Abstract
Background:
The “Dutch hypothesis” suggests that asthma and COPD have common genetic determinants. The serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2) gene previously has been associated with COPD. We sought to determine whether SERPINE2 is associated with asthma and asthma-related phenotypes.
Methods:
We measured the association of 39 SERPINE2 single-nucleotide polymorphisms (SNPs) with asthma-related phenotypes in 655 parent-child trios from the Childhood Asthma Management Program (CAMP), and we measured the association of 19 SERPINE2 SNPs with asthma in a case-control design of 359 CAMP probands and 846 population control subjects. We attempted to replicate primary asthma-related phenotype findings in one independent population and primary asthma affection status findings in two independent populations. We compared association results with CAMP proband expression quantitative trait loci.
Results:
Nine of 39 SNPs had P < .05 for at least one phenotype in CAMP, and two of these replicated in an independent population of 426 people with childhood asthma. Six of 19 SNPs had P < .05 for association with asthma in CAMP/Illumina. None of these replicated in two independent populations. The expression quantitative trait loci revealed that five SNPs associated with asthma in CAMP/Illumina and one SNP associated with FEV1 in CAMP are strongly correlated with SERPINE2 expression levels. Comparison of results to previous COPD studies identified five SNPs associated with both asthma- and COPD-related phenotypes.
Conclusions:
Our results weakly support SERPINE2 as a Dutch hypothesis candidate gene through nominally significant associations with asthma and related traits. Further study of SERPINE2 is necessary to verify its involvement in asthma and COPD.
Asthma and COPD are complex human airway diseases that are influenced by various genetic and environmental susceptibility factors. In 1961, Orie and colleagues1 proposed that these diseases were different expressions of one basic pulmonary disease whose expression in individuals was shaped differently depending on a combination of endogenous (ie, host) and exogenous (ie, environmental) factors. This view, which became known as the “Dutch hypothesis,” suggests that overlapping clinical features of asthma and COPD are partly based on an allergic diathesis and airways hyperresponsiveness (AHR).2 Although there is some opposition to the original Dutch hypothesis, little doubt exists that common mechanisms underlie asthma and COPD.3 Identifying genetic variants that influence some of these common mechanisms would provide important insights into both diseases.
Genetic linkage studies of COPD,4 asthma,5,6 and general population lung function7 have demonstrated linkage peaks on chromosome 2q, suggesting that in this region are good Dutch hypothesis candidate genes. This is further supported by linkage results observed on chromosome 2q for asthma and COPD being strongest for the FEV1/FVC phenotype. A promising Dutch hypothesis candidate gene within this region is the serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2) gene (MIM 177010). SERPINE2 variants have been associated with lung function among patients with COPD.8,9 Additionally, SERPINE2 protein products are present in lung tissue from individuals with asthma and COPD, and there is evidence that SERPINE2 plays a role in the formation and maintenance of mouse lung structure.8 Here, we determine whether SERPINE2 is associated with asthma affection status in three cohorts and with lung function and IgE in two cohorts with the aim to determine whether SERPINE2 is a Dutch hypothesis gene.
Materials and Methods
Subjects
Childhood Asthma Management Program:
Detailed methods are provided in e-Appendix 1. Our primary population consisted of 655 non-Hispanic white subjects from the Childhood Asthma Management Program (CAMP), a multicenter clinical trial that followed 1,041 children with asthma for 4 years and 84% of the original participants for 12 years.10 In addition to asthma diagnosis, we considered phenotypes that capture components of the Dutch hypothesis: FEV1, FEV1/FVC, AHR quantified as the log-transformed percentage dose of methacholine causing a 20% decrease in FEV1, and log-transformed total serum IgE (LogIgE) levels. CAMP participants and their parents provided DNA for the family-based genetic studies. Additionally, blood was drawn from a subset of CAMP participants for gene expression profiling studies of CD4+ T lymphocytes.11
CAMP/Illumina:
In a previous asthma genome-wide association study, CAMP/Illumina, a subset of 359 CAMP probands of the 655 CAMP probands who had genome-wide genotype data available, were genetically matched using the genetic matching algorithm12 with 846 publicly available population control subjects.13 Because CAMP/Illumina is better powered to measure asthma affection status than CAMP parent-child trios, CAMP/Illumina results were used to measure the association of SERPINE2 variants with asthma affection status.
