Abstract
Fgf8 and Fgf4 encode FGF family members that are coexpressed in the primitive streak of the gastrulating mouse embryo. We have analyzed the phenotype of Fgf8−/− embryos and discovered that they fail to express Fgf4 in the streak. In the absence of both FGF8 and FGF4, epiblast cells move into the streak and undergo an epithelial-to-mesenchymal transition, but most cells then fail to move away from the streak. As a consequence, no embryonic mesoderm- or endoderm-derived tissues develop, although extraembryonic tissues form. Patterning of the prospective neuroectoderm is greatly perturbed in the mutant embryos. Anterior neuroectoderm markers are widely expressed, at least in part because the anterior visceral endoderm, which provides signals that regulate their expression, is not displaced proximally in the absence of definitive endoderm. Posterior neuroectoderm markers are not expressed, presumably because there is neither mesendoderm underlying the prospective neuroectoderm nor a morphologically normal node to provide the inductive signals necessary for their expression. This study identifies Fgf8 as a gene essential for gastrulation and shows that signaling via FGF8 and/or FGF4 is required for cell migration away from the primitive streak.
Keywords: anterior visceral endoderm, cell migration, epithelial-to-mesenchymal transition, Fgf4, Fgf8, gastrulation, mouse embryo, neuroectoderm patterning, primitive streak
Gastrulation is the process by which a single layer of epithelial cells is transformed into the three germ layers of the embryo—endoderm, mesoderm, and ectoderm—and the basic body plan is established. In the mouse, the undifferentiated epithelium that will develop into the embryo proper is a cup-shaped sheet known as the epiblast. Gastrulation begins at ∼6.5 days after fertilization [embryonic day (E) 6.5], when cells on opposite sides of the epiblast start to move toward a common region, known as the primitive streak. Its location is defined as the posterior side of the embryo, and anterior is on the opposite side of the epiblast. As cells move into the streak region they undergo an epithelial-to-mesenchymal transition (EMT) and subsequently move away from the streak. The streak is therefore a region through which cells are continually trafficking.
The proximal–distal (P-D) axis of the embryo runs from the rim of the cup-like epiblast to its base (see Fig. 1F). Initially the streak forms proximally. Over the next 24 hr it lengthens as cells located more distally in the epiblast move into the streak, and ultimately the streak occupies the entire P-D length of the posterior side of the embryo. Epiblast cells that traverse the streak are fated to form either mesoderm or definitive endoderm. Their specific fates are correlated with the P-D level at which they move through the streak and the direction they take when they move away from it. In contrast, cells that never traverse the streak are fated to form neuroectoderm and surface ectoderm (for review, see Tam and Behringer 1997).
Figure 1.
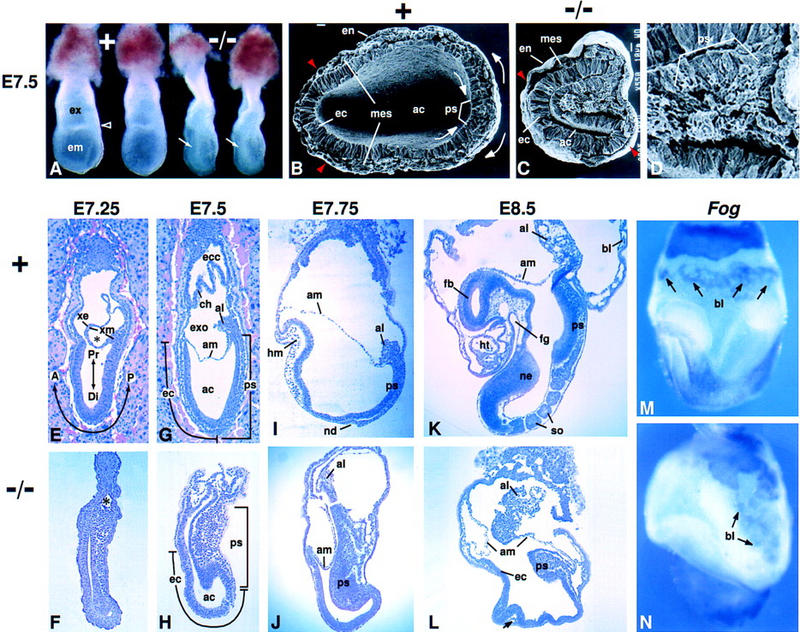
Morphology of Fgf8−/− embryos. In all figures, anterior is to left and posterior to right; intact embryos are viewed laterally, unless otherwise noted. The genotype of each embryo is indicated: Normal embryos were either Fgf8+/− or Fgf8+/+. (A) Offspring at E7.5 of a cross between Fgf8+/− mice. A characteristic bulge of cells in the amniotic cavity (arrow) identifies the two embryos at right as mutant homozygotes. The open arrowhead points to the border between the embryonic (em) and extraembryonic (ex) regions in a normal littermate. (B–D) Scanning electron micrographs of the embryonic region at E7.5. (B) Arrows indicate the direction of epiblast cell movement toward the primitive streak and of mesoderm cell migration away from the streak in a normal embryo. The transverse plane of section was somewhat oblique (higher at right). (C) Fgf8 mutant embryo illustrating the distortion of the primitive streak region, which bulges into the amniotic cavity. Embryos in B and C are shown at the same magnification. (B,C) Red arrowheads indicate the most anterior extent of mesoderm cell migration away from the streak. (D) Higher magnification view of the primitive streak region of the embryo shown in C. Note the accumulation of cells with mesenchymal morphology. (E–L) Sagittal sections of normal and mutant embryos; E and G show embryos sectioned in utero. The times at which they were collected are indicated. The A-P and P-D axes are indicated in E. (E,F) Mid-streak-stage embryos. (*)The nascent exocoelom, which consists of a layer of extraembryonic ectoderm and a layer of mesoderm. (G,H) Late-streak-stage embryos. The extent of the primitive streak and the anterior ectoderm (prospective neuroectoderm) is indicated. (I,J) The normal embryo is at the head-fold stage. In a mutant embryo collected at the same stage, head mesenchyme and a morphologically distinct node are absent. (K,L) The normal embryo contains several somites, a heart, and foregut; the mutant embryo does not contain any such mesoderm- or endoderm-derived tissues. The arrow in L points to a small, isolated patch of mesodermal cells in the mutant embryo. (M,N) Whole-mount in situ hybridization assay for Fog RNA in E8.5 embryos. Signal is detected in blood islands (arrows). (A) Anterior; (ac) amniotic cavity; (al) allantois; (am) amnion; (bl) blood island; (ch) chorion; (Di) distal; (ec) anterior ectoderm (prospective neuroectoderm); (ecc) ectoplacental cone; (em) embryonic region; (en) endoderm; (ex) extraembryonic region; (exo) exocoelom; (fb) forebrain; (fg) foregut; (hm) head mesoderm; (ht) heart; (mes) mesoderm; (nd) node; (ne) neuroectoderm; (P) posterior; (Pr) proximal; (ps) primitive streak; (so) somite; (xe) extraembryonic ectoderm; (xm) extraembryonic mesoderm; (+) normal; (−/−) mutant.
An understanding of mouse gastrulation requires identifying molecules involved in stimulating the movement of epiblast cells towards the streak, inducing them to undergo the EMT, stimulating them to move away from the streak, and determining their fate after they leave the streak. Several lines of evidence suggest that fibroblast growth factors (FGFs) may regulate one or more of these processes. This was first suggested by experiments showing that treatment of Xenopus animal cap explants with FGF2 induced cells that would otherwise adopt an ectodermal fate to form mesoderm (Kimelman and Kirschner 1987; Slack et al. 1987). Subsequent studies have suggested that FGF signaling has multiple functions during gastrulation in Xenopus, including induction of mesoderm, regulation of cell movement, and control of anterior–posterior (A-P) patterning (for review, see Slack et al. 1996). The strongest evidence that FGF signaling is essential for gastrulation in mice comes from loss-of-function studies of FGF receptor 1 (Fgfr1), one of four known vertebrate FGFR genes. Embryos homozygous for null alleles of Fgfr1 fail to gastrulate normally (Deng et al. 1995; Yamaguchi et al. 1995). On the basis of studies of chimeras formed by aggregating Fgfr1−/− cells with wild-type morulae, it was suggested that the primary defect caused by loss of Fgfr1 function is a deficiency in the ability of cells to make the transition from an epithelial to a mesenchymal morphology and thus to traverse the streak (Ciruna et al. 1997).
