibba M, Nilsson-Tillgren T, Calderon IL, Holmberg S. Locus-specific suppression of ilv1 in Saccharomyces cerevisiae by deregulation of CHA1 transcription. Mol & Gen Genet. 1997;255:561–569. doi: 10.1007/s004380050529. [DOI] [PubMed] [Google Scholar]
- Petes TD, Malone RE, Symington LS. Recombination in yeast. In: Pringle JR, Broach JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- Que Q, Jorgensen RA. Homology-based control of gene expression patterns in transgenic petunia flowers. Dev Genet. 1998;22:100–109. doi: 10.1002/(SICI)1520-6408(1998)22:1<100::AID-DVG10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Rabl C. Ueber Zellteilung. Morphol-Jahrbuch. 1885;10:214–330. [Google Scholar]
- Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption replacement and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rose M, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning. A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. J Mol Biol. 1992;223:911–928. doi: 10.1016/0022-2836(92)90252-f. [DOI] [PubMed] [Google Scholar]
- Schena M, Picard D, Yamamoto KR. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- Selker EU. Epigenetic phenomena in filamentous fungi: Useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- Stack SM, Brown WV. Somatic pairing, reduction, and recombination: An evolutionary hypothesis of meiosis. Nature. 1969;222:1275–1276. doi: 10.1038/2221275a0. [DOI] [PubMed] [Google Scholar]
- Staczek P, Higgins NP. Gyrase and Topo IV modulate chromosome domain size in vivo. Mol Microbiol. 1998;29:1435–1448. doi: 10.1046/j.1365-2958.1998.01025.x. [DOI] [PubMed] [Google Scholar]
- West RW, Jr, Yocum RR, Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: Location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984;4:2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wu C-T, Morris J. Transvection and other homology effects. Curr Opin Genet Dev. 1999;9:237–246. doi: 10.1016/S0959-437X(99)80035-5. [DOI] [PubMed] [Google Scholar]