Genetics of Asthma in Costa Rica:
This population consisted of 426 probands from the Genetics of Asthma in Costa Rica study that comprised Costa Rican schoolchildren with asthma and their parents.14,15 For the current study, FEV1, FEV1/FVC, and LogIgE phenotypes were defined as they were in CAMP. For asthma affection status and AHR, we selected available phenotypes most similar to those defined in CAMP: Asthma affection status was defined as the presence of methacholine dose responsiveness (≤ 8.58 μM) or evidence of bronchodilator responsiveness plus the recruitment criteria,15 and AHR was quantified as the log-transformed dose-response slope for methacholine.16
The i2b2 Crimson Asthma Project:
The i2b2 Crimson Asthma Project (iCAP) consists of Partners Healthcare System, Inc (Boston, Massachusetts) patients who were selected based on extracted deidentified electronic medical record (EMR) data.17 In a pilot study of white subjects, cases (n = 220) were defined as those patients whose EMR contained an asthma International Classification of Diseases, Ninth Revision (ICD-9), code and whose medication history included usage of at least one β-agonist or inhaled corticosteroid. Control subjects (n = 853) were selected as those patients who had been seen in the 3 years prior to blood collection in at least one of more than 850 outpatient clinics but did not have any asthma ICD-9 codes.
Ethics Statement:
The CAMP study was approved by the institutional review board of Partners Healthcare (Partners Human Research Committee; Protocol#: 1999-P-001549/29) and by all eight CAMP clinical centers and the CAMP Data Coordinating Center. The Genetics of Asthma in Costa Rica study was approved by the Partners Human Research Committee (Protocol#: 2000-P-001130/55) and the Hospital Nacional de Niños (San José, Costa Rica). The iCAP study was approved by the Partners Human Research Committee (Protocol#: 2007-P-000184/9). Informed consent was obtained for all study participants if they were aged > 18 years. Otherwise, informed consent was obtained from parents of participating children, and the child’s assent was obtained prior to study enrollment. For iCAP subjects, informed consent was obtained at the time of blood collection as part of standard Crimson Project procedures.17
Variant Selection and Genotyping
We selected single-nucleotide polymorphisms (SNPs) within SERPINE2 to be genotyped on the basis of two criteria: (1) previously reported associations in COPD cohorts and (2) capture of maximal variation based on linkage disequilibrium (LD) (e-Table 1). For the first criterion, we selected 14 SNPs that were previously associated with FEV1, FEV1/FVC, or both.8 For the second, we selected 15 SNPs to capture LD across the SERPINE2 gene with an r2 > 0.8 and a minor allele frequency of > 5%. The data for 29 SNPs selected for genotyping in 655 CAMP trios were supplemented by genome-wide association data for 10 additional SNPs available for a cohort of 403 CAMP probands and their parents. Genotyping was performed using four methods. Genotyping pass rates for individual SNPs were > 98%. All SNPs were in Hardy-Weinberg equilibrium among parents or controls of each population (P > .05). A summary of all SNPs genotyped in each population is available in e-Table 2.
Statistical Analysis
Family-based association tests were calculated under an additive inheritance model using Golden Helix PBAT, version 6.4.0 (Golden Helix, Inc; Bozeman, Montana) in CAMP and Costa Rica parent-child trios to measure associations with asthma affection status, LogIgE, FEV1, FEV1/FVC, and AHR.18 No covariates were included in the analysis of asthma affection status or LogIgE. Analyses of FEV1, FEV1/FVC, and AHR included statistical adjustment for age, sex, and height. LD among SNPs was inferred from r2 measures in family data using Haploview.19 The Cochran-Armitage trend test as implemented in PLINK20 was used to measure association of asthma affection status in CAMP/Illumina and iCAP.13 All other statistical analyses were performed in R (R Foundation for Statistical Computing; Vienna, Austria).21 Joint evidence for association across populations was measured by combining P values using the Liptak method.22 In combining P values, hypothesis tests in replication populations had one-sided alternatives (based on the direction of association in CAMP or CAMP/Illumina) so that SNPs with association tests in opposite directions would not produce inappropriately small P values.
Results
There were no significant differences in the sex, age, or IgE distributions between the CAMP and Costa Rica cohorts (Table 1). Despite statistically significant differences in FEV1, FEV1/FVC, and methacholine responsiveness between the two groups, both groups were characterized by a mild decrease in lung function that is not clinically significantly different. The sex distribution of CAMP/Illumina cases is similar to that of probands from the CAMP trios, but opposite that of CAMP/Illumina control subjects (Table 2). The age of CAMP/Illumina control subjects is not known, but all are reported to be adults (ie, aged ≥ 18 years). The largely female iCAP composition partly reflects the sex distribution of patients from Brigham and Women’s Hospital, the primary hospital where iCAP subjects were recruited. Also in contrast to the other populations, the iCAP patients with asthma are adults.
Table 1.