To date, 18 different vertebrate FGF genes have been identified (Ohbayashi et al. 1998 and references therein). Gene expression analyses have shown that five of these genes, Fgf3 (Wilkinson et al. 1988), Fgf4 (Niswander and Martin 1992), Fgf5 (Haub and Goldfarb 1991; Hébert et al. 1991), Fgf8 (Crossley and Martin 1995; Mahmood et al. 1995), and Fgf17 (Maruoka et al. 1998), are expressed in prestreak- and streak-stage embryos. Fgf3 (Mansour et al. 1993), Fgf5 (Hébert et al. 1994), and Fgf17 (J. Xu and D. Ornitz, pers. comm.) are not individually required for gastrulation, as null mutant homozygotes are viable and fertile. Fgf4 is expressed in the primitive streak, but it has not been possible to determine whether it is required for gastrulation, because Fgf4−/− embryos die shortly after implantation, 2 days before streak formation begins (Feldman et al. 1995).
Fgf8 is expressed just prior to streak formation in a patch of epiblast cells on the proximal prospective posterior side of the embryo, as well as in the visceral endoderm (VE), a layer of extraembryonic cells that envelops the epiblast before and during the streak stages of development. Subsequently, its expression is localized to cells within the primitive streak and is down-regulated shortly after they exit it (Crossley and Martin 1995; Mahmood et al. 1995; Maruoka et al. 1998). We described previously the production of mice carrying an allelic series of mutations at the Fgf8 locus (Meyers et al. 1998). Two of the mutant alleles, Fgf8Δ2,3 and Fgf8Δ2,3n, are presumed to be null alleles because they lack exons 2 and 3, which encode most of the ∼120-amino-acid core domain that is conserved in all FGF family members. Sequences within this conserved domain confer an ability to bind to heparin and to signal-transducing FGFRs (for review, see Basilico and Moscatelli 1992). Mice heterozygous for Fgf8Δ2,3n or Fgf8Δ2,3 are viable and fertile, but embryos homozygous for either allele apparently lack all embryonic mesoderm- and endoderm-derived structures and do not survive beyond E9.5. In this study our goal was to determine what aspect of gastrulation is affected in Fgf8 null mutant homozygotes.
Results
Failure of cell migration away from the primitive streak in Fgf8 mutant homozygotes
Embryos identified by PCR analysis as either Fgf8Δ2,3 or Fgf8Δ2,3n homozygotes or as Fgf8Δ2,3/Fgf8Δ2,3n compound heterozygotes appeared similar at all stages examined. The data on embryos of these genotypes, hereafter referred to as Fgf8−/− or mutant embryos, were therefore pooled. Such embryos could be recognized as early as E7.0 (mid-streak stage) by an abnormal thickening on the posterior side (data not shown). At ∼E7.5, when their wild-type or mutant heterozygous littermates (hereafter referred to as Fgf8+ or normal embryos) were at the late-streak/neural plate stage, the mutant embryos were found to be abnormally narrow, with an indentation on the posterior side and a mass of cells in the proamniotic cavity (Fig. 1A). Analysis of transverse sections revealed the nature of these defects (Fig. 1B–D). In Fgf8+ embryos, cells that traverse the distal and middle region of the primitive streak migrate away from it in an anterolateral direction, thus forming two wings of embryonic mesoderm that extend around the circumference of the embryo (Fig. 1B). In mutant embryos, we found that epiblast cells apparently undergo a transition from epithelial to mesenchymal morphology. However, the majority of nascent mesodermal cells fail to migrate away from the streak (Fig. 1C,D). The number that do was found to differ from one embryo to the next and also from the left to the right side of the same embryo (see Figs. 1C and 2F,L). This failure of cell migration and the consequent pile-up of cells in the streak region results in deformation of the epiblast and inward collapse of the posterior side of the embryo, creating a massive bulge that protrudes into the amniotic cavity (Fig. 1C). From an analysis of sagittal as well as transverse sections at various P-D levels we concluded that cell migration away from the streak is affected along the entire length of the primitive streak (Fig. 1E–H; data not shown).
Figure 2.
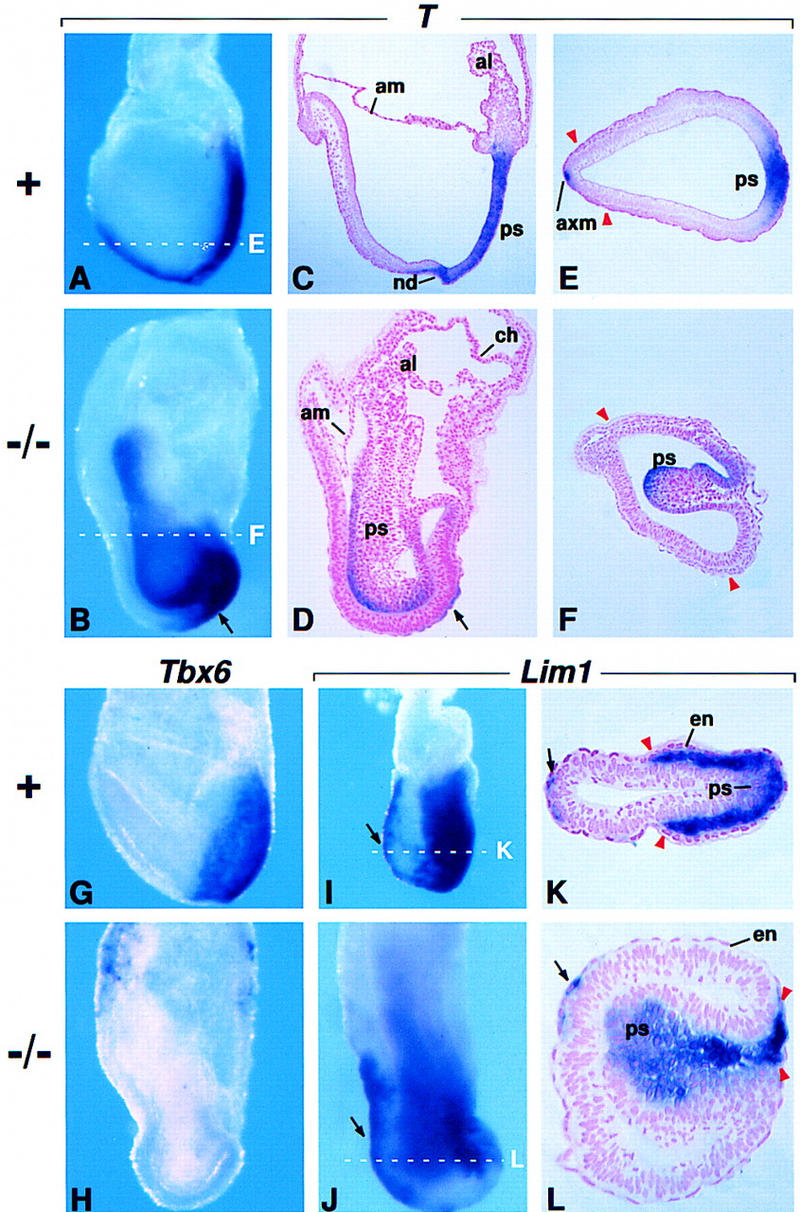
Gene expression in the primitive streak. (A–F) RNA in situ hybridization using a probe for T RNA. (A,B) Embryos at E7.75 in whole mount. (C,D) Sagittal sections of an E7.75 normal embryo stained in whole mount and the mutant embryo shown in B. (E,F) Transverse sections through the normal embryo shown in A and an E7.75 mutant embryo stained in whole mount. The broken white lines in A and B indicate the approximate level of the sections shown in E and F, respectively. Arrows in B and D point to T-expressing cells anterior to the primitive streak. (G,H) Tbx6 expression at E7.75. (I–L) Lim1 expression in an E7.25 normal embryo and an E7.5 mutant embryo. (K,L) Transverse sections through the embryos shown in I and J, respectively. Broken white lines indicate the approximate level of the sections shown. Arrows point to Lim1-expressing cells in the AVE/axial mesoderm. Red arrowheads indicate the most anterior extent of mesoderm cell migration. Abbreviations as in Fig. 1. (axm) Axial mesoderm.
