Abstract
Drosophila Pimples (PIM) and Three rows (THR) are required for sister chromatid separation in mitosis. PIM accumulates during interphase and is degraded rapidly during mitosis. This degradation is dependent on a destruction box similar to that of B-type cyclins. Nondegradable PIM with a mutant destruction box can rescue sister chromatid separation in pim mutants but only when expressed at low levels. Higher levels of nondegradable PIM, as well as overexpression of wild-type PIM, inhibit sister chromatid separation. Moreover, cells arrested in mitosis before sister chromatid separation (by colcemid or by mutations in fizzy/CDC20) fail to degrade PIM. Thus, although not related by primary sequence, PIM has intriguing functional similarities to the securin proteins of budding yeast, fission yeast, and vertebrates. Whereas these securins are known to form a complex with separins, we show that PIM associates in vivo with THR, which does not contain the conserved separin domain.
Keywords: Mitosis, sister chromatid separation, securin, separin, pimples, three rows
Pairs of sister chromatids are generated during the S phase of the eukaryotic cell division cycle. Sister chromatids remain paired throughout the G2 phase and during the initial phase of mitosis (prophase) while chromatin is condensed and the spindle is assembled. However, the cohesion between sister chromatids is ultimately destroyed at the metaphase–anaphase transition allowing their segregation to opposite poles. Considerable progress has been made recently in understanding the molecular basis of cohesion and separation of sister chromatids (for review, see Zachariae and Nasmyth 1999; Nasmyth et al. 2000). Cohesion is known to be dependent on the binding of the cohesin protein complex to nascent sister chromatids during S phase (Guacci et al. 1997; Michaelis et al. 1997; Losada et al. 1998; Uhlmann and Nasmyth 1998; Blat and Kleckner 1999; Tanaka et al. 1999; Toth et al. 1999; Watanabe and Nurse 1999). Separation of sister chromatids in budding yeast mitosis requires the proteolytic processing of the cohesin subunit Scc1p/Mcd1p during the metaphase–anaphase transition (Uhlmann et al. 1999). In vertebrates, cohesin complexes dissociate from chromosomes already during prophase concomitant with chromatin condensation and well before the onset of sister chromatid separation (Losada et al. 1998; Darwiche et al. 1999). Moreover, Scc1p cleavage during prophase is not detectable in Drosophila (S. Heidmann, unpubl.). However, it is not excluded that residual cohesin complexes might persist in particular in the centromeric region of vertebrate chromosomes. The final separation of sister chromatids in higher eukaryotes, therefore, might also result from Scc1p cleavage during the metaphase–anaphase transition.
This hypothesis of a conserved mechanism of sister chromatid separation in eukaryotes is supported by findings concerning the role of the separin and securin proteins (Nasmyth et al. 2000). The separins (Esp1p, Cut1, BimB) were implicated originally in mitosis based on genetic analyses in fungi. Homologous genes have been detected recently in plant and animal species. All these separins share a conserved carboxy-terminal domain, the separin domain. The budding yeast separin Esp1p is known to be required for Scc1p cleavage and sister chromatid separation (Uhlmann et al. 1999). Separins are thought to be activated only during the metaphase–anaphase transition. Premature activation of separins is prevented by securin proteins that accumulate during interphase and bind to the separins. The budding yeast securin Pds1p forms a complex with Esp1p (Ciosk et al. 1998). The fission yeast securin Cut2 binds to Cut1 (Funabiki et al. 1996a; Yanagida 2000). In vertebrates, the protein encoded by the pituitary tumor transforming gene (PTTG) associates with a protein containing the conserved separin domain (Zou et al. 1999). All these securins (Pds1p, Cut2, PTTG) share essentially no sequence similarity except for the presence of at least one destruction box, a nine amino acid consensus motif [RX(A or V or L)LGXXXN] originally defined in B-type cyclins. Securins are therefore degraded rapidly during the metaphase–anaphase transition like mitotic cyclins. Securin proteins with mutations in the destruction box fail to be degraded and inhibit sister chromatid separation in yeast and in Xenopus extracts (Cohen-Fix et al. 1996; Funabiki et al. 1996b, 1997; Zou et al. 1999).
Mitotic proteolysis of destruction box proteins occurs after polyubiquitination resulting from the activation of a special ubiquitin ligase known as anaphase-promoting complex/cyclosome (APC/C). APC/C activation, therefore, is a crucial step in the regulation of the metaphase–anaphase transition (for review, see Zachariae and Nasmyth 1999). This activation process is not yet fully understood. However, it is clear that the WD-40 repeat proteins Fizzy/Cdc20p and Fizzy-related/Hct1p/Cdh1p play important roles in APC/C regulation. These proteins bind to the APC/C in different cell cycle phases and respond to different regulatory inputs. While Drosophila Fizzy-related is known to be essential for the degradation of mitotic cyclins in G1, Fizzy is required for cyclin degradation and sister chromatid separation during mitosis (Sigrist et al. 1995; Sigrist and Lehner 1997). The dependency of sister chromatid separation on Cdc20p function has been explained in budding yeast by the finding that Cdc20p is required for the degradation of the securin Pds1p (Visintin et al. 1997; Lim et al. 1998; Shirayama et al. 1999). Fizzy/Cdc20p is inactivated in the presence of unattached kinetochores and spindle damage by a mitotic checkpoint pathway which results in the binding of the inhibitor Mad2p to the Fizzy/Cdc20p–APC/C complex (Chen et al. 1996; Fang et al. 1998; Hwang et al. 1998; Kallio et al. 1998; Kim et al. 1998; Alexandru et al. 1999; Waters et al. 1999; Zachariae and Nasmyth 1999). This checkpoint pathway therefore assures that sister chromatid separation and exit from mitosis occur only when all chromosomes have acquired the correct bipolar orientation within a functional spindle.
With the exception of securins, all the components involved in the control of sister chromatid separation that have been introduced above are highly conserved in eukaryotes. Interestingly, we have identified previously two nonconserved Drosophila genes, pimples (pim) and three rows (thr), which are both required specifically for sister chromatid separation during mitosis (D'Andrea et al. 1993; Stratmann and Lehner 1996). We show that the Pimples protein (PIM) shares extensive functional similarities with securin proteins and in particular with Cut2 from fission yeast. Moreover, we demonstrate that PIM is found in a complex with Three rows protein (THR). Our results indicate that the regulation of sister chromatid separation in Drosophila involves securin-like proteins that associate with proteins lacking the evolutionary conserved separin domain.
Results
PIM and THR are present in a complex
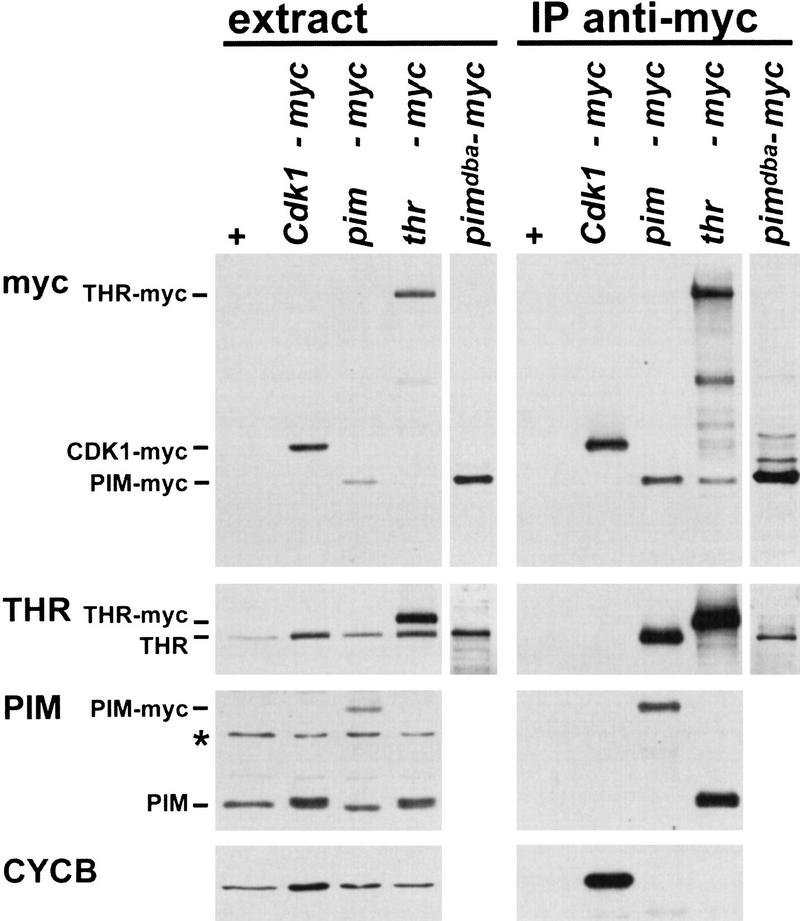
Neither PIM nor THR share significant sequence similarities with known proteins, and their biochemical function is not known. However, the indistinguishable phenotypes resulting from null mutations in pim and thr suggested that the corresponding gene products might function in a complex. Therefore, we analyzed PIM–THR complex formation by coimmunoprecipitation. Extracts were prepared from embryos carrying transgenes (gpim–myc or gthr–myc) allowing expression of either PIM protein with a carboxy-terminal extension of six myc epitope copies or THR protein with a carboxy-terminal extension of 10 myc epitope copies under the control of the corresponding genomic promoters. These myc-tagged proteins are functional because the transgenes can rescue pim and thr mutants, respectively. Anti-myc immunoprecipitates of PIM–myc were found to contain THR (Fig. 1). Conversely, immunoprecipitates of THR–myc contained PIM (Fig. 1). Control immunoprecipitates of CDK1–myc contained Cyclin B, as expected, but did not contain PIM or THR (Fig. 1), indicating that coimmunoprecipitation of PIM and THR is specific. Our coimmunoprecipitation experiments also indicated that the PIM–THR complex does not contain multiple copies of PIM and THR. In case of complexes with multiple copies, PIM–myc and THR–myc immunoprecipitates would be expected to contain wild-type PIM and THR, respectively. However, the products expressed from the endogenous loci were not coimmunoprecipitated by the myc-tagged transgene products (Fig. 1).