—Characteristics of Children in Family-Based Cohorts
| Variable | CAMP (n = 655) | Costa Rica (n = 426) | P Value |
| Sex | .44 | ||
| Male | 394 (0.60) | 267 (0.63) | |
| Female | 261 (0.40) | 159 (0.37) | |
| Age, y | 8.9 ± 2.1 | 9.1 ± 1.8 | .077 |
| FEV1 % predicted | 94.3 ± 14.2 | 99.9 ± 15.9 | 6.6 × 10−9 |
| FEV1/FVC | 79.7 ± 8.3 | 82.6 ± 7.1 | 9.8 × 10−10 |
| Methacholine responsea | 1.08 (0.48-2.77) | 1.34 (0.71-3.37) | .026 |
| Total serum IgE | 407 (159-1072) | 426 (121-977) | .28 |
Data are presented as No. (%), mean ± SD, or median (interquartile range). CAMP = Childhood Asthma Management Program.
Measured as methacholine provocation concentration causing a 20% fall in FEV1 in CAMP and methacholine provocation dose causing a 20% fall in FEV1 in Costa Rica.
Table 2.
—Characteristics of Case-Control Cohorts
| CAMP/Illumina |
iCAP |
|||
| Variable | Case Subjects (n = 359) | Control Subjects (n = 846) | Case Subjects (n = 223) | Control Subjects (n = 858) |
| Sex | ||||
| Male | 222 (62) | 309 (37) | 43 (19) | 136 (16) |
| Female | 137 (38) | 537 (63) | 180 (81) | 722 (84) |
| Age, y | 8.8 ± 2.1 | … | 30 ± 4 | 29 ± 6 |
Data are presented as No. (%) or mean ± SD. iCAP = i2b2 Crimson Asthma Project. See Table 1 legend for expansion of other abbreviation.
Based on results in CAMP trios, nine of 39 SNPs had nominally significant associations (ie, P < .05) for at least one asthma-related phenotype, but none were below the multiple comparisons correction threshold of 2.9 × 10−4 (ie, 0.05/175, where 175 was the number of measurements made [(39 SNPs × 4 phenotypes) + (19 SNPs × 1 phenotype)]. Eight of these nine SNPs were successfully genotyped in Costa Rica (Table 3). The LD patterns among the eight SNPs genotyped in CAMP and Costa Rica were similar (e-Figure 1). Association results for these SNPs with asthma-related phenotypes are shown in (Table 4). Of the five SNPs that were nominally associated with FEV1/FVC in CAMP and genotyped in Costa Rica, only one (rs6738983) supported the CAMP findings (combined P = 2.8 × 10−3). One SNP (rs10164837) was nominally associated with FEV1 in both the CAMP and the Costa Rica cohorts, with a combined P = 2.3 × 10−3. The other nominally significant CAMP associations failed to replicate in the Costa Rica cohort.
Table 3.
—SNPs Genotyped in CAMP and Costa Rica to Measure Association With Asthma-Related Phenotypes
| CAMP |
Costa Rica |
||||||
| SNP | CHR | BP | Allele | MAF | Parent MAF | MAF | Parent MAF |
| rs6734100 | 2 | 224550239 | G | 0.14 | 0.12 | 0.12 | 0.12 |
| rs7597833 | 2 | 224550394 | T | 0.49 | 0.49 | 0.61 | 0.60 |
| rs10164837 | 2 | 224555512 | C | 0.30 | 0.29 | 0.19 | 0.18 |
| rs6712954 | 2 | 224564894 | A | 0.07 | 0.07 | 0.26 | 0.24 |
| rs6738983 | 2 | 224566994 | T | 0.32 | 0.33 | 0.45 | 0.44 |
| rs6715768 | 2 | 224570735 | A | 0.19 | 0.20 | … | … |
| rs3795879 | 2 | 224571065 | T | 0.19 | 0.20 | 0.23 | 0.24 |
| rs6747096 | 2 | 224571086 | G | 0.19 | 0.20 | 0.20 | 0.20 |
| rs3795877 | 2 | 224574421 | G | 0.19 | 0.20 | 0.20 | 0.20 |
BP = base pair; CHR = chromosome; MAF = minor allele frequency; SNP = single-nucleotide polymorphism. See Table 1 legend for expansion of other abbreviation.
Table 4.