Normally, as the streak elongates, the cell population that was initially localized in the distal epiblast is displaced proximally and expands into the area occupied previously by cells that entered the streak (Lawson et al. 1991; Quinlan et al. 1995). In mutant embryos at ∼E7.5, the streak appeared to be relatively normal in overall length, particularly when distortions resulting from the failure of mesoderm migration were taken into account (Fig. 1G,H). However, despite its relatively normal length, the streak was never found to extend to the distal tip of mutant embryos (Fig. 1H; see also Figs. 2D and 3E). Concomitantly, the length of the embryonic ectoderm anterior to the distal end of the primitive streak appeared to be greater in mutant than in Fgf8+ embryos (Fig. 1G,H). One possible explanation for these observations is that movement of cells into the streak is initiated normally, but the rate of cell entry subsequently decreases because cells are failing to exit the streak region; consequently, the morphogenetic movements that reshape the distal/anterior portion of the epiblast are inhibited in Fgf8−/− embryos.
Figure 3.
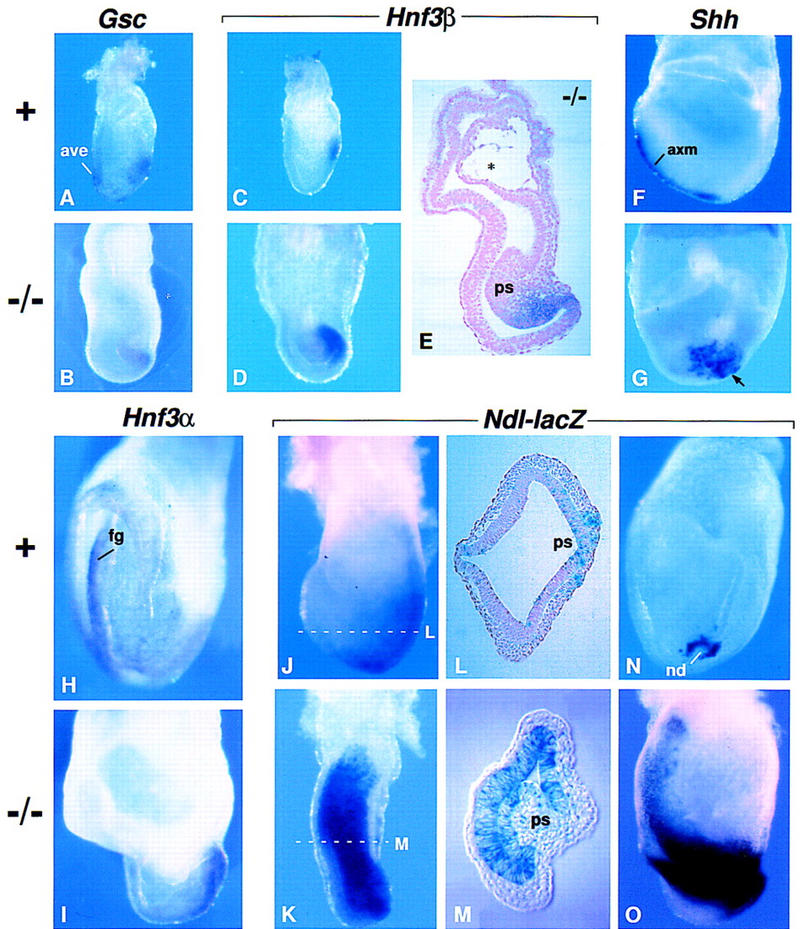
Analysis of markers for the anterior streak and its derivatives. (A,B) Gsc expression in a normal embryo at E7.0 and a mutant embryo at E7.25. Expression was detected in the AVE of the normal but not the mutant embryo. (C,D) Hnf3β expression in a normal embryo at E6.75 and a mutant embryos at E7.5. (E) Parasagittal section of the mutant embryo shown in D. The asterisk (*) indicates the nascent exocoelom. (F,G) Shh expression in a normal (lateral view) and a mutant (posterior view) embryo at E8.0. The arrow points to Shh-expressing cells in the mutant embryo that remain near the distal end of the primitive streak. In most mutant embryos examined there were fewer Shh-expressing cells than are seen here. (H,I) Hnf3α expression in a normal (anterior view) and a mutant (lateral view) embryo at E8.25. Expression is detected in the definitive endoderm of the normal embryo. (J–O) Ndl expression as detected by β-gal staining. (J,K) Normal and mutant embryos at E7.5. (L,M) Transverse sections of the embryos shown in J and K, respectively. The broken white lines indicate the approximate level of the sections. Note that Ndl continues to be detected in the ectoderm on the anterior side of the mutant embryo. (N,O) A normal (posterior view) and a mutant (lateral view) embryo at E8.0. Abbreviations as in Figs. 1 and 2.
Visual inspection of sagittal sections suggested that despite their morphogenetic abnormalities, mutant embryos contain approximately the same total number of cells as their normal littermates. To investigate this point, we collected a litter of eight conceptuses at ∼E7.75, disaggregated them individually, and counted the total number of cells (embryonic and extraembryonic) in each one. The three that were identified as mutant by morphological criteria contained 45,900 ± 1011 cells. Their five morphologically normal littermates contained 50,020 ± 2274 cells. Using Student’s t-test, these numbers are not statistically different. These data suggest that loss of Fgf8 function has little effect on cell proliferation and/or survival during the primitive streak stages of development.
As the mutant embryos developed beyond the streak stages, we observed expansion of the amniotic cavity and elongation along the A-P axis, with the bulging primitive streak region remaining at the caudal end of the embryo (Fig. 1I–L). Nothing resembling the node was detected in mutant embryos, and little if any mesoderm was detected anterior to the streak region. By E8.5, when their Fgf8+ littermates had reached the early somite stage, the Fgf8−/− embryos were found to be significantly smaller than normal and to lack all embryonic mesodermal tissues, such as the somites and heart, as well as all endodermal tissues, such as the gut. However, a few small, isolated patches of mesodermal cells were detected in serial sections of the mutant embryos at E8.5–E9.5 (arrow in Fig. 1L; data not shown). The ectoderm on the anterior side of the embryo, which normally develops into the brain, appeared to be undifferentiated and failed to form a neural tube. The mutant embryos thus show no morphological manifestation of brain development (Fig. 1K,L). By E9.5, the mutant embryos began to die, most likely because the heart did not form.
Development of the extraembryonic region in Fgf8−/− embryos
Although the extraembryonic region is histologically abnormal, the morphogenetic processes that result in the formation of the extraembryonic tissues occur in Fgf8−/− embryos. In the normal embryo, cells that traverse the proximal portion of the primitive streak move into the extraembryonic region (Lawson et al. 1991). Subsequently, lacunae form within the extraembryonic mesodermal cell population, particularly on the posterior side, gradually expand, and coalesce to form the exocoelom (asterisk in Fig. 1E). During this process, the extraembryonic mesoderm, as well as the extraembryonic ectoderm overlying it, is stretched and thinned, like the wall of an expanding balloon (Fig. 1E). By the late-streak stage, the net effect is the formation of the amnion and chorion, which divide the central space of the conceptus into three cavities: the amniotic cavity, the exocoelom, and the ectoplacental cavity (Fig. 1G). In Fgf8−/− mutants, mesoderm cells were found in the extraembryonic region, and we observed evidence of lumen formation and expansion within the extraembryonic mesoderm (Fig. 1F, see also 3E). Subsequently, a chorion and amnion could usually be identified, but they were often tortuous, particularly the amnion (Figs. 1J,L and 2D).