Figure 1.
PIM and THR form a complex in vivo. Extracts (extract) prepared from embryos expressing either no transgene (+), or Cdk1–myc (Cdk1–myc), gpim–myc (pim–myc), gthr–myc (thr–myc), or pimdba–myc (pimdba–myc), as well as anti-myc immunoprecipitates isolated from these extracts (IP anti-myc), were analyzed by immunoblotting with antibodies against the myc epitope (myc), THR (THR), PIM (PIM), or Cyclin B (CYCB). (*) Crossreaction of the antibodies against PIM with an unknown protein.
Mitotic PIM degradation depends on a destruction box and is required for sister chromatid separation
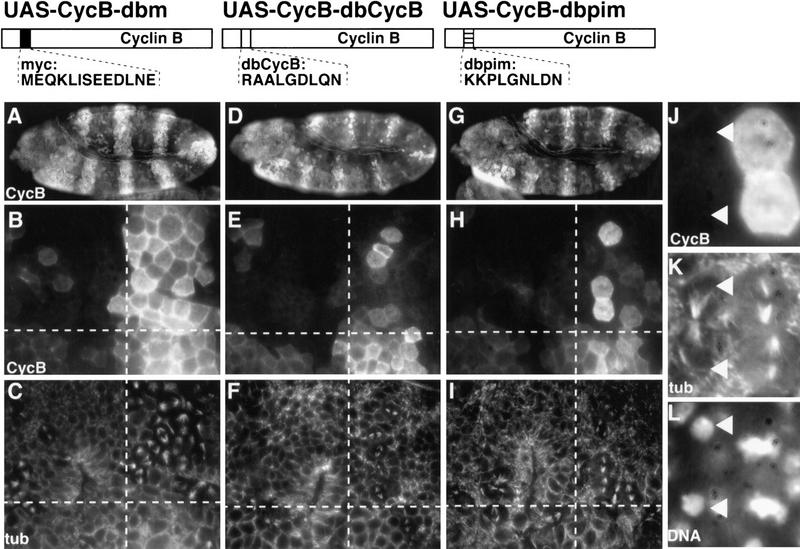
By immunolabeling we have shown previously that PIM–myc is cleared from mitotic cells after the metaphase–anaphase transition similar to Cyclin B (Stratmann and Lehner 1996). The mitotic degradation of Cyclin B and other mitotic regulators is dependent on the presence of a destruction box motif in the amino-terminal region (Peters et al. 1998). Drosophila Cyclin B lacking this destruction box cannot be degraded during mitosis and blocks exit from mitosis (Rimmington et al. 1994; Sigrist et al. 1995; Fig. 2A–C). Although PIM does not have a motif that fits the RX(A or V or L)LGXXXN consensus sequence of mitotic destruction boxes (King et al. 1996; Peters et al. 1998; Zou et al. 1999), it contains the related sequence KKPLGNLDN. To determine whether this sequence variant can function as a destruction box, we expressed a mutant Cyclin B protein in Drosophila embryos that had this PIM motif instead of the Cyclin B destruction box. The PIM motif conferred mitotic instability indistinguishable from wild-type Cyclin B and did not result in a mitotic arrest (Fig. 2, cf. D–F with G–L). When the PIM motif was mutated from KKPLGNLDN to AKPAGNLDA (dba), it was no longer able to functionally replace the destruction box in Cyclin B (data not shown).
Figure 2.
The PIM destruction box variant can replace the Cyclin B destruction box. prd–GAL4, which directs UAS target gene expression in alternating segments starting before mitosis 15 of Drosophila embryogenesis, was used to express either Cyclin B with the myc epitope in place of the destruction box (UAS–CycB–dbm; A–C), or Cyclin B with the endogenous destruction box (UAS–CycB–dbCycB; D–F), or Cyclin B with the PIM destruction box (UAS–CycB–dbpim; G–L). Embryos (A,D,G) were fixed during the stage of mitosis 15 and double-labeled with antibodies against Cyclin B (CycB; A,B,D,E,G,H,J), tubulin (tub; C,F,I,K) and a DNA stain (DNA; L). Higher magnification views of the embryonic epidermis (B,C,E,F,H,I) are shown with the regions of UAS target gene expression to the right of the dashed vertical lines. UAS target gene expression is absent from the regions on the left of the dashed vertical line. These regions express only endogenous Cyclin B and serve as internal control for progression through mitosis 15. Progression through mitosis 15 is accompanied by degradation of Cyclin B protein when carrying a functional degradation box and occurs in a segmentally repeated pattern (Foe et al. 1993) first in the dorsal epidermis (above the horizontal dashed line) and only later in the ventral epidermis (below the horizontal dashed line). At the stage shown, mitosis 15 is largely completed in the dorsal epidermis and just starting in the ventral epidermis. Nondegradable Cyclin B with the myc epitope in place of the destruction box blocks exit from mitosis and results in an enrichment of mitotic figures (C, upper right) in cells that are labeled by anti-Cyclin B (B, upper right). In contrast, Cyclin B with the PIM motif in place of the destruction box does not block exit from mitosis and is degraded during late mitosis as illustrated in the regions shown at even higher magnification (J–L). Arrowheads mark a telophase cell that is not labeled with anti-Cyclin B (J) while the neighboring metaphase cells (right) are strongly labeled. The structure of the different UAS-transgenes is schematically illustrated above the panels with the corresponding results.
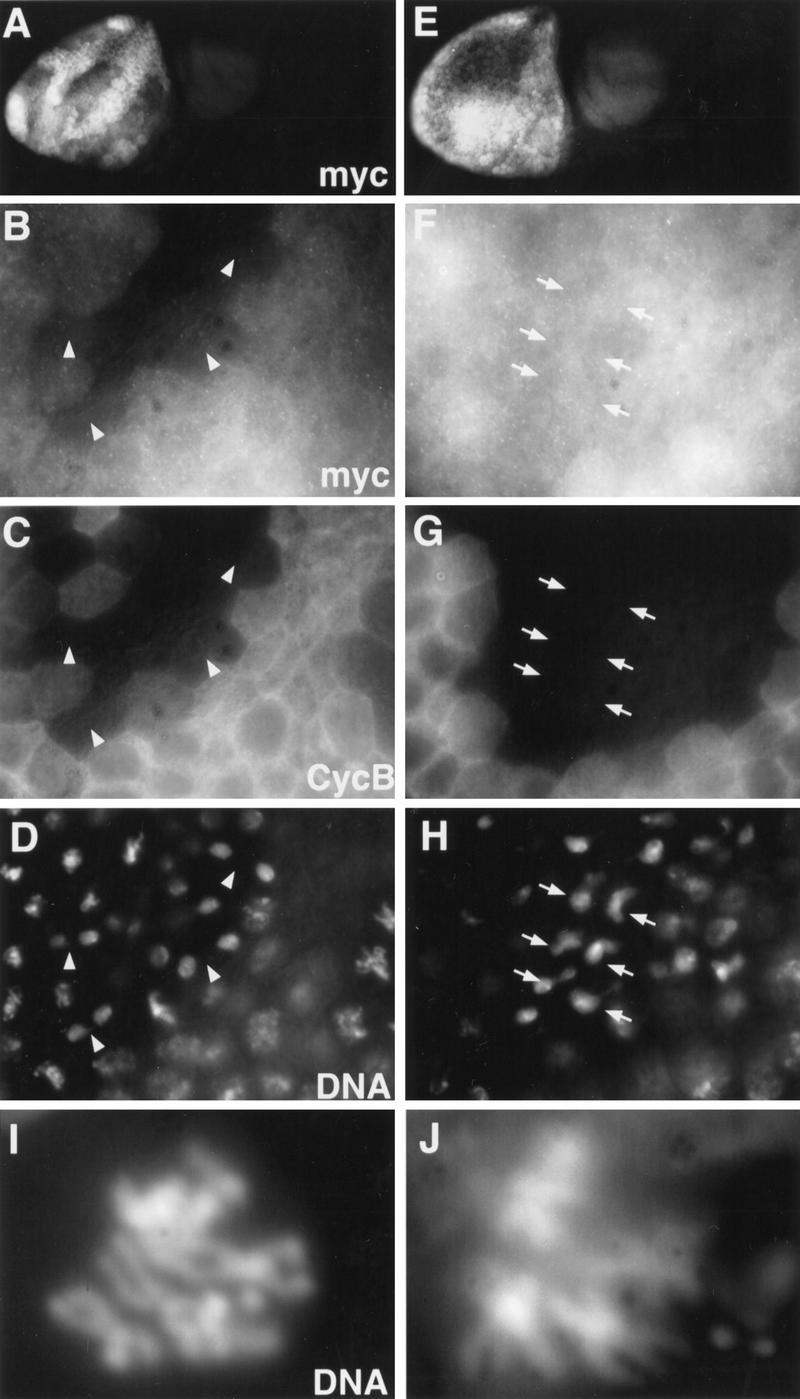
To determine whether the KKPLGNLDN sequence is required for PIM degradation during mitosis, we introduced the dba mutation into a pim transgene (UAS–pimdba–myc). The mutant PIMdba–myc protein product was found to be stable during mitosis, while wild-type PIM–myc expressed from an analogous transgene (UAS–pim–myc) was degraded normally (Fig. 3, cf. A,B with E,F).