—Association Results for Asthma-Related Phenotypes in CAMP and Costa Rica
| LogIgE |
FEV1 |
FEV1/FVC |
AHR |
||||||
| SNP | Allele | CAMP | Costa Rica | CAMP | Costa Rica | CAMP | Costa Rica | CAMP | Costa Rica |
| rs6734100 | G | .70 | .69 | .43 | .61 | .83 | .76 | −.038a | −.36 |
| rs7597833 | T | .018a | .74 | .11 | .09 | −.49 | −.10 | −.82 | −.50 |
| rs10164837 | C | .20 | .89 | .042a | .011a | .33 | .79 | .20 | .98 |
| rs6712954 | A | 2.4 × 10−3a | .63 | −.49 | −.09 | −1.3 × 10−3a | −.31 | −.15 | −.76 |
| rs6738983 | T | .15 | .15 | −.66 | −.11 | −.038a | −.016a | −.20 | −.83 |
| rs6715768 | A | .12 | … | .96 | … | −.027a | … | .55 | … |
| rs3795879 | T | .17 | .41 | .72 | .78 | −.040a | −.47 | .62 | .91 |
| rs6747096 | G | .17 | .45 | .84 | .82 | −.020a | −.25 | .45 | .96 |
| rs3795877 | G | .13 | .39 | −.69 | −.18 | −.025a | −.17 | .66 | .91 |
Minus signs before P values indicate direction of effect in the case where increased dosage of allele is associated with decreased phenotype. AHR = airways hyperresponsiveness; LogIgE = log-transformed total serum IgE. See Table 1 and 3 legends for expansion of other abbreviations.
Nominally significant P < .05.
Because CAMP/Illumina was better powered to measure association with asthma affection status than CAMP trios,17 we used CAMP/Illumina as the primary population to study affection status. Of 19 SNPs with association data available, six showed nominally significant (ie, P < .05) association with asthma affection status, but none was below the multiple comparisons correction threshold of 2.9 × 10−4. Replication of the nominally significant associations was attempted in the Costa Rica and iCAP cohorts (Table 5). Three of these six SNPs were successfully genotyped in the Costa Rica cohort, and four of six were genotyped in iCAP. Measures of LD among the six SNPs revealed that there were three pairs of SNPs with r2 ≥ 0.94 in CAMP trios (e-Figure 2, Table 5), and hence, the associations in this region could be captured by a subset of three SNPs. At least one SNP from each strong-LD pair was genotyped in the Costa Rica and iCAP cohorts. The CAMP/Illumina findings were not replicated in the Costa Rica or iCAP cohorts.
Table 5.
—Association Results for Asthma Affection Status
| CAMP/Illumina |
Costa Rica |
iCAP |
|||||||||||||
| SNP | CHR | BP | Minor Allele | MAF CaseSubjects | MAF Control Subjects | OR | P Value | MAF | Num Fam | P Value | MAF CaseSubjects | MAF Control Subjects | OR | P Value | LD Pairs |
| rs7590948 | 2 | 224581934 | G | 0.44 | 0.50 | 0.77 | 3.8 × 10−3 | … | … | … | 0.48 | 0.46 | 1.10 | .41 | 1 |
| rs2037755 | 2 | 224591549 | C | 0.44 | 0.50 | 0.79 | 9.4 × 10−3 | 0.47 | 375 | .96 | … | … | … | … | 1 |
| rs10194024 | 2 | 224599084 | C | 0.42 | 0.48 | 0.78 | 4.6 × 10−3 | 0.44 | 382 | −.88 | 0.51 | 0.44 | 1.30 | .017 | 2 |
| rs920251 | 2 | 224601189 | A | 0.32 | 0.37 | 0.79 | .011 | … | … | … | 0.39 | 0.32 | 1.36 | 6.7 × 10−3 | 3 |
| rs1438831 | 2 | 224614559 | T | 0.31 | 0.36 | 0.79 | .011 | 0.29 | 328 | −.87 | 0.38 | 0.31 | 1.38 | 4.1 × 10−3 | 3 |
| rs282254 | 2 | 224615591 | C | 0.42 | 0.48 | 0.77 | 3.6 × 10−3 | … | … | … | … | … | … | … | 2 |
P values for CAMP/Illumina and iCAP correspond to allelic association. P values for Costa Rica correspond to family-based association test additive model association. LD pairs refers to pairs of SNPs that are in tight LD in CAMP (e-Figure 2). LD = linkage disequilibrium; Num Fam = number of informative families. See Table 1, 2, and 3 legends for expansion of other abbreviations.
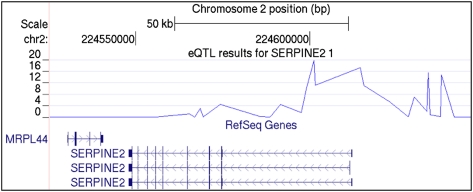
To determine whether the CAMP and CAMP/Illumina associations correspond to potential functional loci, we referenced results from a recent genome-wide survey for cis-acting expression quantitative trait loci (eQTL) in peripheral blood CD4+ lymphocytes performed in a subset of 200 CAMP subjects. There was strong evidence for regulatory genetic variation in a SERPINE2 region spanning the putative promoter region and first intron of all three RefSeq SERPINE2 isoforms (Fig 1). The strongest eQTL observed was that of the CAMP/Illumina asthma-associated SNP rs920251, where the minor allele conferred reduced SERPINE2 transcript abundance relative to the major allele and explained 42% of the total variation in transcript abundance (P = 1.5 × 10−20) (Table 6). Several additional asthma-associated SNPs demonstrated similar evidence of being regulatory variants, as did one SNP (rs3795877; eQTL P = 2.8 × 10−5; proportion variation explained, 14.3%) that was associated with FEV1/FVC in CAMP.