In the normal conceptus, a population of extraembryonic mesoderm cells localized on the posterior side develops into the allantois, which grows across the exocoelom and eventually fuses with the chorion to become part of the placenta. In the mutants an allantois was present and usually relatively normal in size, but it was often displaced anteriorly (Fig. 1J,L). Its abnormal location, as well as the general disorganization of the extraembryonic region, appeared to be due, at least in part, to the accumulation of nascent mesoderm and the characteristic inward collapse of the posterior side of the embryo. Despite these abnormalities, cell types that are normally found in the extraembryonic region were detected in the mutants. For example, germ cells, which are derived from proximal epiblast cells and are marked by alkaline phosphatase (AP) activity, are normally found near the base of the allantois (Ginsburg et al. 1990). In the mutants, AP-positive cells were detected in the extraembryonic region, usually near the base of the allantois (data not shown). Hematopoietic and endothelial cells, which are marked by the expression of Fog (Tsang et al. 1997) and platelet endothelial cell adhesion molecule (PECAM; Baldwin et al. 1994), respectively, normally develop in a network distributed throughout the extraembryonic mesoderm lining the lateral walls of the exocoelom. Fog- and PECAM-expressing cells were detected in the mutants, but they appeared to be present in smaller numbers than normal and were localized exclusively on the posterior side of the extraembryonic region (Fig. 1M,N; data not shown).
Analysis of gene expression in cells traversing the primitive streak
The observations described above demonstrate that cells move into the primitive streak and undergo an EMT in Fgf8−/− embryos and that cells traversing the proximal primitive streak differentiate into the normal complement of extraembryonic cell types. To investigate the development of the streak region further, we performed an in situ hybridization analysis using probes for genes that are normally expressed throughout the streak and in its embryonic derivatives. In the normal embryo beginning at ∼E6.5, expression of the T gene specifically marks all cells in the primitive streak. At E7.75 and later stages, it also marks the axial mesoderm cell population that extends rostrally from the anterior (distal) end of the primitive streak (Fig. 2A,C,E; Wilkinson et al. 1990). In mutant embryos, we detected T RNA in cells with an epithelial morphology along the P-D length of the bulge that protrudes into the amniotic cavity (Fig. 2B,D). Interestingly, T RNA was not detected in most of the cells with a mesenchymal morphology within the bulge (Fig. 2D,F). Although T RNA was not detected in the anterior midline (Fig. 2B,F), it was consistently observed in the region just anterior to the streak (Fig. 2B). Analysis of sagittal sections indicated that these T-expressing cells were localized on the outer surface of the embryo (Fig. 2D); such cells may represent axial mesoderm cells emerging from the distal end of the streak that failed to migrate appropriately. Evx1 expression, which is detected in normal embryos in a decreasing proximal-to-distal gradient within the primitive streak and nascent mesoderm (Dush and Martin 1992), was detected in the streak region of Fgf8−/− embryos (data not shown). In contrast, expression of Tbx6, which normally marks cells in and exiting from the primitive streak beginning at ∼E7.0 (Fig. 2G; Chapman et al. 1996), was not detected in Fgf8−/− embryos (Fig. 2H).
We next assayed for Lim1 RNA, which is normally detected in midstreak-stage embryos at low levels in cells in the primitive streak and at much higher levels in mesodermal cells migrating away from the streak. It is also detected in the VE on the anterior side of the embryo (anterior VE, AVE) as well as in mesendoderm migrating rostrally from the anterior end of the streak (Fig. 2I,K; Shawlot and Behringer 1995; Belo et al. 1997). However, by ∼E7.5 (late-streak/neural plate stage) Lim1 expression is restricted to the node (Barnes et al. 1994). In E7.5 mutant embryos, the Lim1 expression pattern was found to be similar to that in Fgf8+ embryos at an earlier stage. Lim1 RNA was detected at low levels in cells just entering the streak and at higher levels in cells with mesodermal morphology within and exiting the streak region. It was also detected in cells in the anterior midline (Fig. 2J,L). These data show that cells traversing the streak in mutant embryos express at least one marker characteristic of nascent mesodermal cells.
Regionalization of the primitive streak in Fgf8−/− embryos
In the normal embryo, the primitive streak is regionalized with respect to cell fate: Cells that traverse the proximal-most (posterior) end of the streak become extraembryonic mesoderm, whereas those that traverse the distal-most (anterior) end become axial mesoderm and definitive endoderm (mesendoderm) (Lawson et al. 1991). Our finding that extraembryonic mesoderm derivatives such as allantois, blood, and endothelial cells are formed in Fgf8−/− embryos indicates that regionalization of the proximal end of the streak is relatively normal in the absence of FGF8. Consistent with this conclusion, Bmp4 expression, which is detected in the amnion, allantois, and posterior streak of the wild-type embryo (Winnier et al. 1995), appeared normal in the mutants (data not shown). To determine whether the anterior end of the streak is regionalized we assayed for the expression of several genes. In normal embryos at ∼E7, Goosecoid (Gsc) and Hnf3β RNAs are detected in cells at the anterior end of the primitive streak and their derivatives, as well as in the AVE, although the latter expression domain is sometimes difficult to detect (Fig. 3A,C; Blum et al. 1992; Ang et al. 1993; Sasaki and Hogan 1993; Belo et al. 1997). In mutant embryos at E7.5, Gsc and Hnf3β RNAs were detected in cells at the distal tip of the bulging primitive streak region (Fig. 3B,D,E; and data not shown). These data suggest that the anterior end of the primitive streak is correctly patterned in Fgf8−/− embryos. During normal development, Hnf3β is later detected in the node and the mesendoderm on the anterior side of the normal embryo (Ang et al. 1993; Sasaki and Hogan 1993). Although we detected Hnf3β RNA in these tissues in E8.0 Fgf8+ embryos, in their mutant littermates Hnf3β RNA continues to be detected at the distal end of the streak and just anterior to it, but was never detected in any cells on the anterior side (data not shown). At E7.5 Hnf3α is also expressed in cells at the distal end of the normal primitive streak, and subsequently in definitive endodermal cells, such as those in the foregut (Fig. 3H; Monaghan et al. 1993; Sasaki and Hogan 1993). In mutant embryos at E8.0, Hnf3α RNA was detected in cells just anterior to the streak region but not in cells on the anterior side of the embryo (Fig. 3I). Finally, we assayed for the expression of Sonic hedgehog (Shh), which is first detected in the normal embryo at the late-streak stage, exclusively in axial mesoderm extending rostrally from the distal end of the streak. By E8.0 it is also detected in the node (Fig. 3F; Echelard et al. 1993). In mutant embryos at E8.0, Shh RNA was usually detected in a patch of cells clustered near the distal tip of the bulging streak region (Fig. 3G). Together these data indicate that mesendoderm cells are specified in Fgf8−/− embryos, but that they fail to migrate rostrally.
We also examined the pattern of expression of Nodal (Ndl) in Fgf8−/− embryos heterozygous for NdllacZ, an allele in which lacZ disrupts the Nodal gene and functions as a reporter for Ndl expression (Collignon et al. 1996). At all stages examined, Fgf8−/−;NdllacZ/+ embryos were indistinguishable from their Fgf8−/−;Ndl+/+ littermates, indicating that a decrease in Ndl expression does not increase the severity of the Fgf8−/− mutant phenotype. In the normal early embryo Ndl expression is very dynamic and involves a step-wise restriction of the expression domain, which is initially widespread in the epiblast at E6.5 and then localized around the node by E8.0 (Fig. 3J,L,N; Varlet et al. 1997). In mutant embryos at E7.5, Ndl expression, as reported by staining for β-gal activity, was detected throughout the streak region and in the anterior embryonic ectoderm near the embryonic/extraembryonic (em/ex) border (Fig. 3K,M) and persisted until at least E8.25 (Fig. 3O). Because the mutant embryos are not significantly retarded in their development during the streak stages, these data suggest that the mechanism by which Ndl expression becomes restricted is defective in Fgf8−/− embryos.
Anterior neuroectodermal markers are expressed but not regionally restricted in Fgf8−/− embryos
We next sought to determine how loss of Fgf8 function affected the development of the anterior side of the embryo. We first assayed for expression of Hex, a gene that marks the AVE beginning at the prestreak stage, when this cell population is located at the distal tip of the embryo. Hex expression continues in the AVE as it moves proximally, spreading along the P-D length of the anterior side of the embryo (Thomas et al. 1998). As the definitive endoderm is formed and moves rostrally from the distal end of the streak and node, it displaces most of the VE, including the AVE, proximally into the extraembryonic region (Lawson et al. 1991; Thomas and Beddington 1996). Because Hex expression also marks the definitive endoderm population, it continues to be detected on the anterior side of the embryo. However, the Hex expression domain becomes progressively more restricted and by ∼E7.5 is normally detected in a small group of definitive endoderm cells just distal to the em/ex border (Fig. 4A; Thomas et al. 1998). In Fgf8−/− embryos at that stage, Hex RNA was detected in cells on the surface of the embryo in the anterior midline. However, this expression domain was not compact, as in the normal embryo, but was spread along the anterior side of the embryo (Fig. 4B). This is reminiscent of the Hex expression domain in the AVE of normal embryos at the midstreak stage and presumably reflects a lack of proximal displacement of AVE cells caused by the failure of definitive endoderm cells to migrate rostrally in the mutant embryos.