Figure 3.
PIM with mutations in the destruction box motif is stable in mitosis and inhibits sister chromatid separation. nos–GAL4–GCN4–bcd3′UTR was used to express either wild-type PIM with carboxy-terminal myc epitopes (UAS–pim–myc; A–D) or PIM with carboxy-terminal myc epitopes and a mutant destruction box (UAS–pimdba–myc; E–H) in the anterior region of gastrulating embryos. Embryos (A,E) were fixed and labeled with antibodies against the myc epitope (myc; A,B,E,F), Cyclin B (CycB; C,G) and a DNA stain (DNA; D,H). Arrowheads in the high magnification views of a head region indicate normal anaphase and telophase figures (B–D), while arrows mark abnormal “metaphase” plates with decondensing chromosomes (E–H) in regions that lack anti-Cyclin B labeling and thus have progressed beyond the metaphase–anaphase transition. UAS–pimdba–myc I.1; UAS–pimdba–myc III.1 embryos for control (I) and UAS–pimdba–myc I.1/+; UAS–pimdba–myc III.1/da–GAL4 embryos (J), in which mitosis 15 is the first division affected by the expression of nondegradable PIM, were incubated in colcemid at the stage of mitosis 16 before preparation of mitotic chromosome spreads stained for DNA. Diplo-chromosomes (J) indicating the failure of sister chromatid separation during mitosis 15 were not observed in controls (I).
PIMdba–myc did not block the mitotic degradation of Cyclin A (data not shown) and Cyclin B (Fig. 3G) indicating that it does not inhibit the APC/C-dependent degradation pathway. Interestingly, however, PIMdba–myc was found to block sister chromatid separation. In UAS–pimdba–myc-expressing embryos, we observed only abnormal, decondensing metaphase plates in the regions without Cyclin B labeling (Fig. 3H, see arrows) instead of anaphase and telophase figures which are abundant in those regions of control embryos that have degraded Cyclin B and thus have progressed beyond the metaphase–anaphase transition (Fig. 3D, see arrowheads). Early mitotic figures (prophase and metaphase) were normal in UAS–pimdba–myc-expressing embryos, and tubulin labeling revealed the presence of mitotic spindles (Fig. 4E; data not shown). The observation that congression of mitotic chromosomes into the metaphase plate occurred normally indicated that PIMdba–myc does not interfere with spindle function.
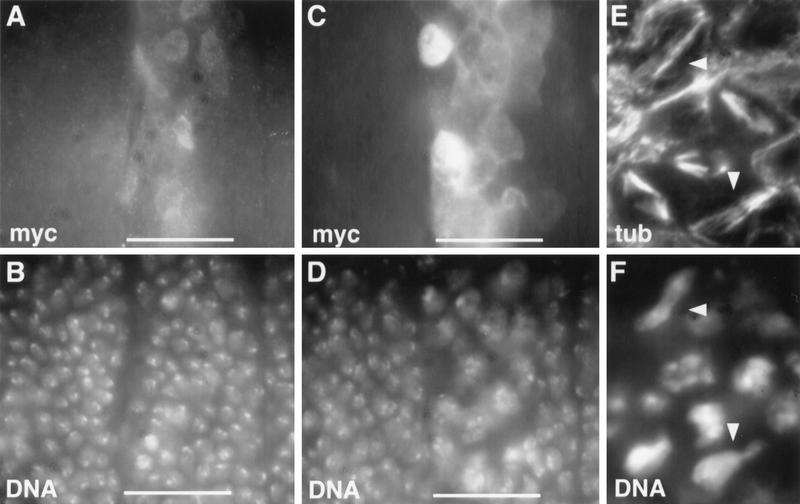
Figure 4.
Overexpression of wild-type PIM inhibits sister chromatid separation. prd–GAL4 (A–D) or da–GAL4 (E,F) was used for overexpression of wild-type PIM with carboxy-terminal myc epitopes from either one (A,B) or two (C–F) UAS–pim–myc transgene copies. Embryos were fixed either after mitosis 16 (A–D) or during mitosis 16 (E,F) and labeled with antibodies against the myc epitope (myc; A,C), tubulin (tub; E) and with a DNA stain (DNA; B,D,F). Overexpression from one UAS–pim–myc copy does not affect progression through the sixteenth embryonic division. The normal nuclear density is therefore observed in the prd–GAL4-expressing segments (A,B, white horizontal bars). In contrast, overexpression from two UAS–pim–myc copies results in inhibition of sister chromatid separation during mitosis 16. Arrowheads in E and F indicate cells during telophase of mitosis 16 with unseparated chromosomes. As a consequence, cytokinesis fails as well, but exit from mitosis 16 occurs normally. This failure of sister chromatid separation and cytokinesis is evidenced by the lower density of interphase nuclei in the prd–GAL4-expressing segments (C,D, white bars) after mitosis 16.
The nos–GAL4–GCN4–bcd3′UTR transgene used in these experiments to drive UAS–pimdba–myc expression resulted in a graded expression with a maximum at the anterior pole of the embryo. Whereas an apparently complete block of sister chromatid separation occurred in regions with high levels of expression (Fig. 3F–H), only a partial inhibition was observed in regions with lower expression levels. In these regions, aberrant anaphase and telophase figures with chromatin bridges were frequent (data not shown).
To confirm that high levels of UAS–pimdba–myc expression abolished sister chromatid separation specifically and not other processes during cell cycle progression, we analyzed mitotic chromosomes from UAS–pimdba–myc I.1; UAS–pimdba–myc III.1/da–GAL4 embryos after treatment with the microtubule destabilizing drug colcemid (demecolcine) during the stage of mitosis 16. In these embryos, sister chromatid separation appeared to be inhibited completely during mitosis 15 which follows after the onset of da–GAL4-driven UAS-transgene expression (data not shown). Thus, after nondisjunction of sister chromatids during mitosis 15 and re-replication during S phase 16, diplochromosomes would be expected to be present during the colcemid-arrested mitosis 16. In fact, whereas we observed only normal mitotic chromosomes in control embryos (Fig. 3I), mitotic cells with a normal number of chromosomes that had twice as many arms than normal chromosomes were present in the UAS–pimdba–myc-expressing embryos (Fig. 3J). The presence of these diplochromosomes demonstrates that UAS–pimdba–myc expression specifically blocks sister chromatid separation.
The finding that sister chromatid separation was inhibited by the nondegradable PIMdba–myc protein suggested that this process is dependent on mitotic PIM degradation. High levels of wild-type PIM resulting from overexpression, therefore, might inhibit sister chromatid separation equally. In fact, sister chromatid separation failed when two copies of the UAS–pim–myc transgene were expressed during the embryonic mitoses using the da–GAL4 or prd–GAL4 transgenes (Fig. 4C–F). Expression of one UAS–pim–myc copy did not inhibit sister chromatid separation (Fig. 4A,B). Quantitative immunoblotting experiments indicated that ubiquitous expression of two UAS–pim–myc copies with da–GAL4 resulted in about five-fold higher levels of expression compared to wild type (data not shown). Although this level of overexpression inhibited sister chromatid separation, it did not interfere with mitotic cyclin destruction. Moreover, UAS–pim–myc overexpression in endoreduplicating salivary gland cells throughout late embryogenesis and larval development had no effect, whereas it resulted in severe phenotypic abnormalities in mitotically proliferating imaginal disc cells (data not shown). Overexpression of wild-type pim, therefore, is not generally cytotoxic and inhibits sister chromatid separation specifically.
Interestingly, the phenotype resulting from UAS–pimdba–myc and UAS–pim–myc overexpression is identical to the phenotype observed in mutant embryos lacking pim function (Stratmann and Lehner 1996). It appears, therefore, that both the accumulation of PIM during interphase as well as the subsequent degradation during mitosis are important for sister chromatid separation.
PIM degradation is regulated by the spindle checkpoint
Mitotic degradation of Cyclins A, B, and B3 requires Fizzy/Cdc20p, an activator of APC/C-dependent ubiquitination (Dawson et al. 1995; Sigrist et al. 1995). To evaluate whether Fizzy is also involved in PIM degradation during mitosis, we analyzed the consequences of UAS–pim–myc expression in fizzy mutants. The maternal fizzy contribution present in fizzy mutants is sufficient for progression through all of the 16 embryonic divisions in the dorsal epidermis when UAS–pim–myc is not expressed (Sigrist et al. 1995). However, when UAS–pim–myc was expressed, sister chromatid separation was found to be inhibited in the dorsal epidermis of fizzy mutants during mitosis 16, while exit from this mitosis 16 still occurred. Importantly, in contrast to the results observed in wild-type embryos (Fig. 4A,B), expression of just one UAS–pim–myc copy was already sufficient for inhibition of sister chromatid separation in the dorsal region of fizzy mutants (Fig. 5A,B) and resulted in a phenotype that was only observed in wild-type embryos when two UAS–pim–myc copies were expressed (Fig. 4C,D).