Figure 1.
SERPINE2 eQTL. The top displays the negative log P values for the association of single-nucleotide polymorphisms with SERPINE2 transcript levels. The three known SERPINE2 transcripts, according to RefSeq, demonstrate that single-nucleotide polymorphisms between the promoter region and first intron are those that are most strongly associated with SERPINE2 transcript levels. bp = base pair; eQTL = expression quantitative trait loci; MRPL44 = mitochondrial ribosomal protein L44; SERPINE2 = serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2.
Table 6.
—SNPs With eQTL Results
| SNP | CHR | BP | eQTL P Value | Proportion of Variation Explained | CAMP or CAMP/Illumina Phenotype(s) With Nominal Association P Value |
| rs6754561 | 2 | 224547940 | .95 | 0 | … |
| rs6734100 | 2 | 224550239 | … | … | AHR |
| rs7597833 | 2 | 224550394 | .96 | 0 | LogIgE |
| rs10164837 | 2 | 224555512 | … | … | FEV1 |
| rs975278 | 2 | 224555951 | .97 | 0 | … |
| rs6712954 | 2 | 224564894 | … | … | LogIgE, FEV1/FVC |
| rs10191694 | 2 | 224565510 | .064 | 0.052 | … |
| rs6738983 | 2 | 224566994 | .81 | −0.007 | FEV1/FVC |
| rs13396978 | 2 | 224568460 | 8.3 × 10−4 | −0.11 | … |
| rs11695803 | 2 | 224569405 | 0.77 | 0.009 | … |
| rs3795879 | 2 | 224571065 | … | … | FEV1/FVC |
| rs6747096 | 2 | 224571086 | … | … | FEV1/FVC |
| rs3795877 | 2 | 224574421 | 2.8 × 10−5 | 0.14 | FEV1/FVC |
| rs7590948 | 2 | 224581934 | … | … | Asthma |
| rs7563931 | 2 | 224585805 | 0.65 | −0.015 | … |
| rs6436459 | 2 | 224588580 | 0.93 | 0.002 | … |
| rs2037755 | 2 | 224591549 | 3.0 × 10−5 | −0.14 | Asthma |
| rs13008520 | 2 | 224597627 | 0.019 | −0.069 | … |
| rs10194024 | 2 | 224599084 | 3.0 × 10−11 | 0.27 | Asthma |
| rs920251 | 2 | 224601189 | 1.4 × 10−20 | −0.42 | Asthma |
| rs6719480 | 2 | 224601673 | 7.3 × 10−12 | 0.28 | … |
| rs1438831 | 2 | 224614559 | 4.7 × 10−18 | 0.39 | Asthma |
| rs282254 | 2 | 224615591 | 8.9 × 10−12 | −0.28 | Asthma |
eQTL entries containing an ellipse were not available. Negative values for proportion of variation explained denote that the minor allele confers reduced SERPINE2 transcript abundance relative to major allele. SERPINE2 = serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2. See Table 1, 3, and 4 legends for expansion of other abbreviations.
Discussion
SERPINE2 encodes a serpin that is synthesized and secreted by many cell types. Its protein product is an inhibitor of several biologically important serine proteases, including thrombin and urokinase-type plasminogen activator.23‐25 As such, it has been implicated in many cell processes, including coagulation, cell movement, and fibrinolysis. Following reports of SERPINE2 association with quantitative phenotypes of lung function among patients and COPD8,9 and its presence in lung tissue from individuals with asthma and COPD,8 SERPINE2 also has become an attractive Dutch hypothesis candidate gene. In the current analysis, we found mixed evidence that SERPINE2 is associated with asthma and asthma-related phenotypes.