Figure 4.

Patterning of the neuroectoderm in Fgf8−/− embryos. (A,B) Hex expression in normal and mutant embryos at E7.5. (Arrows) Hex-expressing AVE cells. (C,D) Hesx1 expression at E8.0. (E,F) Six3 expression in a normal embryo at E7.75 and a mutant embryo at E8.25. (G,H) Otx2 expression at E7.5. (I,J) Gbx2 expression at E7.75.
Hesx1 (also known as Rpx; Hermesz et al. 1996; Thomas and Beddington 1996) is another gene that is initially expressed in the AVE and subsequently in the definitive endoderm. Beginning at the late-streak stage, Hesx1 expression is induced in the prospective neuroectoderm overlying the Hesx1-positive domain in the endoderm near the em/ex border. Hesx1 expression subsequently intensifies and spreads laterally in the neuroectoderm but remains restricted to the most anterior portion of the developing brain until ∼E9.5 (Fig. 4C; Hermesz et al. 1996; Thomas and Beddington 1996). In Fgf8−/− embryos, Hesx1 RNA was detected throughout the ectoderm, in a domain stretching caudally from the ex/em border on the anterior side of the embryo to the primitive streak (Fig. 4D). A similar expression pattern was observed in mutant embryos for Six3, a gene whose expression domain is normally restricted to the most anterior neuroectoderm from ∼E7.75 through E8.5 (Fig. 4E,F; Oliver et al. 1995). These data indicate that genes whose expression normally marks the onset of neurulation in the anterior neuroectoderm are expressed in Fgf8−/− embryos, but that their expression domains are abnormally expanded.
Otx2 is another gene whose expression is normally detected in the anterior neuroectoderm at the late-streak stage, in the region encompassing the prospective forebrain and midbrain (Fig. 4G; Simeone et al. 1993; Ang et al. 1994). However, unlike Hesx1 and Six3 expression, which is initially induced in the anterior neuroectoderm, Otx2 expression is first detected throughout the epiblast at the prestreak stage and is gradually restricted to the anterior neuroectoderm. In Fgf8 mutant embryos at the late-streak stage, Otx2 expression was detected throughout the embryonic ectoderm anterior to the primitive streak. Together these data show that the signals necessary to regionally restrict gene expression in the prospective forebrain and midbrain are not produced in Fgf8−/− embryos.
We also assayed for Gbx2, a gene required for patterning the anterior hindbrain (Wassarman et al. 1997). It is normally expressed in all three germ layers at the late-streak/neural plate stage, in a domain that extends caudally from the prospective midbrain/hindbrain boundary through the primitive streak to the posterior end of the embryo (Fig. 4I; Bouillet et al. 1995; Wassarman et al. 1997). No Gbx2 RNA was detected in the mutant embryos assayed at E7.5–E8.0 (Fig. 4J). Likewise, Hoxb1 RNA, which is normally expressed in a similar domain but with a rostral limit at the prospective boundary between rhombomeres 3 and 4 (Frohman et al. 1990), was not detected in mutant embryos (data not shown). These results show that markers of the prospective posterior brain and the mesoderm underlying it are not expressed in Fgf8−/− embryos.
FGF8 is required for expression of Fgf4 in the primitive streak
Previous studies have shown that Fgf3 expression, which is normally detected transiently in embryonic mesodermal cells exiting the primitive streak and in the extraembryonic mesoderm (Fig. 5A,C,E; Wilkinson et al. 1988; Niswander and Martin 1992), is not detected in embryos homozygous for a null allele of Fgfr1. However, we detected Fgf3 RNA in both the embryonic and extraembryonic region of Fgf8−/− embryos (Fig. 5B,D,F). Thus, FGF8 is not required for expression of Fgf3. Fgf5 expression was also detected in mutant embryos, in the normal pattern throughout the epiblast up to the late-streak stage (Fig. 5G,H; Haub and Goldfarb 1991; Hébert et al. 1991). We also assayed at E7.5 for Fgf8 expression using a riboprobe that hybridizes to Fgf8 sequences present in the null alleles. Fgf8 RNA was detected in the bulge of cells within the amniotic cavity of the mutant embryos (data not shown), indicating that the Fgf8 gene does not require FGF8 protein for its own transcription in the early mouse embryo. In contrast, Fgf4 expression, which is normally restricted to the primitive streak (Fig 5I; Niswander and Martin 1992) was not detected in the mutant embryos from E7.25 to E8.5 (Fig. 5J; data not shown). These results indicate that the expression of the Fgf4 gene in the primitive streak depends, directly or indirectly, on the presence of a functional Fgf8 gene.
Figure 5.

Expression of FGF family members in Fgf8−/− embryos. (A–F) Fgf3 expression in normal and mutant embryos at E7.5. Broken lines in A and B indicate the levels of the sections shown in C, E, and D, F, respectively. (G,H) Fgf5 expression at E7.25. (I,J) Fgf4 expression at E7.5. As illustrated in J, Fgf4 RNA is not detected in mutant embryos.
Discussion
During mouse gastrulation, Fgf8 and Fgf4 are coexpressed in the primitive streak: Fgf4 is highly expressed at the distal end and barely detectable at the proximal end, whereas Fgf8 is expressed in an opposite gradient throughout the streak. Surprisingly, Fgf4 is not expressed in the streak of Fgf8−/− embryos. Presumably it is expressed prior to E6.5, otherwise Fgf8 mutants would die at the time of implantation, as Fgf4−/− embryos do (Feldman et al. 1995). Thus, elimination of Fgf8 function in effect mimics a primitive streak-specific knock-out of Fgf4, making possible an analysis of the combined function of FGF8 and FGF4 (FGF8/4) in the primitive streak. The most striking feature of this double loss-of-function phenotype is a massive accumulation of cells on the posterior side of the embryo, which results from the failure of most cells that have entered the streak and appear to have made the EMT to migrate away from the streak region. Consequently, the embryonic portion of the conceptus develops in the almost complete absence of mesoderm- and definitive endoderm-derived tissues. In turn, this greatly perturbs the patterning of the prospective neuroectoderm.
FGF signaling is required for cell migration away from the primitive streak
The primary defect in Fgf8 mutant embryos is a failure of cell movement away from the streak. Although there is virtually no development of embryonic tissues, the extraembryonic region apparently contains all the structures and cell types normally found there: The amnion and chorion are formed, the allantois develops, and hematopoietic, endothelial, and presumptive germ cells are detected, but they are present in smaller numbers than normal and are abnormally localized. Thus, the prospective extraembryonic cells, which traverse the proximal portion of the streak, are able to exit it and move proximally into the extraembryonic region, whereas most prospective embryonic mesoderm and endoderm cells, which traverse more distal regions, are unable to exit the streak.
One possible explanation for this difference is that there is regional variation in the mechanism by which cells move away from the streak. Time-lapse micrography of gastrulating mouse embryos has shown that migration away from the distal region of the streak is an active process (Nakatsuji et al. 1986). Our data demonstrate that some aspect of this active process is dependent on FGF8- and/or FGF4-mediated signal transduction. The fact that a small fraction of cells can move away from the streak in Fgf8−/− embryos may reflect functional redundancy between FGF8/4 and FGF17, which is also produced in the streak (Maruoka et al. 1998), or some other FGF family member that has yet to be identified. In contrast, cell movement into the extraembryonic region may be a more passive process that does not require FGF signaling, and once there, cells may be displaced to the anterior side without active migration. For example, the cells lining the wall of the exocoelom are apparently spread anteriorly in the normal conceptus by a process involving expansion of the fluid-filled exocoelomic cavity in which both the extraembryonic mesoderm and ectoderm cells are thinned and stretched. Such a process, which does not appear to require active cell movement, could account for the presence of mesoderm on the anterior side of the extraembryonic region in Fgf8 mutant embryos. On the other hand, the observation that blood and endothelial cells are localized exclusively on the posterior side (Fig. 1, cf. M and N) suggests that FGF-dependent cell movements may be required for the normal distribution of some extraembryonic mesodermal cell types.