Figure 5.
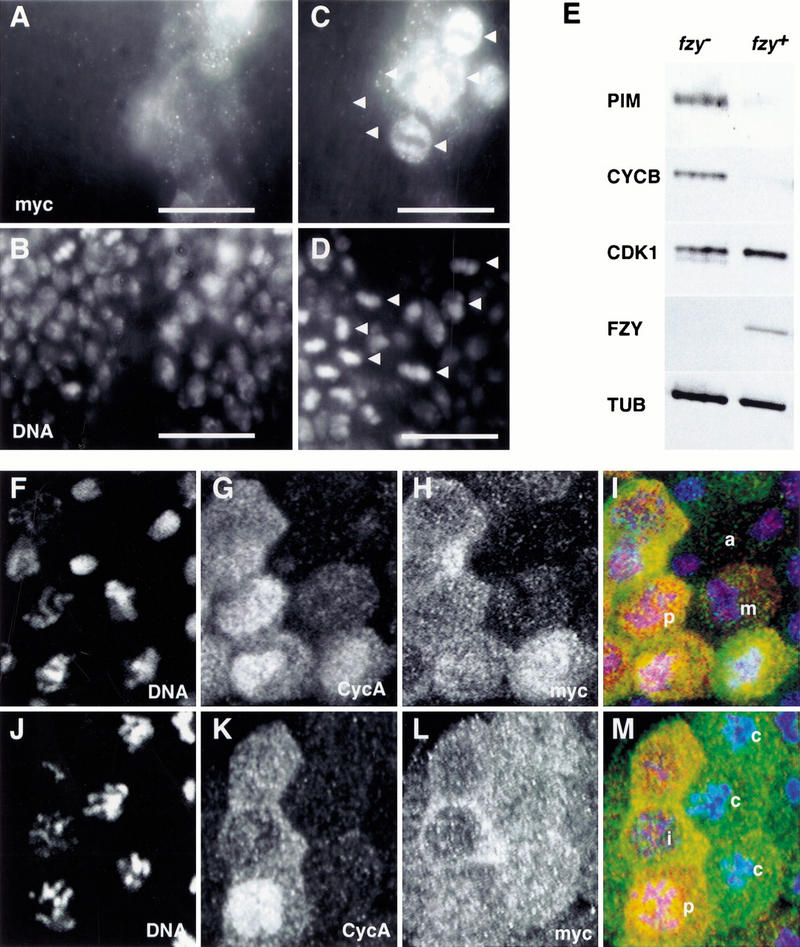
PIM persists during the mitotic arrest caused by colcemid or lack of Fizzy. (A–D) prd–GAL4 was used to express one UAS–pim–myc copy in fizzy mutant embryos. Embryos were fixed at a stage where mitosis 16 is completed during wild-type development and labeled with anti-myc (A,C), a DNA stain (B,D), and anti-β-galactosidase for the identification of fizzy homozygotes (data not shown). High magnification views of the dorsal epidermis (A,B) illustrate that one UAS–pim–myc copy is sufficient to inhibit sister chromatid separation during mitosis 16 in fizzy mutants, leading to the reduced nuclear density in the prd–GAL4-expressing regions (white bar). High magnification views of the ventral epidermis (C,D) illustrate the persistence of PIM–myc in cells arrested in metaphase 16 because of lack of Fizzy (arrowheads), leading to the intense anti-myc labeling in the arrested cells within the prd–GAL4-expressing region (white bar). (E) Progeny from fizzy/CyO parents was aged to the stage where mitosis 16 is completed during wild-type development. fizzy homozygous embryos (fzy−) were sorted from sibling embryos (fzy+) and analyzed by immunoblotting with antibodies against PIM (PIM), Cyclin B (CYCB), Cdk1 (CDK1), FZY (FZY), and tubulin (TUB). (F–M) nos–GAL4–GCN4–bcd3′ UTR was used to express PIM with carboxy-terminal myc epitopes (UAS–pim–myc). Embryos at the stage of mitosis 14 were permeabilized and incubated for 25 min either in the absence (F–I) or presence (J–M) of colcemid before fixation and labeling with antibodies against the myc epitope (myc; H,L), Cyclin A (CycA; G,K) and a DNA stain (DNA; F,J). The merged panels (I,M) show labeling of DNA in blue, Cyclin A in red, and PIM–myc in green. (i) Interphase; (p) prophase; (m) metaphase; (a) anaphase; (c) colcemid-arrested cells.
In the ventral region of fizzy homozygotes, the maternal fizzy contribution is not sufficient to allow completion of mitosis 16. Therefore, a large fraction of ventral cells become arrested during metaphase 16 in fizzy mutants (Dawson et al. 1995; Sigrist et al. 1995). When UAS–pim–myc was expressed in fizzy mutants, we observed very strong anti-myc labeling in the arrested cells of the ventral region (Fig. 5C). This labeling was much more intense than in the dorsal UAS–pim–myc expressing cells that were not arrested (Fig. 5A). The persistence of PIM–myc during metaphase arrest resulting from lack of fizzy function was also observed when expression was directed at lower levels by transgenes under the control of the pim+ regulatory region (data not shown). Moreover, immunoblotting experiments confirmed that the endogenous PIM protein is also stabilized in fizzy homozygotes (Fig. 5E). We conclude, therefore, that fizzy is required for PIM degradation during mitosis.
Spindle defects result in a mitotic checkpoint arrest during which sister chromatids do not separate, possibly because PIM is not degraded. To evaluate whether PIM is stable during a mitotic checkpoint arrest, we treated UAS–pim–myc-expressing embryos with colcemid (Fig. 5J–M) and used mock-treated embryos as control (Fig. 5F–I). These embryos were subsequently labeled with a DNA stain (Fig. 5F,J) to identify arrested cells and with anti-myc antibodies (Fig. 5H,L) to monitor the presence of PIM–myc. We also labeled the embryos with an antibody against Cyclin A (Fig. 5G,K) which is known to be degraded in colcemid-arrested cells in contrast to Cyclin B (Whitfield et al. 1990). PIM–myc remained clearly detectable in mitotic domains of arrested cells with condensed chromosomes and without Cyclin A labeling (Fig. 5L,M, see cells labeled “c”). These observations demonstrate that PIM is not degraded in cells arrested by the spindle checkpoint pathway.
Sister chromatid separation in the presence of low levels of nondegradable PIM
The PIM persistence in cells arrested by colcemid or lack of fizzy, as well as the inhibition of sister chromatid separation resulting from UAS–pimdba–myc and UAS–pim–myc expression, were consistent with the notion that sister chromatid separation is strictly dependent on PIM degradation during mitosis. However, the experiments with UAS–pimdba–myc and UAS–pim–myc involved overexpression. To analyze the effects of physiological levels of PIMdba–myc, we constructed a transgene with the pim+ regulatory region directing PIMdba–myc expression (gpimdba–myc). Interestingly, we were able to establish transgenic lines indicating that expression of a single gpimdba–myc copy is tolerated in a pim+ background. However, when present in two copies, transgene insertions resulted in complete lethality (five out of eight lines) or severe morphological abnormalities (rough eyes, notched wings, sterility) in rare escapers (three out of eight lines). The analysis of heterozygous combinations of different transgene insertions indicated that these phenotypes were not caused by transgene insertion position effects. The phenotypes indicated clearly that PIMdba–myc is highly toxic.
The fact that we were able to isolate transgenic lines with gpimdba–myc insertions suggested that sister chromatid separation is not absolutely dependent on complete PIM degradation during each mitosis. However, gpimdba–myc expression might occur only at very low levels as a result of a selection against insertions generating normal expression levels during transgene establishment. Moreover, wild-type PIM might compete with PIMdba–myc and thereby protect cells. Therefore, we addressed gpimdba–myc expression levels in immunoblotting experiments (Fig. 6) and analyzed the consequences of gpimdba–myc expression in pim mutants (Fig. 7).
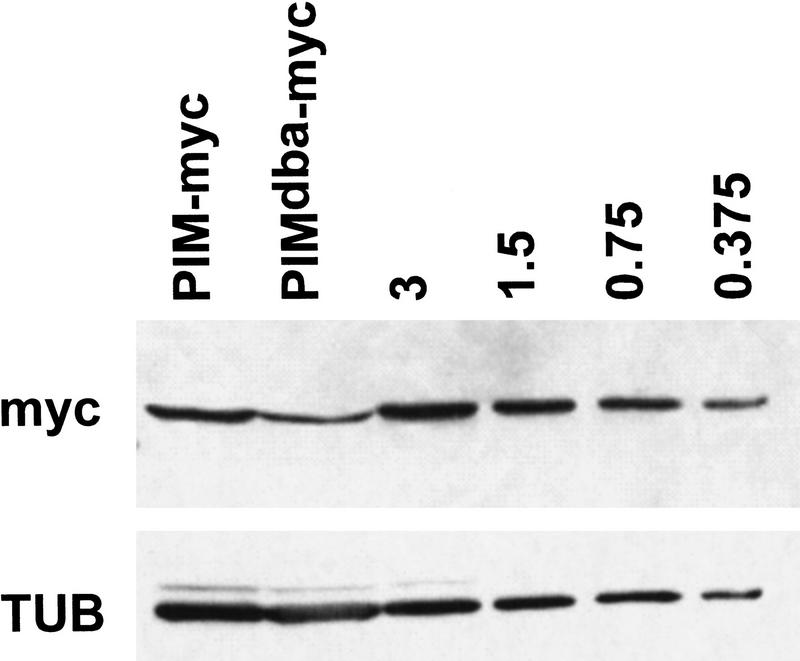
Figure 6.