Seven of the eight SNPs that were genotyped in the CAMP and Costa Rica cohorts were analyzed in at least one of two previous COPD studies.8,9 The SNP (rs6738983) associated with FEV1/FVC in CAMP and Costa Rica cohorts was not genotyped in either COPD study. The SNP (rs10164837) associated with FEV1 in CAMP and Costa Rica was genotyped in one previous COPD study8 but was not significantly associated with any phenotype. Three SNPs (rs3795879, rs6747096, and rs3795877) associated with FEV1/FVC in CAMP were previously associated with postbronchodilator FEV1/FVC in one COPD cohort.8 Further, two of these SNPs (rs6747096 and rs3795877) were associated with postbronchodilator FEV1, whereas two (rs3795879 and rs6747096) were associated with COPD.8 The SNP (rs6734100) associated with AHR in CAMP was previously associated with COPD, COPD severity, and postbronchodilator FEV1 and FEV1/FVC.8,9 Finally, the SNP (rs7597833) associated with LogIgE in CAMP was found to be associated with postbronchodilator FEV1 and COPD severity in one COPD study.8 Thus, in the present study, we found evidence that some SERPINE2 SNPs are associated with pulmonary function phenotypes in both asthma and COPD, whereas other SERPINE2 SNPs are only associated in one disease.
Our eQTL results demonstrate that some SERPINE2 SNPs have a large influence on gene expression levels of SERPINE2, making it more likely that these variants (or ones in LD to them) play a functional role in SERPINE2 biology. Many of the eQTL SNPs, which are near the 5′ end of the gene, are associated with asthma in CAMP/Illumina (association P values between 3.6 × 10−3 and .011). Most of the SNPs associated with asthma-related phenotypes were not eQTL, except for rs3795877, which had an eQTL P = 2.8 × 10−5. This SNP also was associated with postbronchodilator FEV1 and FEV1/FVC in a previous COPD study.8 Overall, the eQTL results strengthen some observed associations and suggest that further study of some SERPINE2 SNPs may yield insights into their role in gene function.
The present study is limited by the small population sample sizes, and consequently, low power, to detect associations. We chose to pursue replication of nominally associated SNPs despite being below the multiple comparisons threshold for two reasons. First, some of our findings were supportive of previous COPD studies, and if they had been strict replications, a nominal significance threshold of .05 would have been considered adequate. Second, the strict correction was conservative in that it assumes independence of measurements, whereas the LD among the genotyped SNPs demonstrates that many are not independent. Another limitation is cohort heterogeneity. Although CAMP and Costa Rica cohorts are similar populations (Table 1) that comprise children with asthma who were carefully ascertained for asthma studies, they are ethnically different and have different environmental exposures. Although the LD among the eight SNPs genotyped in the CAMP and Costa Rica cohorts is generally similar, there are some differences that could have affected our ability to replicate results (e-Figure 1). In contrast to CAMP and Costa Rica cohorts, the iCAP cohort is a general clinical population identified on the basis of ICD-9 codes for asthma and medication history extracted from deidentified EMRs.
One physiologic process through which SERPINE2 could have an influence on asthma and COPD susceptibility and severity is airway remodeling, the process by which injury and inflammation leads to changes in the cellular and structural architecture of the airways.26‐29 Evidence that modulation of airway remodeling could be used to treat asthma has been suggested by studies in which inhalation of urokinase-type plasminogen activator reduced airway remodeling in a mouse model of asthma.30 In COPD, the involvement of airway remodeling genes is supported by a gene expression study in which extracellular matrix synthesis and degradation pathway genes were related to a forced expiratory flow between 25% and 75% of FEV.31 In a study that evaluated the relationship between the expression of 54 tissue-repair genes and COPD progression, SERPINE2 was found to have increased expression in the small airways as FEV1 declined.32 Thus, it is plausible that genetic variants in SERPINE2 influence asthma and COPD through modulation of airway remodeling processes.
In conclusion, the current results weakly support SERPINE2 as a Dutch hypothesis candidate gene through nominally significant associations with asthma and related traits. Further study of SERPINE2 is necessary to verify whether it is involved in pathophysiological processes common to asthma and COPD.
Supplementary Material
Acknowledgments
Author contributions: Drs Himes, Silverman, Weiss, and DeMeo had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Dr Himes: contributed to the study design, analysis of the genetics data, drafting of the manuscript, and approval of the final version of the manuscript.
Dr Klanderman: contributed to the genotype data collection and approval of the final version of the manuscript.
Mr Ziniti: contributed to the genotype data collection and approval of the final version of the manuscript.
Ms Senter-Sylvia: contributed to the genotype data collection and approval of the final version of the manuscript.
Dr Soto-Quiros: contributed to the Costa Rica cohort data collection and approval of the final version of the manuscript.
Dr Avila: contributed to the Costa Rica cohort data collection and approval of the final version of the manuscript.
Dr Celedón: contributed to the Costa Rica cohort data collection and to the revision and approval of the final version of the manuscript.
Dr Lange: contributed statistical expertise in the analysis of the genetics data and to the revision and approval of the final version of the manuscript.
Dr Mariani: contributed to the revision and approval of the final version of the manuscript.
Dr Lasky-Su: contributed statistical expertise in the analysis of the genetics data and to the revision and approval of the final version of the manuscript.