An important question is whether the mutant phenotype described here is due to the absence of FGF8, FGF4, or both. Studies of embryos homozygous for a null allele of eed, a gene homologous to Drosophila extra sex combs (Schumacher et al. 1996), suggest that FGF4 is required for cell migration away from the streak. In eed mutant embryos, Fgf4 is not expressed in the primitive streak (Faust et al. 1995), but Fgf8 expression appears to be normal (T. Magnuson, pers. comm.). An elegant lineage analysis has demonstrated that loss of eed function causes epiblast cells to preferentially make mesoderm, which is then unable to migrate anteriorly and laterally away from the primitive streak and becomes mislocalized to the extraembryonic region (Faust et al. 1998). It is tempting to speculate that the migration defects in the eed mutant embryos are due to the absence of FGF4, thus accounting for the many similarities in the eed and Fgf8 mutant phenotypes. Features that are specific to the eed mutants, including excess extraembryonic mesoderm and failure of the primitive streak to elongate normally, are presumably caused by loss of eed function per se. A test of the hypothesis that FGF4 is required for cell migration from the primitive streak will require streak-specific inactivation of Fgf4 to circumvent the early lethality of Fgf4−/− embryos. Because the absence of FGF4 does not affect Fgf8 expression, at least in eed mutant embryos, it should be possible to determine the specific function of Fgf4 by inactivating it in the streak.
Mesodermal cells also accumulate in the streak region of Fgfr1−/− embryos (Deng et al. 1995; Yamaguchi et al. 1995), suggesting that the Fgf8/4 signal required for cell migration is transduced by FGFR1. However, the Fgfr1 mutant phenotype is less severe than that of Fgf8−/− embryos: More cells migrate away from the streak region and thus there is substantially more development of mesoderm- and endoderm-derived tissues in many of the Fgfr1−/− embryos. This raises the possbility that loss of Fgfr1 function only partially inhibits FGF signaling via FGF8/4 and that another FGF receptor functions in conjunction with FGFR1 during gastrulation. Additional expression and genetic analysis will need to be carried out to test this hypothesis.
Other functions of FGF signaling during gastrulation
Studies on Xenopus animal caps have suggested that FGF signaling plays a role in mesoderm induction (Kimelman and Kirschner 1987; Slack et al. 1987). However, interference with FGFR function in Xenopus embryos (Amaya et al. 1991) and loss of Fgfr1 function in the mouse (Deng et al. 1995; Yamaguchi et al. 1995) have no obvious effect on the initiation of gastrulation. Although the Fgf8 mutant embryos display some morphological abnormalities suggestive of a slower than normal rate of cell entry into the streak, it seems likely that such defects are a secondary consequence of the ‘traffic jam’ in the streak.
Two lines of evidence suggest that FGF signaling may play a role in the EMT at gastrulation: Treatment of mouse anterior epiblast explants with FGF2 causes them to undergo an EMT (Burdsal et al. 1993), and mutant cells accumulate in the epithelial portion of the streak in Fgfr1−/− ↔ Fgfr+/+ chimeras. This led Ciruna et al. (1997) to suggest that the primary defect associated with loss of FGFR1 function is a deficiency in the ability to make the EMT. Although the epiblast is deformed in the streak region of Fgf8 mutant embryos, this does not seem to be because of an excess of epithelial cells, but rather because of the accumulation of cells that have completed the EMT but are then unable to migrate away from the streak region (see Fig. 1C,D). Thus, if some aspect of the EMT is dependent on FGF signaling, FGF8/4 are unlikely to be the ligands involved. Likewise, signaling via FGF8/4 cannot be responsible for the lack of Fgf3 expression in Fgfr1−/− embryos (Yamaguchi et al. 1995) because Fgf3 is expressed in its normal domains in Fgf8−/− embryos. Genetic analysis of the functions of other FGF family members, alone and in various combinations, should lead to an identification of the ligands involved in these processes.
There is also some evidence that FGF signaling may be required for the specification of mesodermal cell fate. Analysis of marker gene expression in developmentally advanced Fgfr1−/− embryos showed a remarkable excess of axial mesoderm and a paucity of paraxial mesoderm (Yamaguchi et al. 1995). It has been suggested that this phenotype is secondary to reduced cell movement through the streak (Ciruna et al. 1997). However, we found that primitive streak cells in Fgf8 mutant embryos fail to express Tbx6, a gene required for the specification of posterior paraxial mesoderm (Chapman and Papaioannou 1998). This raises the possibility that in addition to effects on cell migration, FGF8/4 signaling, perhaps via FGFR1, has a role in regulating the expression of genes involved in cell fate determination.
Fgfr1−/− embryos were reported to be significantly smaller than their normal littermates, and the outgrowths of Fgfr1−/− embryos in vitro were consistently smaller than those formed by normal embryos (Deng et al. 1995; Yamaguchi et al. 1995), suggesting that FGFR1 signaling may stimulate cell proliferation or survival in the gastrulating embryo. On the other hand, when Fgfr1−/− or Fgfr1−/+ embryonic stem (ES) cells were combined with wild-type embryos, the ES cells of the two genotypes made similar contributions to the resulting chimeric embryos, arguing against a role for FGFR1 in cell proliferation or survival (Ciruna et al. 1997). Our data suggest that signaling via FGF8/4 is not required for these processes, although we cannot rule out the possibility that the proliferation rate in specific regions of the embryo, such as the streak itself, is abnormal in Fgf8−/− embryos.
How does FGF signaling control cell migration?
In invertebrates, FGF signaling is also necessary for cell migration. For example, in Drosophila it is required for migration and spreading of the embryonic mesoderm over the ectoderm and for branching morphogenesis of the tracheal system, and in Caenorhabditis elegans it is required for sex myoblast migration. In both organisms, ectopic expression experiments have suggested that FGFs can function as attractants for cell migration (for review, see Skaer 1997; Chen and Stern 1998). By analogy, one might argue that FGF8 produced in the VE acts as an attractant for cell migration away from the streak. We tested this hypothesis by injecting wild-type embryonic stem cells into Fgf8−/− blastocysts, producing chimeras in which the VE presumably contained only Fgf8−/− cells, whereas the epiblast contained a mixture of wild-type and mutant cells. No defects in gastrulation were detected in four such chimeras, in which at least 25% of the embryonic cell population was derived from wild-type ES cells. In these embryos, Fgf8−/− cells contributed to all tissues, including somites, head mesenchyme, and foregut (data not shown). These results indicate that lack of FGF8 in the VE is not responsible for the gastrulation defects in Fgf8 mutant embryos. Instead, FGF signaling appears to be required in the primitive streak itself, presumably to regulate the production of proteins necessary for cell migration.
Genes that encode molecules involved in adhesive interactions between cells and their surrounding extracellular matrix (ECM) are obvious candidates for the downstream targets affected by loss of Fgf8 function. Mutational analysis has shown that there is a deficit of mesoderm in embryos homozygous for null alleles of Fibronectin (George et al. 1993), Integrin α5 (Yang et al. 1993), and Focal adhesion kinase (Furuta et al. 1995), which encode a component of the ECM, part of the receptor for Fibronectin, and a nonreceptor tyrosine kinase thought to mediate Integrin signaling, respectively. This suggests that those genes might be required for cell migration away from the primitive streak. However, abnormalities in the mutant embryos are not detected until at least the late headfold stage. It therefore seems unlikely that the more severe phenotype of Fgf8−/− embryos is due to effects of FGF8/4 signaling on any one of these genes, although it remains possible that the defects are due to simultaneous effects on more than one such gene.