Analysis of PIMdba–myc expression levels. Extracts from embryos with either the gpim–myc (PIM–myc) or the gpimdba–myc (PIMdba–myc) transgene were analyzed by immunoblotting with antibodies against the myc epitope (myc) or tubulin (TUB), which served as a loading control. In addition, to allow quantitative comparisons, we also analyzed an extract from embryos with a 3× higher gpim–myc transgene dose (3) as well as twofold serial dilutions of this extract (1.5; 0.75; 0.375).
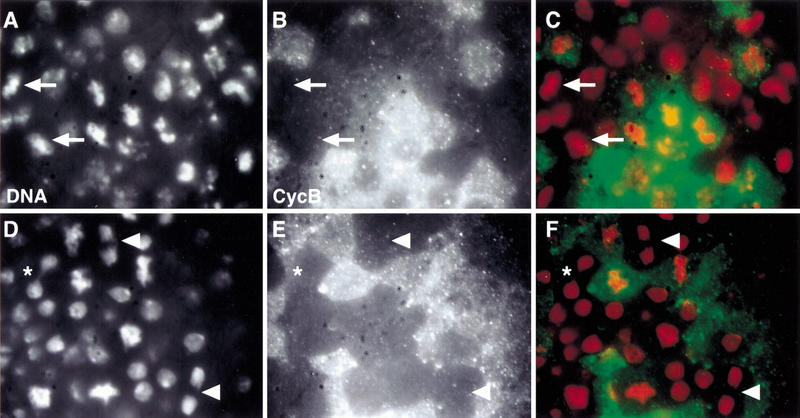
Figure 7.
Low levels of PIMdba–myc expression allow sister chromatid separation in pim mutants. pim1/ pim1 (A–C) or pim1/ pim1, gpimdba–myc (D–F) embryos at the stage of mitosis 15 were labeled with a DNA stain (DNA; A,D) and antibodies against Cyclin B (CycB; B,E). Regions of the dorsal epidermis are shown. In the merged panels (C,F) DNA labeling is shown in red and Cyclin B in green. Arrows in A–C indicate cells that have failed to separate sister chromatids even though they have progressed beyond the metaphase–anaphase transition as evidenced by lack of anti-Cyclin B labeling. Arrowheads in D–F indicate cells that have separated sister chromatids successfully after the metaphase–anaphase transition. (*, D–F) Cell with an anaphase bridge.
The insertion gpimdba–myc II.5, which resulted in lethality when homozygous, was found to result in expression levels that were only ∼25% lower as those of the endogenous locus (Fig. 6). In these experiments, protein products resulting from early zygotic expression during < 2 embryonic cell cycles were compared (see Materials and Methods). Comparison of protein levels that had accumulated during this brief phase was chosen as the difference in PIM–myc and PIMdba–myc levels is likely to increase with every cell cycle due to the differential stability during mitosis. Moreover, the maternal pim+ contribution is known to be exhausted in pim mutants at this stage (Stratmann and Lehner 1996; see also Fig. 7A–C). Thus the consequences of gpimdba–myc expression on progression through mitosis in the absence of wild-type pim+ were also analyzed at this stage (Fig. 7D–F). For this analysis, gpimdba–myc II.5 was recombined with a mutant pim allele (pim1) which abolishes expression from the endogenous locus (data not shown). Analysis of mitosis 15 in pim1/pim1, gpimdba–myc embryos (Fig. 7D–F) and in pim1, gpimdba–myc/pim1, gpimdba–myc embryos (data not shown) indicated that sister chromatid separation occurred almost normally. Moreover, the same observations were also made during mitosis 16. As in wild-type embryos, anaphase and telophase figures were observed readily in these embryos in cells lacking Cyclin B labeling (Fig. 7D–F). However, a significant fraction of anaphase and telophase figures (∼10%) had chromatin bridges (Fig. 7D, see asterisk) suggesting that sister chromatid separation was not always normal. Moreover, gpimdba–myc failed to rescue the development of pim1 homozygotes to the adult stage, whereas the lethality associated with pim1 is prevented by gpim–myc. Nevertheless, the very significant rescue of sister chromatid separation during the embryonic divisions obtained with gpimdba–myc in pim1 mutants, demonstrated that PIMdba–myc can still provide some positive function required for sister chromatid separation. In addition, coimmunoprecipitation experiments indicated that PIMdba–myc is associated with THR (Fig. 1). The dba mutation therefore appears to interfere specifically with mitotic degradation. The observation that sister chromatid separation is not inhibited in the presence of near physiological levels of nondegradable PIMdba–myc argues strongly that sister chromatid separation is likely to be controlled by other mechanisms operating in addition to PIM degradation.
Discussion
PIM is degraded during the metaphase–anaphase transition. This mitotic degradation of PIM is dependent on a destruction box motif that deviates at the first invariant position of the hitherto established destruction box consensus. While all previously characterized destruction boxes start with an arginine (King et al. 1996; Peters et al. 1998), we find a lysine in the PIM motif. However, this PIM variant can replace the destruction box of Cyclin B. Moreover, we find that Fizzy, an activator of the APC/C that is required for the degradation of mitotic cyclins (Dawson et al. 1995; Sigrist et al. 1995), is also required for PIM degradation. We assume, therefore, that PIM is degraded by the proteasome after APC/C-dependent polyubiquitination just like the mitotic cyclins.
PIM degradation during mitosis appears to be an important step for sister chromatid separation. Overexpression of wild-type PIM and nondegradable PIM results in a complete inhibition of sister chromatid separation. We have shown previously that sister chromatid separation is equally defective in the absence of PIM (Stratmann and Lehner 1996). PIM therefore can act as both an activator and an inhibitor of sister chromatid separation.
Inhibitors of sister chromatid separation which are degraded during the metaphase–anaphase transition by the APC/C pathway, and which function to a variable extent as activators of sister chromatid separation, have been described previously in yeast and vertebrates (Cohen-Fix et al. 1996; Funabiki et al. 1996b; Zou et al. 1999). These securin proteins (S. cerevisiae Pds1p, S. pombe Cut2, vertebrate PTTG) have an additional property in common. They all bind to proteins containing a conserved carboxy-terminal separin domain (S. cerevisiae Esp1p, S. pombe Cut1, vertebrate Esp1p). The separin proteins play a crucial role for sister chromatid separation. The functional characterization of budding yeast separin Esp1p has suggested that it may function as a protease which cleaves a cohesin subunit and thereby causes the dissolution of sister chromatid cohesion at the metaphase–anaphase transition (Uhlmann et al. 1999; Nasmyth et al. 2000). In addition, separin proteins might also be involved in the regulation of spindle function (Yanagida 2000). Premature activation of separin activity is restricted by the securin proteins (Ciosk et al. 1998; Kumada et al. 1998; Uhlmann et al. 1999).
We do not know whether PIM binds to a protein with a separin domain. The Drosophila genome sequence predicts the existence of a protein with a separin domain (Gadfly gene number CG 10583). Interestingly, this Drosophila separin homolog exhibits less sequence conservation than all the other known separin proteins from fungi, plants, nematodes, and vertebrates. The Drosophila protein is highly divergent in one of the conserved motifs within the carboxy-terminal separin domain and has a relatively small amino-terminal domain (H. Jäger, S. Heidmann and C.F. Lehner, unpubl.). The amino-terminal regions of separin proteins generally show very little sequence conservation, but the fission yeast securin has been shown to bind to this nonconserved region. Although the separin proteins share at least a related carboxy-terminal domain, securin proteins do not display significant sequence similarity. The only conserved feature of the securins is the distribution of charged residues. The amino-terminal region is highly basic and the carboxy-terminal region highly acidic. This charge distribution is also present in PIM. It is therefore entirely possible that PIM represents a securin protein involved in the regulation of a Drosophila separin.
Although we do not yet know whether PIM binds to a separin protein, we can clearly demonstrate that PIM associates with THR in vivo. Like PIM, THR is also required for sister chromatid separation and shows no significant similarity to known proteins (D'Andrea et al. 1993). A further detailed characterization of the PIM–THR complex is underway including an analysis of its relationship to separin complexes. Three main hypotheses will have to be addressed:
The separin gene might have broken apart during the evolution of Drosophila melanogaster resulting in the thr gene encoding the nonconserved amino-terminal region and a distinct gene encoding the conserved carboxy-terminal separin domain which might also be part of the PIM–THR complex. The PIM–THR complex might therefore be largely equivalent to the securin/separin complex.
Instead of playing the role of the amino-terminal separin region, THR might be a novel separin-associated protein. Unknown separin-associated proteins in addition to the known securins were revealed by affinity purification in Xenopus (Zou et al. 1999) but not in budding yeast (Ciosk et al. 1998).