Dr Hersh: contributed statistical expertise in the analysis of the genetics data and to the revision and approval of the final version of the manuscript.
Dr Raby: contributed to the analysis of the eQTL data and to the revision and approval of the final version of the manuscript.
Dr Silverman: contributed to the study concept and design and to the revision and approval of the final version of the manuscript.
Dr Weiss: contributed to the study concept and design and to the revision and approval of the final version of the manuscript.
Dr DeMeo: contributed to the study concept and design and to the revision and approval of the final version of the manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following conflicts of interest: Dr Avila participates in speaking activities for AstraZeneca. Dr Silverman received grant support and consulting fees from GlaxoSmithKline for studies of COPD genetics and has received honoraria and consulting fees from AstraZeneca. The remaining authors have reported that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: The sponsors had no role in the design of the study, the collection and analysis of the data, or in the preparation of the manuscript.
Other contributions: We thank the CAMP subjects for their ongoing participation in this study. We acknowledge the CAMP investigators and research team, supported by the National Heart, Lung, and Blood Institute, for collection of CAMP Genetic Ancillary Study data. We thank Andy Liu, MD, and Stanley Szefler, MD, (National Jewish; Denver, Colorado); Nadia Hansel, MD, MPH, Greg Diette, MD, MHS, and N. Franklin Adkinson Jr, MD (Johns Hopkins; Baltimore, Maryland); and Karen DeMuth, MD, Robert Strunk, MD, and Mario Castro, MD, MPH (Washington University; St. Louis, Missouri) for their assistance in collection of CD4+ lymphocytes and RNA extraction. We give special thanks to Anne Plunkett, RN, Teresa Concordia, Debbie Bull, LPN, Denise Rodgers, RPFT, and D. Sundstrom, BA, for their assistance with sample collection; Huiqing Yin-DeClue, PhD, Michael McLane, PhD, and Chris Allaire, BSc, for their assistance with T-cell isolations and RNA preparation; and Ankur Patel, BSc, for his assistance with running the microarrays. This work was performed at the Channing Laboratory, Brigham and Women’s Hospital, under appropriate CAMP policies and human subject protections.
Additional information: The e-Appendix, e-Table, and e-Figures can be found in the Online Supplement at http://chestjournal.chestpubs.org/content/140/3/667/suppl/DC1.
Abbreviations
- AHR
airways hyperresponsiveness
- CAMP
Childhood Asthma Management Program
- EMR
electronic medical record
- eQTL
expression quantitative trait loci
- iCAP
i2b2 Crimson Asthma Project
- ICD-9
International Classification of Diseases, Ninth Revision
- LD
linkage disequilibrium
- LogIgE
log-transformed total serum IgE
- SERPINE2
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
- SNP
single nucleotide polymorphism
Footnotes
Funding/Support: The CAMP Genetics Ancillary Study is supported by the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH) [Grants U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601, and T32 HL07427]. Additional support was provided by the NIH [Grants R37 HL066289, HL04370, R01 HL086601, R01 HL087680 (NHLBI)] and National Library of Medicine [2T15LM007092-16]. Dr DeMeo is supported in part by a Doris Duke Clinical-Scientist Development Award.
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians (http://www.chestpubs.org/site/misc/reprints.xhtml).
References
- 1.Orie N, Sluiter H, DeVries K, Tammeling G, Witkop J. The host factor in bronchitis. In: Orie N, Sluiter H, editors. Bronchitis. Assen, The Netherlands: Royal Van Gorcum; 1961. pp. 43–59. [Google Scholar]
- 2.Postma DS, Boezen HM. Rationale for the Dutch hypothesis. Allergy and airway hyperresponsiveness as genetic factors and their interaction with environment in the development of asthma and COPD. Chest. 2004;126(suppl 2):96S–104S. doi: 10.1378/chest.126.2_suppl_1.96S. [DOI] [PubMed] [Google Scholar]
- 3.Barnes PJ. Against the Dutch hypothesis: asthma and chronic obstructive pulmonary disease are distinct diseases. Am J Respir Crit Care Med. 2006;174(3):240–244. doi: 10.1164/rccm.2604008. [DOI] [PubMed] [Google Scholar]
- 4.DeMeo DL, Celedón JC, Lange C, et al. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170(12):1294–1301. doi: 10.1164/rccm.200404-524OC. [DOI] [PubMed] [Google Scholar]