Another type of molecule that appears to play some role in cell migration away from the primitive streak is the transcription factor T. When the behavior of T null homozygous cells is monitored in chimeras, they are found to accumulate in the mesodermal layer of the streak region, but this effect is not evident until the headfold stage (Wilson et al. 1995). Moreover, T null mutant embryos do not show any obvious defects at the primitive streak stages (Chesley 1935). The fact that Fgf8 mutant embryos display a more severe phenotype argues against interference with T expression as the primary cause of the defect in cell migration. However, other genes related to T might be the downstream targets of FGF signaling required for cell migration away from the streak. Consistent with this hypothesis, we have found that FGF8/4 signaling regulates expression of at least some T-related genes. For example, Tbx6 is not expressed in Fgf8−/− embryos. Furthermore, although T expression is detected in epithelial cells in the mutant streak region, it is not detected in nascent mesenchymal cells that have traversed the streak and accumulated there (Fig. 2D,F). In contrast, T expression is detected in both the epithelial and mesenchymal portions of the streak in Fgf8+ embryos, and even in cells a short distance away from the streak (Fig. 2E). These observations are consistent with some aspects of the positive-feedback loop model proposed for regulation of the expression of Xbra and eFgf, the Xenopus orthologs of T and Fgf4, respectively (for review, see Smith et al. 1997) and the finding that in zebrafish, expression of T and two T-related genes, spadetail and Tbx6 is regulated by FGF signaling (Griffin et al. 1998).
Patterning defects in the prospective neuroectoderm
In Fgf8 mutant embryos, there is widespread expression throughout the anterior ectoderm of genes whose expression domains are normally restricted within the prospective anterior neuroectoderm, as well as a lack of expression of genes that normally mark the prospective hindbrain. These abnormalities arise from the failure of different aspects of the neuroectoderm patterning process.
One source of signals that pattern the prospective neuroectoderm is the AVE (Beddington and Robertson 1998, 1999). Precursors of the AVE are initially localized in the VE at the distal tip of the embryo, but just prior to streak formation they become distributed along the future anterior midline and are later displaced proximally into the extraembryonic region by definitive endoderm migrating anteriorly from the anterior streak. When AVE progenitors remain distal, as in embryos homozygous for a null allele of Cripto, gene expression normally restricted to the anterior neuroectoderm is detected throughout the distal ectoderm (Ding et al. 1998). In Fgf8 mutant embryos, expression of Lim1 and Hex, two markers of the AVE, persists along the anterior midline of the mutant embryos (Figs. 2J and 4B). This indicates that the AVE is not displaced proximally, presumably because there is no anterior movement of definitive endoderm cells. Therefore the entire anterior ectoderm remains under the influence of the AVE. This provides an explanation for our finding that Hesx1 and Six3 are expressed throughout the ectoderm rather than in restricted domains in the prospective forebrain. Our data thus suggest that the AVE is sufficient to induce anterior neural markers and support the hypothesis that the early proximal movement of the AVE is critical in orchestrating the normal patterning of the neuroectoderm.
Mesoderm and definitive endoderm are also a source of neuroectoderm patterning signals (for review, see Doniach 1993; Ruiz i Altaba 1993). For example, explant-recombination experiments have shown that streak-derived tissue from the posterior side produces signals that can repress Otx2 expression and that anterior mesendoderm produces signals that can induce and maintain Otx2 expression. Together, these signals appear to be responsible for the progressive restriction of the Otx2 expression domain to the forebrain and midbrain (Ang et al. 1994). In Fgf8−/− embryos, Otx2 expression was detected throughout the anterior ectoderm. This failure to become restricted within the anterior ectoderm is most likely due to the absence of streak-derived tissue underlying the prospective neuroectoderm resulting from the lack of cell migration away from the streak. The lack of mesoderm, which is thought to be a potent source of vertical signals that pattern the prospective hindbrain and spinal cord, is also likely to explain the lack of expression of posterior neuroectoderm markers such as Hoxb1 and Gbx2. It is also possible that the absence of a normal node, from which planar signals to the neuroectoderm are thought to emanate, contributes to the abnormalities detected in the anterior ectoderm of Fgf8 mutant embryos.
Conclusion
The data reported here identify FGF8 as a factor required for cells that have undergone the EMT to move away from the streak, possibly because it is necessary for expression of Fgf4. However, in other developmental settings in the vertebrate embryo these same ligands appear to play a role in controlling cell proliferation, cell survival, and embryonic patterning. One of the major challenges for the future will be to understand what determines the specific downstream responses to signaling by a particular FGF ligand and how that leads to a specific biological response such as cell migration.
Materials and methods
Production and analysis of mutant embryos
The Fgf8Δ2,3 and Fgf8Δ2,3n alleles (Meyers et al. 1998) were maintained on a mixed genetic background. Mutant homozygotes were obtained from appropriate crosses of heterozygous mice. They were identified either by their characteristic morphology or by PCR analysis. PCR amplification of DNA fragments unique to Fgf8Δ2,3 or Fgf8Δ2,3n was performed using the primer pairs 5′-CTTAGGGCTATCCAACCCATC-3′ and 5′-AGCTCCCGCTGGATTCCTC-3′ or 5′-GTTCTAAGTACTGTGGTTTCC-3′ and 5′-AGCTCCCGCTGGATTCCTC-3′, respectively. Standard protocols were used, with an annealing temperature of 54°C. Normal embryos used for comparison were either mutant heterozygous or wild-type littermates of the Fgf8 mutant homozygotes or were stage-matched wild-type embryos obtained from crosses of outbred mice.
Noon of the day on which the vaginal plug was detected was considered as E0.5 in the timing of embryo collection. All dissections were performed in phosphate buffered saline (PBS) and embryos were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. Samples for histological analysis were embedded in plastic resin (JB-4 catalyzed resin, Polysciences, Inc., Warrington, PA.) according to the manufacturer’s protocol. Sections were cut at 5 μm and stained with hematoxylin and eosin. Samples for scanning electron microscopy were prepared according to standard protocols; the embryos were bisected after fixation but prior to dehydration. Cell number in individual embryos was determined using a hemacytometer. The embryos were disaggregated by incubating them in 0.05% trypsin for 20 min at 37°C.
Whole-mount RNA in situ hybridization analysis was carried out as described previously (Neubüser et al. 1997) using riboprobes prepared from plasmids described in references cited for each gene. For sectioning after staining, the embryos were postfixed in 4% PFA with 0.1% glutaraldehyde and embedded in plastic resin as described above. Sections were counterstained with 0.1% nuclear fast red.
Ndl expression was analyzed by staining for β-gal activity in embryos carrying the NdllacZ allele (Collignon et al. 1996). Fgf8Δ2,3n /+;NdllacZ/+ double heterozygotes are normal and fertile. They were crossed to Fgf8Δ2,3n /+ mice to generate Fgf8−/−; NdllacZ/+ embryos.
Acknowledgments
We thank S.-L. Ang, R. Beddington, R. Behringer, E. De Robertis, P. Gruss, B. Herrmann, A. McMahon, K. Mahon, S. Orkin, V. Papaioannou, and J. Rossant for providing the probes and E. Robertson for providing the NdllacZ mice used in this study. We are grateful to M. Flannery and R. Pederson for providing the wild-type embryo sections shown in Figure 1E,G; to D. Ilic and C. Damsky for helpful advice; and to M. Embry, A. Gannon, and L. Prentice for excellent technical assistance. We thank P. Tam and T. Magnuson for helpful discussion. We are also grateful to our laboratory colleagues for critical readings of the manuscript. X.S. is the recipient of a postdoctoral fellowship from the American Cancer Society. E.N.M. is the recipient of an MCSD award (HD01216) from the National Institutes of Health (NIH). This work was supported by NIH grant RO1 HD34380 (to G.R.M.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gmartin@itsa.ucsf.edu; FAX (415) 476-3493.