The PIM–THR complex might be distinct from securin/separin complexes. We emphasize that the role of higher eukaryote separin proteins have not yet been studied in detail and that the vertebrate homolog of the budding yeast cohesin subunit Scc1p dissociates from chromatin already during prophase, presumably as a requirement for chromatin condensation (Nasmyth et al. 2000). It remains a possibility therefore, that additional mechanisms have evolved in higher eukaryotes to maintain sister chromatid cohesion until the metaphase–anaphase transition, in particular in the centromeric region. The PIM–THR complex might be involved in the dissolution of this residual cohesion maintained in the centromeric region of higher eukaryote mitotic chromosomes. Maintenance of cohesion in the centromeric region is also of particular importance during the first meiotic division and a gene specifically required for this maintenance has been identified in Drosophila. Interestingly, this gene (Mei-S332) has no obvious homologs in other species, and it has been proposed to be involved in maintaining cohesion not only during the first meiotic division but also during mitotic divisions (Tang et al. 1998).
Apart from their shared functions (inhibition of sister chromatid separation, separin binding, APC/C substrate) the securins Pds1p, Cut2, and PTTG differ with regard to their role as positive regulators of sister chromatid separation and their involvement in checkpoint mechanisms. The following comparisons indicate that PIM is functionally most similar to fission yeast Cut2. Both proteins provide a positive and essential function. They are absolutely required for sister chromatid separation (Uzawa et al. 1990; Stratmann and Lehner 1996). In contrast, budding yeast Pds1p is clearly not required for sister chromatid separation in unperturbed cells (Yamamoto et al. 1996a; Alexandru et al. 1999). Pds1p is only essential at high temperatures and in the presence of DNA damage, lagging chromosomes and spindle damage. Moreover, while Cut2 appears to function only in mitotic checkpoint control, Pds1p has been implicated in both DNA damage and mitotic checkpoint pathways (Yamamoto et al. 1996a,b; Cohen-Fix and Koshland 1997, 1999; Alexandru et al. 1999; Tinker-Kulberg and Morgan 1999). Overexpression of nondegradable mutant Pds1p inhibits both sister chromatid separation and exit from mitosis in budding yeast (Cohen-Fix and Koshland 1999; Tinker-Kulberg and Morgan 1999). In contrast, only the former process appears to be blocked by nondegradable mutant Cut2 in fission yeast (Funabiki et al. 1996b). This differential involvement in checkpoint regulation might reflect a fundamental physiological difference between budding and fission yeast. DNA damage causes fission yeast (and animal cells) primarily to block entry into mitosis by preventing Cdk1 activation. In contrast, DNA damage inhibits the metaphase–anaphase transition in budding yeast and Pds1p is of major importance for this DNA damage checkpoint arrest. It is not known whether PIM and vertebrate PTTG are involved in a DNA damage checkpoint pathway. However, based on the effects caused by the expression of nondegradable mutant proteins, PIM and vertebrate PTTG appear to be more similar to Cut2 than to Pds1p. Nondegradable PTTG and PIM inhibit only sister chromatid separation but not the degradation of mitotic cyclins and exit from mitosis (Zou et al. 1999; this paper).
The mitotic checkpoint pathway which delays the metaphase–anaphase transition in the presence of unattached kinetochores prevents the activation of APC/C by Fizzy/Cdc20p (Zachariae and Nasmyth 1999). As a consequence, mitotic cyclins and securins persist in the arrested cells. In budding yeast, the persistence of Pds1p has been demonstrated to be required for the inhibition of sister chromatid separation by elegant and conclusive genetic experiments involving cells lacking PDS1 (Yamamoto et al. 1996b; Zachariae et al. 1998; Alexandru et al. 1999). Although not proven, the persistence of fission yeast Cut2 and vertebrate PTTG in checkpoint arrested cells is also thought to be responsible for the inhibition of sister chromatid separation. We show that PIM persists in mitotic checkpoint-arrested cells as well. As in the case of Cut2, however, the essential positive role of PIM in sister chromatid separation makes it impossible to demonstrate that this PIM persistence is responsible for the inhibition of sister chromatid separation in the same elegant way realized in budding yeast. It is clear, however, that PIM levels are of crucial importance for sister chromatid separation. Our results demonstrate that modest overexpression of nondegradable PIM leads to a complete inhibition of sister chromatid separation. Moreover, higher but still relatively modest levels of wild-type PIM overexpression (∼fivefold) inhibit as well. In this context, we consider it very likely that overexpression of the vertebrate securin PTTG results in chromosome instability, explaining its oncogenic potential (Pei and Melmed 1997; Zhang et al. 1999).
Even though our findings demonstrate clearly the importance of PIM degradation, they cannot answer the question whether PIM protein persistence is responsible for the inhibition of sister chromatid separation in cells arrested by the spindle checkpoint. In this case, one would expect physiological levels of nondegradable PIM to inhibit sister chromatid separation completely. However, most cells with near physiological levels of mutant, nondegradable PIM protein (∼75% of wild type) instead of normal PIM appear to separate sister chromatids without major problems. Although not excluded, it appears unlikely, therefore, that the persistence of PIM protein during a mitotic checkpoint arrest is solely responsible for the block of sister chromatid separation. PIM persistence might be only one of several measures that cooperatively prevent premature sister chromatid separation in the presence of spindle damage. However, we emphasize that it remains to be shown that the myc epitopes present at the carboxyl terminus of the nondegradable PIM protein expressed in our experiments do not reduce the anaphase inhibitor function of PIM.
The almost normal progression through mitosis in the presence of near physiological levels of nondegradable PIM suggests that the onset of sister chromatid separation is not determined exclusively by the kinetics of PIM degradation. We have not detected an increase in the fraction of cells with metaphase plates in the presence of nondegradable PIM and careful examination by confocal microscopy did not reveal any residual partial degradation of the mutant PIM protein. The notion that the timing of sister chromatid separation during mitosis can be controlled by pathways that are independent of PIM degradation is also supported by the observation that expression of nondegradable Cyclin A clearly delays the metaphase–anaphase transition (Sigrist et al. 1995) even though it does not appear to result in a delay of PIM degradation (O. Leismann and C.F. Lehner, unpubl.). The timing of sister chromatid separation under normal conditions in budding yeast, when spindle or DNA damage checkpoints are not activated, is also not controlled by Pds1p degradation, because it occurs with normal kinetics in cells lacking Pds1p (Alexandru et al. 1999).
We conclude that PIM levels that are controlled by mitotic degradation are of crucial importance for sister chromatid separation. PIM therefore shares extensive similarities with securin proteins. Although its role in the regulation of Drosophila separin remains to be analyzed, it is clear that it associates with THR, a protein that is equally important for sister chromatid separation and that does not contain a separin domain.
Materials and methods
Drosophila stocks
The alleles fzy1, fzy3, and pim1 were used in our experiments (Nüsslein-Volhard et al. 1984; Dawson et al. 1993; Stratmann and Lehner 1996). Second site lethal mutation present on the original pim1 chromosome were removed by meiotic recombination. The lethality resulting from homozygosity of the pim1 chromosome which was used in this study was completely prevented by a pim+ transgene (Stratmann and Lehner 1996), indicating the absence of other lethal mutations except for the mutation in pim.
For expression of UAS transgenes, we used the following GAL4 transgenes: prd–GAL4 (Brand and Perrimon 1993), da–GAL4 G32 (Wodarz et al. 1995), arm–GAL4 (Sanson et al. 1996), F4, which results in expression in salivary glands (Weiss et al. 1998), and nos–GAL4–GCN4–bcd3′UTR (kindly provided by N. Dostatni, LGPD, IBDM, Marseille, France). A nos+ promoter directs transcription of the nos–GAL4–GCN4–bcd3′ UTR transgene during oogenesis. The 3′ UTR from bcd which is present in the resulting transcripts leads to mRNA localization to the anterior end of the egg. The GAL4–GCN4 fusion protein translated from this mRNA, therefore, forms a concentration gradient with maximal concentrations at the anterior pole and results in graded expression of UAS target genes starting during cellularization in cell cycle 14 of embryogenesis.
The UAS–CycB (Weiss et al. 1998) and UAS–Cdk1–myc (Meyer et al. 2000) lines have been described previously. The UAS–Cdk1–myc transgene allows Gal4p-dependent expression of functional Drosophila Cdk1 with a carboxy-terminal extension consisting of six myc epitope copies. UAS–CycA–Δ170 (TF73) was kindly provided by Frank Sprenger (University of Cologne, Germany). The UAS–CycA–Δ170 transgene allows expression of Drosophila Cyclin A-lacking amino acids 1–170 which contains the signals required for mitotic destruction (Sigrist et al. 1995; Sprenger et al. 1997).
Lines with UAS–CycB transgenes allowing expression of Drosophila Cyclin B with alterations in the destruction box region were obtained after P element-mediated germline transformation with pUAST (Brand and Perrimon 1993) constructs following standard procedures. In an initial step of the generation of these constructs, we deleted the region encoding the destruction box in a Drosophila Cyclin B cDNA (Lehner and O'Farrell 1990) by inverse polymerase chain reaction (PCR) using the primers 5′-CATGGTACCTTTTGTTGTTGCCTCCATGG-3′ and 5′-GACGGTACCCGCGGCATAAGTCGTCCC-3′. Ligation of the amplification product after digestion with KpnI resulted in a mutant cDNA plasmid that contained a KpnI restriction site instead of the destruction box. A double-stranded oligonucleotide encoding the myc epitope (MEQKLISEEDLNE) with compatible ends was inserted into this KpnI site for the construction of the UAS–CycB–dbm transgene. The compatible ends resulted in two additional codons on either side of the myc sequence. The mutant cDNA, therefore, coded for GT MEQKLISEEDLNE RT in place of the destruction box (RAALGDLQN). For the construction of the UAS–CycB–dbpim transgene, we inserted a different oligonucleotide encoding the PIM destruction box with compatible ends (GT KKPLGNLDN GT). An oligonucleotide encoding a mutant PIM destruction box (GT AKPAGNLDA GT) was used for the construction of UAS–CycB–dbapim. For control experiments, we also inserted an oligonucleotide restoring the Cyclin B destruction box flanked by the extra amino acids resulting from the compatible ends on either side (GT RAALGDLQN GT) to yield UAS–CycB–dbCycB. The Cyclin B cDNA fragments with the different destruction box regions were excised with XhoI and XbaI and inserted into the corresponding sites of pUAST.