- 5.Postma DS, Meyers DA, Jongepier H, Howard TD, Koppelman GH, Bleecker ER. Genomewide screen for pulmonary function in 200 families ascertained for asthma. Am J Respir Crit Care Med. 2005;172(4):446–452. doi: 10.1164/rccm.200407-864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hersh CP, Soto-Quirós ME, Avila L, et al. Genome-wide linkage analysis of pulmonary function in families of children with asthma in Costa Rica. Thorax. 2007;62(3):224–230. doi: 10.1136/thx.2006.067934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra A, Peiffer AP, Ryujin DT, et al. Further evidence for the role of genes on chromosome 2 and chromosome 5 in the inheritance of pulmonary function. Am J Respir Crit Care Med. 2003;168(5):556–561. doi: 10.1164/rccm.200303-410OC. [DOI] [PubMed] [Google Scholar]
- 8.Demeo DL, Mariani TJ, Lange C, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet. 2006;78(2):253–264. doi: 10.1086/499828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu G, Warren L, Aponte J, et al. International COPD Genetics Network (ICGN) Investigators The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176(2):167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 10.Childhood Asthma Management Program Research Group The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 11.Murphy A, Chu JH, Xu M, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19(23):4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luca D, Ringquist S, Klei L, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82(2):453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himes BE, Hunninghake GM, Baurley JW, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84(5):581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunninghake GM, Soto-Quiros ME, Avila L, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol. 2007;119(3):654–661. doi: 10.1016/j.jaci.2006.12.609. [DOI] [PubMed] [Google Scholar]
- 15.Hunninghake GM, Soto-Quirós ME, Avila L, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol. 2007;120(1):84–90. doi: 10.1016/j.jaci.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor GT, Sparrow D, Weiss ST. A prospective longitudinal study of methacholine airway responsiveness as a predictor of pulmonary-function decline: the Normative Aging Study. Am J Respir Crit Care Med. 1995;152(1):87–92. doi: 10.1164/ajrccm.152.1.7599868. [DOI] [PubMed] [Google Scholar]
- 17.Himes BE, Lasky-Su J, Wu AC, et al. Asthma-susceptibility variants identified using probands in case-control and family-based analyses. BMC Med Genet. 2010;11(1):122. doi: 10.1186/1471-2350-11-122. www.biomedcentral.com/1471-2350/11/122. Accessed June 30, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74(2):367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 22.Liptak T. On the combination of independent tests. Magyar Tud Akad Mat Kutato Int Kozl. 1958;3:171–197. [Google Scholar]
- 23.Evans DL, McGrogan M, Scott RW, Carrell RW. Protease specificity and heparin binding and activation of recombinant protease nexin I. J Biol Chem. 1991;266(33):22307–22312. [PubMed] [Google Scholar]
- 24.Bouton MC, Venisse L, Richard B, Pouzet C, Arocas V, Jandrot-Perrus M. Protease nexin-1 interacts with thrombomodulin and modulates its anticoagulant effect. Circ Res. 2007;100(8):1174–1181. doi: 10.1161/01.RES.0000265066.92923.ee. [DOI] [PubMed] [Google Scholar]
- 25.Carter RE, Cerosaletti KM, Burkin DJ, et al. The gene for the serpin thrombin inhibitor (PI7), protease nexin I, is located on human chromosome 2q33-q35 and on syntenic regions in the mouse and sheep genomes. Genomics. 1995;27(1):196–199. doi: 10.1006/geno.1995.1025. [DOI] [PubMed] [Google Scholar]
- 26.Bai TR. Evidence for airway remodeling in chronic asthma. Curr Opin Allergy Clin Immunol. 2010;10(1):82–86. doi: 10.1097/ACI.0b013e32833363b2. [DOI] [PubMed] [Google Scholar]
- 27.Davies DE. The role of the epithelium in airway remodeling in asthma. Proc Am Thorac Soc. 2009;6(8):678–682. doi: 10.1513/pats.200907-067DP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macnee W. Pathogenesis of chronic obstructive pulmonary disease. Clin Chest Med. 2007;28(3):479–513. doi: 10.1016/j.ccm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Postma DS, Timens W. Remodeling in asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3(5):434–439. doi: 10.1513/pats.200601-006AW. [DOI] [PubMed] [Google Scholar]
- 30.Kuramoto E, Nishiuma T, Kobayashi K, et al. Inhalation of urokinase-type plasminogen activator reduces airway remodeling in a murine asthma model. Am J Physiol Lung Cell Mol Physiol. 2009;296(3):L337–L346. doi: 10.1152/ajplung.90434.2008. [DOI] [PubMed] [Google Scholar]
- 31.Wang IM, Stepaniants S, Boie Y, et al. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med. 2008;177(4):402–411. doi: 10.1164/rccm.200703-390OC. [DOI] [PubMed] [Google Scholar]
- 32.Gosselink JV, Hayashi S, Elliott WM, et al. Differential expression of tissue repair genes in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181(12):1329–1335. doi: 10.1164/rccm.200812-1902OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.