References
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Ang S-L, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Ang S-L, Conlon RA, Jin O, Rossant J. Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development. 1994;120:2979–2989. doi: 10.1242/dev.120.10.2979. [DOI] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin HS, Shen HM, Yan HC, DeLisser HM, Chung A, Mickanin C, Trask T, Kirschbaum NE, Newman PJ, Albelda SM, Buck CA. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): Alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–2553. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- Barnes JD, Crosby JL, Jones CM, Wright CV, Hogan BL. Embryonic expression of Lim-1, the mouse homolog of Xenopus Xlim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev Biol. 1994;161:168–178. doi: 10.1006/dbio.1994.1018. [DOI] [PubMed] [Google Scholar]
- Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- Beddington RSP, Robertson EJ. Anterior patterning in mouse. Trends Genet. 1998;14:277–284. doi: 10.1016/s0168-9525(98)01499-1. [DOI] [PubMed] [Google Scholar]
- ————— Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- Belo JA, Bouwmeester T, Leyns L, Kertesz N, Gallo M, Follettie M, De Robertis EM. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech Dev. 1997;68:45–57. doi: 10.1016/s0925-4773(97)00125-1. [DOI] [PubMed] [Google Scholar]
- Blum M, Gaunt SJ, Cho KW, Steinbeisser H, Blumberg B, Bittner D, De Robertis EM. Gastrulation in the mouse: The role of the homeobox gene goosecoid. Cell. 1992;69:1097–1106. doi: 10.1016/0092-8674(92)90632-m. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Chazaud C, Oulad-Abdelghani M, Dolle P, Chambon P. Sequence and expression pattern of the Stra7 (Gbx-2) homeobox-containing gene induced by retinoic acid in P19 embryonal carcinoma cells. Dev Dynamics. 1995;204:372–382. doi: 10.1002/aja.1002040404. [DOI] [PubMed] [Google Scholar]
- Burdsal CA, Damsky CH, Pedersen RA. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829–844. doi: 10.1242/dev.118.3.829. [DOI] [PubMed] [Google Scholar]
- Chapman D, Papaioannou V. Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature. 1998;391:695–697. doi: 10.1038/35624. [DOI] [PubMed] [Google Scholar]
- Chapman D, Agulnik I, Hancock S, Silver L, Papaioannou V. Tbx6, a mouse T-box gene implicated in paraxial mesoderm formation at gastrulation. Dev Biol. 1996;180:534–542. doi: 10.1006/dbio.1996.0326. [DOI] [PubMed] [Google Scholar]
- Chen EB, Stern MJ. Understanding cell migration guidance: Lessons from sex myoblast migration in C. elegans. Trends Genet. 1998;14:322–327. doi: 10.1016/s0168-9525(98)01507-8. [DOI] [PubMed] [Google Scholar]
- Chesley P. Development of the short-tailed mutant in the house mouse. J Exp Zool. 1935;70:429–459. [Google Scholar]
- Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: A role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Deng C-X, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial formation. Genes & Dev. 1995;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- Doniach T. Planar and vertical induction of anteroposterior pattern during the development of the amphibian central nervous system. J Neurobiol. 1993;24:1256–1275. doi: 10.1002/neu.480241003. [DOI] [PubMed] [Google Scholar]
- Dush M, Martin GR. Analysis of mouse Evx genes: Evx-1 displays graded expression in the primitive streak. Dev Biol. 1992;151:273–287. doi: 10.1016/0012-1606(92)90232-6. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohle RJ, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Boyle M, Martin GR. Isolation of the mouse Hox-2.9 gene: Analysis of embryonic expression suggests that positional information along the anterior posterior axis is specified by mesoderm. Development. 1990;110:589–607. doi: 10.1242/dev.110.2.589. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Ilic D, Kanazawa S, Takeda N, Yamamoto T, Aizawa S. Mesodermal defect in late phase of gastrulation by a targeted mutation of focal adhesion kinase, FAK. Oncogene. 1995;11:1989–1995. [PubMed] [Google Scholar]
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- Ginsburg M, Snow M, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Griffin K, Amacher S, Kimmel C, Kimelman D. Molecular identification of spadetail: Regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development. 1998;125:3379–3388. doi: 10.1242/dev.125.17.3379. [DOI] [PubMed] [Google Scholar]
- Haub O, Goldfarb M. Expression of the fibroblast growth factor-5 gene in the mouse embryo. Development. 1991;112:397–406. doi: 10.1242/dev.112.2.397. [DOI] [PubMed] [Google Scholar]
- Hébert JM, Boyle M, Martin GR. mRNA localization studies suggest that murine fibroblast growth factor-5 plays a role in gastrulation. Development. 1991;112:407–415. doi: 10.1242/dev.112.2.407. [DOI] [PubMed] [Google Scholar]
- Hébert J, Rosenquist T, Götz J, Martin G. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Hermesz E, Mackem S, Mahon KA. Rpx: A novel anterior-restricted homeobox gene progressively activated in the prechordal plate, anterior neural plate and Rathke’s pouch of the mouse embryo. Development. 1996;122:41–52. doi: 10.1242/dev.122.1.41. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner MW. Synergistic induction of mesoderm by FGF and TGF-β and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell. 1987;51:869–877. doi: 10.1016/0092-8674(87)90110-3. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- Mahmood R, Bresnick J, Hornbruch A, Mahony C, Morton N, Colquhoun K, Martin P, Lumsden A, Dickson C, Mason I. A role for FGF-8 in the initiation and maintenance of vertebrate limb bud outgrowth. Curr Biol. 1995;5:797–806. doi: 10.1016/s0960-9822(95)00157-6. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Maruoka Y, Ohbayashi N, Hoshikawa M, Itoh N, Hogan B, Furuta Y. Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mech Dev. 1998;74:175–177. doi: 10.1016/s0925-4773(98)00061-6. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kaestner KH, Grau E, Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Nakatsuji N, Snow MH, Wylie CC. Cinemicrographic study of the cell movement in the primitive-streak-stage mouse embryo. J Embryol Exp Morphol. 1986;96:99–109. [PubMed] [Google Scholar]
- Neubüser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: A mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Hoshikawa M, Kimura S, Yamasaki M, Fukui S, Itoh N. Structure and expression of the mRNA encoding a novel fibroblast growth factor, FGF-18. J Biol Chem. 1998;273:18161–18164. doi: 10.1074/jbc.273.29.18161. [DOI] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Quinlan G, Williams E, Tan S, Tam P. Neuroectodermal fate of epiblast cells in the distal region of the mouse egg cylinder: Implication for body plan organization during early embryogenesis. Development. 1995;121:87–98. doi: 10.1242/dev.121.1.87. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Induction and axial patterning of the neural plate: Planar and vertical signals. J Neurobiol. 1993;24:1276–1304. doi: 10.1002/neu.480241004. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;384:19–26. doi: 10.1038/384648a0. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D’Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaer H. Morphogenesis: FGF branches out. Curr Biol. 1997;7:238–241. doi: 10.1016/s0960-9822(06)00110-2. [DOI] [PubMed] [Google Scholar]
- Slack JMW, Darlington BG, Heath JK, Godsave SF. Mesoderm induction in early Xenopus embryos by heparin-binding growth factors. Nature. 1987;326:197–200. doi: 10.1038/326197a0. [DOI] [PubMed] [Google Scholar]
- Slack J, Issacs H, Song J, Durbin L, Pownall M. The role of fibroblast growth factors in early Xenopus development. Biochem Soc Symp. 1996;62:1–12. [PubMed] [Google Scholar]
- Smith JC, Armes NA, Conlon FL, Tada M, Umbhauer M, Weston KM. Upstream and downstream from Brachyury, a gene required for vertebrate mesoderm formation. Cold Spring Harb Symp Quant Biol. 1997;62:337–346. [PubMed] [Google Scholar]
- Tam PPL, Behringer RR. Mouse gastrulation: The formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Thomas P, Beddington R. Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr Biol. 1996;6:1487–1496. doi: 10.1016/s0960-9822(96)00753-1. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: A homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Tsang A, Visvader J, Turner C, Fujiwara Y, Yu C, Weiss M, Crossley M, Orkin S. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development. 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- Wassarman K, Lewandoski M, Campbell K, Joyner AL, Rubenstein JLR, Martinez S, Martin GR. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development. 1997;124:2923–2934. doi: 10.1242/dev.124.15.2923. [DOI] [PubMed] [Google Scholar]
- Wilkinson D, Peters G, Dickson C, McMahon A. Expression of the FGF-related proto-oncogene int-2 during gastrulation and neurulation in the mouse. EMBO J. 1988;7:691–695. doi: 10.1002/j.1460-2075.1988.tb02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Wilson V, Manson L, Skarnes WC, Beddington RS. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development. 1995;121:877–886. doi: 10.1242/dev.121.3.877. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & Dev. 1995;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Embryonic mesodermal defects in alpha 5 integrin-deficient mice. Development. 1993;119:1093–1105. doi: 10.1242/dev.119.4.1093. [DOI] [PubMed] [Google Scholar]