To analyze the toxicity of Cyclin B containing either a myc epitope tag, or the PIM destruction box, or the mutant PIM destruction box instead of the normal Cyclin B destruction box, we expressed the appropriate transgenes (UAS–CycB–dbm III.1, UAS–CycB–dbm III.2, UAS–CycB–dbpim II.1, UAS–CycB–dbpim II.2, UAS–CycB–dbapim II.1, UAS–CycB–dbapim II.2, UAS–CycB–dbCycB II.1, UAS–CycB–dbCycB III.1, UAS–CycB II.2, UAS–CycB III.3) ubiquitously using da–GAL4 G32. UAS–CycB–dbm and UAS–CycB–dbapim expression resulted in complete embryonic lethality in these experiments. In contrast, UAS–CycB–dbpim, UAS–CycB–dbCycB, and UAS–CycB did not affect embryonic viability.
Lines with transgenes resulting in the expression of PIM protein with a carboxy-terminal extension of six myc epitopes under the control of the pim+ regulatory region (gpim–myc) have been described previously (Stratmann and Lehner 1996). For the construction of gpimdba–myc transgenes, in which the pim+ regulatory region directed expression of myc-tagged PIM protein with a mutant destruction box (AKPAGNLDA), we started with the removal of an XbaI–BglII fragment encompassing the pim+ coding region from pKS + gpim–myc, a cloning intermediate that had been used already for the construction of gpim–myc. Insertion of an XbaI–BglII replacement fragment including the mutant destruction box region resulted in pKS + gpimdba–myc. For the construction of this replacement fragment, we amplified a first PCR fragment using primer 1 (5′-CCATCTCTAGAAAAGTGCCGC-3′) and primer 2 (5′-ACCTGCCGGTTTGGCCAATACGGAATTTGTAGG-3′) from pKS + gpim–myc. In addition, using primer 3 (5′-ATTGGCCAAACCGGCAGGTAACCTTGACGCTGTGATGCACCAAACTCCT-3′) and primer 4 (5′-GATCTAAAATAGAAGATCTGAATT-3′) we amplified a second PCR fragment from pKS + gpim–myc. The intended mutations in the destruction box were introduced by primers 2 and 3 (bold print). The two PCR fragments were digested with BglI and ligated. The final replacement fragment was obtained after digestion of the resulting ligation product with XbaI and BglII. In a final step, the insert was excised from pKS + gpimdba–myc using NotI and KpnI and inserted into the corresponding sites of pCaSpeR 4 (Pirrotta 1988).
The pUAST construct used for the generation of lines allowing Gal4p-dependent expression of PIM protein with a carboxy-terminal extension of six myc epitopes (UAS–pim–myc) contained an insert fragment obtained from pKS + gpim–myc by PCR. Primer 5 (5′- GGACGGCCGAAGTGCCGCTCGTTT-3′) and primer 6 (5′-GCATCTAGAAGTTTTTATAGTTGCTTTAATTC-3′) were used for amplification. The resulting fragment was digested with EagI and XbaI and inserted into the corresponding sites of pUAST. For the construction of UAS–pimdba–myc, we inserted a different EagI–XbaI fragment including the mutant destruction box region into pUAST. For the generation of this fragment, we amplified a first fragment from pKS + gpim–myc with primers 5 and 2. In addition, using primers 3 and 6 we amplified a second PCR fragment from pKS + gpim–myc. The two PCR fragments were digested with BglI and ligated. The final insert fragment was obtained after digestion of the resulting ligation product with EagI and XbaI.
Lines allowing expression of THR protein with a carboxy-terminal extension of 10 myc epitopes (gthr–myc) were obtained using a pCaSpeR 4 construct. In a first construction phase, we introduced restriction sites immediately downstream of the initiation codon (SalI) and immediately upstream of the stop codon (NcoI) into a 9.2-kb genomic XbaI/XhoI fragment that contains all of the sequences required for thr+ function (M. Sadler, S. Heidmann, and C. Lehner, unpubl.). An NcoI/AflIII PCR fragment encoding 10 myc epitope copies was inserted into the NcoI site. The modified XbaI/XhoI fragment was subsequently transferred into the corresponding sites of pCaSpeR 4 and used to establish the gthr–myc lines.
Antibodies
We used mouse monoclonal antibodies against a myc epitope (Evan et al. 1985), the PSTAIRE epitope present in Cdk1 (Yamashita et al. 1991), β-galactosidase (Promega), α-tubulin (Amersham), and Drosophila Cyclin B (Knoblich and Lehner 1993). In addition, we used rabbit polyclonal antibodies against Drosophila Cyclin A (Lehner and O'Farrell 1989), Cyclin B (Jacobs et al. 1998), FZY (Sigrist et al. 1995), PIM (Stratmann and Lehner 1996), and THR. The rabbit antiserum against THR was induced with a 45-kD hexahistidine tagged carboxy-terminal fragment that was expressed in bacteria and purified using Ni2+–NTA affinity chromatography (Qiagen).
Immunoprecipitation
For the coimmunoprecipitation experiments, we collected eggs from either w1, or arm–GAL4, UAS–Cdk1–myc II.2, or gpim–myc 3A, or gthr–myc III.1 flies. In addition, we collected eggs from a cross of da–GAL4 females with UAS–pimdba–myc III.1 males. Eggs were collected for 3 hr on apple juice agar plates and aged for 3 hr at 25°C before extract preparation. Extracts were prepared by homogenization in lysis buffer (50 mm HEPES at pH 7.5, 60 mm NaCl, 3 mm MgCl2 1 mm CaCl2, 0.2% Triton X-100, 0.2% Nonidet NP-40, 10% glycerol, 1 mm DTT, 2 mm Pefabloc, 2 mm Benzamidin, 10 μg/ml Aprotinin, 2 μg/ml Pepstatin A, 10 μg/ml Leupeptin). For immunoprecipitation from the cleared homogenates, we used the anti-myc antibody cross-linked (Harlow and Lane 1988) to Protein A–Sepharose 6MB beads (Pharmacia). The immunoprecipitates were analyzed by immunoblotting using ECL (Amersham). Analysis of the immunoprecipitates by silver staining indicated the presence of many nonspecifically precipitated proteins obscuring the specifically coimmunoprecipitated proteins.
Immunolabeling
Fixation of embryos and immunolabeling was performed as described previously (Lehner and O'Farrell 1989). Secondary antibodies against rabbit or mouse IgG were conjugated to Alexa488 (Molecular Probes), Cy3, or Cy5 (Dianova). DNA was labeled by propidium iodide for analysis by confocal microscopy (Leica TCS-SP) or by Hoechst 33258 for conventional fluorescence microscopy (Zeiss Axiophot equipped with a Photometrics Nu200A cooled CCD camera).
For the analysis of the mitotic degradation of Cyclin B with various destruction boxes, we collected embryos for immunolabeling experiments from crosses of prd–GAL4 females with either UAS–CycB–dbm III.2, or UAS–CycB–dbpim II.1, or UAS–CycB–dbCycB II.1, or UAS–CycB–dbapim II.1. Eggs were collected for 4 hr and aged for 4 hr at 25°C before fixation and labeling.
For the comparison of the mitotic degradation of PIM–myc and PIMdba–myc, we collected embryos from crosses of nos–GAL4–GCN4–bcd3′UTR females and either UAS–pim–myc III.3 or UAS–pimdba–myc I.1 males. Eggs were collected for 2 hr and aged for 2 hr at 25°C before fixation and immunolabeling.
For the inhibition of sister chromatid separation during mitosis 15 and cytological analysis of chromosomes in the subsequent mitosis 16, we crossed da–GAL4 G32 females to UAS–pimdba–myc I.1; UAS–pimdba–myc III.1 males. Eggs were collected for 1 hr and aged for 6 hr at 25°C before permeabilization and colcemid treatment (Sigrist et al. 1995). Mitotic chromosome spreads were prepared from these embryos as described previously (Sigrist et al. 1995).
The consequences of pim overexpression were analyzed in progeny collected from crosses of prd–GAL4 females with either UAS–pim–myc III.3 or UAS–pim–myc II.2; UAS pim–myc III.3 males.
To analyze UAS–pim–myc expression in fizzy mutant embryos, we collected eggs from a cross of fzy3/CyO, P[w +, ftz–lacZ]; prd–GAL4/+ females with fzy3/CyO, P[w +, ftz–lacZ]; UAS–pim–myc III.3/+ males. Embryos homozygous for fzy3 could be identified because they were lacking anti-β-galactosidase labeling.
To analyze PIM behavior during spindle checkpoint arrest, we collected eggs from a cross of nos–GAL4–GCN4–bcd3′UTR females and UAS–pim–myc III.3 males. Eggs were collected for 30 min and aged for 165 min at 25°C. After chorion removal in 5% sodium hypochlorite (50% Klorix, Palmolive), we incubated the embryos in a 1:1 mixture of octane and Schneider's Drosophila cell culture medium containing 10 μm demecolcine (Sigma) for 25 min at room temperature on a rotating wheel. For fixation, the cell culture medium was replaced by phosphate buffered saline containing 4% formaldehyde and further processing for immunolabeling was as described previously (Lehner et al. 1991).
To analyze the function of nondegradable PIM in pim mutants, we constructed a pim1 chromosome with the gpimdba–myc II.5 insertion by meiotic recombination. We collected eggs from parents carrying this chromosome over CyO [gpimdba–myc II.5, pim1/CyO, P(w +, ftz–lacZ)]. In addition, we also analyzed progeny from a cross of gpimdba–myc II.5, pim1/CyO, P[w +, ftz–lacZ] females and pim1/CyO, P[w +, ftz–lacZ] males. Moreover, for control experiments we collected eggs from pim1/CyO, P[w +, ftz–lacZ] flies and from pim1/CyO, P[w +, ftz–lacZ]; gpim–myc III.1 flies. Progeny homozygous for pim1 could be identified because they were lacking anti-β-galactosidase labeling.
Immunoblotting
Eggs collected from fzy1/CyO flies were aged to stage 12 and fixed as described previously (Edgar et al. 1994). After DNA labeling, homozygous fzy1 embryos were sorted from sibling embryos using an inverted epifluorescence microscope (Zeiss Axiovert). Extracts were prepared from pooled embryos and analyzed by immunoblotting as described previously (Edgar et al. 1994).
To analyze pim transgene expression levels by immunoblotting with anti-myc, we crossed pim1/CyO, P[w +, ftz–lacZ] virgin females with gpimdba–myc II.5, pim1/CyO P[w +, ftz–lacZ] males. In parallel, we crossed females of the same genotype also with males carrying either only the gpim–myc III.1 transgene insertion [pim1/CyO, P(w +, ftz–lacZ); gpim–myc III.1/+] or multiple gpim–myc transgene insertions [gpim–myc I.1/Y; pim1/CyO, P(w +, ftz–lacZ); gpim–myc III.3/gpim–myc III.3]. Progeny from these crosses were collected for 30 min on apple juice agar plates and aged for 4 hr at 25°C before preparation of total embryo extracts in SDS–gel sample buffer. The embryos used for extract preparation, therefore, were between 4 and 4.5 hr old, i.e., at the stage when the fifteenth round of embryonic mitoses starts. The embryos contained only zygotically expressed transgene products, because all transgenes were of paternal origin. Zygotic pim+ expression starts during cycle 14 of embryogenesis and reaches significant levels only during cycle 15. Zygotic expression of gpim–myc directed by the pim+ regulatory region present in our transgenes results in levels comparable to those expressed from the endogenous pim+ gene (data not shown). The fraction of unfertilized eggs was controlled after DNA labeling and microscopic inspection of eggs collected immediately after the eggs used for the immunoblotting experiments.
The quantification of PIMdba–myc levels relative to PIM–myc levels by immunoblotting with anti-myc antibodies instead of using anti-PIM antibodies and comparing transgene product levels relative to the endogenous pim+ gene products was chosen because the reactivity of our anti-PIM antibodies was found to be strongly decreased by the carboxy-terminal myc epitope extension present in the transgene products. Moreover, it is not possible to distinguish the early zygotic from the maternal expression of the endogenous pim+ gene when using the anti-PIM antibodies.
Acknowledgments
We acknowledge the contribution of Carola Weise, Malte Siedler, and Andreas Weiss in the initial PIM–THR coimmunoprecipitation experiments, the initial characterization of the thr genomic region, and the generation of the UAS–CycBdbm lines, respectively. We thank Rob Saint for thr cDNA and genomic clones, and Frank Sprenger for the UAS–CycA–Δ170 lines. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG Le 987/2-1 and 3-1).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chle@uni-bayreuth.de; FAX 49-921-55-2710.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.176700.
References
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Pds1p of budding yeast has dual roles: Inhibition of anaphase initiation and regulation of mitotic exit. Genes & Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- D'Andrea RJ, Stratmann R, Lehner CF, John UP, Saint R. The three rows gene of Drosophila melanogaster encodes a novel protein that is required for chromosome disjunction during mitosis. Mol Biol Cell. 1993;4:1161–1174. doi: 10.1091/mbc.4.11.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwiche N, Freeman LA, Strunnikov A. Characterization of the components of the putative mammalian sister chromatid cohesion complex. Gene. 1999;233:39–47. doi: 10.1016/s0378-1119(99)00160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson IA, Roth S, Akam M, Artavanis-Tsakonas S. Mutations of the fizzy locus cause metaphase arrest in Drosophila melanogaster embryos. Development. 1993;117:359–376. doi: 10.1242/dev.117.1.359. [DOI] [PubMed] [Google Scholar]
- Dawson IA, Roth S, Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH. Distinct molecular mechanisms regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes & Dev. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo: Point and counterpoint. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 149–300. [Google Scholar]
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996a;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996b;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, Yanagida M. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jacobs HW, Knoblich JA, Lehner CF. Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes & Dev. 1998;12:3741–3751. doi: 10.1101/gad.12.23.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Lehner CF. Synergistic action of Drosophila cyclin A and cyclin B during the G2-M transition. EMBO J. 1993;12:65–74. doi: 10.1002/j.1460-2075.1993.tb05632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada K, Nakamura T, Nagao K, Funabiki H, Nakagawa T, Yanagida M. Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr Biol. 1998;8:633–641. doi: 10.1016/s0960-9822(98)70250-7. [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. Expression and function of Drosophila cyclin A during embryonic cell cycle progression. Cell. 1989;56:957–968. doi: 10.1016/0092-8674(89)90629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The roles of Drosophila cyclin A and cyclin B in mitotic control. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, Yakubovich N, O'Farrell PH. Exploring the role of Drosophila cyclin A in the regulation of S-phase. Cold Spring Harb Symp Quant Biol. 1991;56:465–475. doi: 10.1101/sqb.1991.056.01.053. [DOI] [PubMed] [Google Scholar]
- Lim HH, Goh PY, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, Lehner CF. Drosophila. 2000. Cdk4 is required for normal growth and dispensable for cell cycle progression. EMBO J. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosomes: Cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Roux's Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Mol Endocrinol. 1997;11:433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- Peters JM, King RW, Deshaies RJ. Cell cycle control by ubiquitin-dependent proteolysis. In: Peters R, et al., editors. Ubiquitin and the biology of the cell. New York, NY: Plenum Press; 1998. pp. 345–387. [Google Scholar]
- Pirrotta V. Vectors for P-element transformation in Drosophila. In: Rodriguez RL, Denhardt DT, editors. Vectors. A survey of cloning vectors and their uses. Boston, MA: Butterworths; 1988. pp. 437–456. and London, UK. [Google Scholar]
- Rimmington G, Dalby B, Glover DM. Expression of N-terminally truncated cyclin-B in the Drosophila larval brain leads to mitotic delay at late anaphase. J Cell Sci. 1994;107:2729–2738. doi: 10.1242/jcs.107.10.2729. [DOI] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent JP. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Sigrist S, Jacobs H, Stratmann R, Lehner CF. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J. 1995;14:4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger F, Yakubovich N, O'Farrell PH. S phase function of Drosophila cyclin A and its downregulation in G1 phase. Curr Biol. 1997;7:488–499. doi: 10.1016/s0960-9822(06)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann R, Lehner CF. Separation of sister chromatids in mitosis requires the Drosophila pimples product, a protein degraded after the metaphase anaphase transition. Cell. 1996;84:25–35. doi: 10.1016/s0092-8674(00)80990-3. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tang TTL, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes & Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes & Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes & Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uzawa S, Samejima I, Hirano T, Tanaka K, Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990;62:913–925. doi: 10.1016/0092-8674(90)90266-h. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Gorbsky GJ, Salmon ED, Nicklas RB. Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr Biol. 1999;9:649–652. doi: 10.1016/s0960-9822(99)80287-5. [DOI] [PubMed] [Google Scholar]
- Weiss A, Herzig A, Jacobs H, Lehner CF. Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr Biol. 1998;8:239–242. doi: 10.1016/s0960-9822(98)70090-9. [DOI] [PubMed] [Google Scholar]
- Whitfield WGF, Gonzalez C, Maldonado-Codina G, Glover DM. The A-type and B-type cyclins of Drosophila are accumulated and destroyed in temporally distinct events that define separable phases of the G2-M transition. EMBO J. 1990;9:2563–2572. doi: 10.1002/j.1460-2075.1990.tb07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor for faithful execution of anaphase in the yeast Saccharomyces cerevisiae. J Cell Biol. 1996a;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996b;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Yoshikuni M, Hirai T, Fukada S, Nagahama Y. A monoclonal antibody against the PSTAIR sequence of p34cdc2, catalytic subunit of maturation-promoting factor and key regulator of the cell cycle. Dev Growth Differ. 1991;33:617–624. doi: 10.1111/j.1440-169X.1991.00617.x. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Cell cycle mechanisms of sister chromatid separation; Roles of Cut1/separin and Cut2/securin. Genes Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJR, Mann M, Nasmyth K. Mass spectrometric analysis of the Anaphase-Promoting Complex from yeast: Identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- Zhang X, Horwitz GA, Heaney AP, Nakashima M, Prezant TR, Bronstein MD, Melmed S. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab. 1999;84:761–767. doi: 10.1210/jcem.84.2.5432. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]