Abstract

The lengths of G-tracts and their connecting loop sequences determine G-quadruplex folding and stability. Complete understanding of the sequence–structure relationships remains elusive. Here, single-loop G-quadruplexes were investigated using explicit solvent molecular dynamics (MD) simulations to characterize the effect of loop length, loop sequence, and G-tract length on the folding topologies and stability of G-quadruplexes. Eight loop types, including different variants of lateral, diagonal, and propeller loops, and six different loop sequences [d0 (i.e., no intervening residues in the loop), dT, dT2, dT3, dTTA, and dT4] were considered through MD simulation and free energy analysis. In most cases the free energetic estimates agree well with the experimental observations. The work also provides new insight into G-quadruplex folding and stability. This includes reporting the observed instability of the left propeller loop, which extends the rules for G-quadruplex folding. We also suggest a plausible explanation why human telomere sequences predominantly form hybrid-I and hybrid-II type structures in K+ solution. Overall, our calculation results indicate that short loops generally are less stable than longer loops, and we hypothesize that the extreme stability of sequences with very short loops could possibly derive from the formation of parallel multimers. The results suggest that free energy differences, estimated from MD and free energy analysis with current force fields and simulation protocols, are able to complement experiment and to help dissect and explain loop sequence, loop length, and G-tract length and orientation influences on G-quadruplex structure.
Introduction
G-quadruplexes are a class of non-canonical four-stranded helical structures formed from guanine-rich DNA and RNA sequences.1−3 Potential quadruplex-forming sequences exist in the telomere and promoter regions,4−7 and G-quadruplexes have aroused great interest as potential anti-cancer targets8−18 and function as a global gene regulation mechanism.19−23 In G-quadruplexes, four guanine bases associate through eight hydrogen bonds to form the characteristic planar structural unit called a G-quartet.(24) In bimolecular or unimolecular G-quadruplexes, the intervening sequences between G-rich repeats form different types of loops: lateral (L, also known as edgewise) loops connect adjacent strands at one terminal quartet, diagonal (D) loops span across the top of the quartet, and propeller (P, also known as double-chain reversal) loops connect opposite ends of the quadruplex.(25) Depending on the loop length, loop sequences, G-tract length and orientation, and choice of monovalent ion or interacting ligands, a diversity of folding topologies is observed.25−29 To better understand the biological roles of G-quadruplexes and their sequence-dependent structures, accurate prediction of the predominant G-quadruplex conformations for a given sequence is of high importance. In a previous study, we used molecular dynamics (MD) simulations with subsequent free energy analyses to derive intrinsic energy preference of the four sequential base-pair step glycosidic bond orientation patterns (i.e., anti-anti, syn-anti, anti-syn, or syn-syn) along the strands in G-DNA stems, and the results help explain how G-tract lengths determine the glycosidic patterns that directly influence the polymorphism of anti-parallel G-quadruplex structures.(30) In the present work, we aim to evaluate the effects of loop length and sequence on G-quadruplex structures through estimating the relative stabilities of different loop types for sets of different loop sequences.
Various techniques have been applied previously to investigate the influence of loops on G-quadruplex folding and stability,31−44 and some general trends emerge: (1) parallel structures are favored by short loops; (2) longer loops favor anti-parallel structures; (3) sequences with single-nucleotide loops usually show extremely high melting temperatures; and (4) increases in the loop length usually lead to decreased melting temperatures. However, a single guanine-rich sequence may populate multiple folding conformations in solution, and sometimes two or more G-quadruplexes may associate to form higher order structures (multimers).29,38,45 Such structural polymorphisms complicate kinetic and thermodynamic analyses and sometimes make the experimental results difficult to interpret. Biomolecular simulation methods, including MD simulations, can aid interpretations by providing detailed information on 3D structures, dynamics, and energetics at the atomic level for single or small sets of molecules near their initial geometry. These technologies have been widely applied to provide useful adjunct information to experiment for a variety of G-quadruplexes.30,46−51 For example, we previously investigated various putative intermediate structures in the folding of tetrameric parallel G-quadruplexes and proposed formation pathways(46) that are consistent with more recent mass spectrophotometry experiments(52) and other solution studies.53,54 Additionally, with combined MD and enhanced sampling methods, Hazel et al. predicted the favorable topologies and loop conformations of dimeric quadruplexes with T2 or T3 loops.(48)
To precisely describe the folding topologies of G-quadruplex molecules, a formalism has been proposed by da Silva.(55) In this work, rather than defining the loop direction relative to the first stem or G-tract of the molecule,(55) we define the loop direction relative to the starting G-tract strand of the loop: facing a strand with its 5′→3′ direction pointing upward, if the strand turns right to connect to the next strand, the loop formed is defined as a right loop, and if the strand turns left, the loop is defined as a left loop. In this way, a single loop property can be analyzed in a simpler and more independent way. If a unimolecular G-quadruplex starts from a right loop, the molecule progresses anti-clockwise; if starts from a left one, the molecule progresses clockwise.(55) As observed, propeller loops always span medium-sized grooves,(55) and theoretically these loops can either be right (Figure 1a) or left loops (Figure 1b), even though only right P loops have been reported to date. Based on our observations, lateral wide (Lw) loops are always right loops (Figure 1c), and lateral narrow (Ln) loops are always left loops (Figure 1d). Note also that a propeller loop may span a different number of quartet layers, and Pn denotes a propeller loop spanning n quartet layers. The propeller loops in parallel and anti-parallel G-quadruplexes are different in the glycosidic angle conformation of the stem residue adjacent to the 3′-end of the P loop: anti for parallel G-quadruplex and syn for anti-parallel G-quadruplex. These two types of P loops are termed parallel P loop (denoted as Pnp) and anti-parallel P loop (denoted as Pna) respectively.
Figure 1.
Schematic illustration of definitions and nomenclature used in this work: (a) right propeller loop, (b) left propeller loop, (c) right lateral loop = lateral wide (Lw) loop, (d) left lateral loop = lateral narrow (Ln) loop.
Distinct from the previously published MD simulations on G-quadruplex structures that contain multiple loop sequences, in the current work we systematically built a series of putative single-loop models (Figure 2). This allows a more straightforward and independent or dissected evaluation of the propensity to form a certain loop type for a given loop sequence. Six loop sequences [d0 (i.e., direct bonding of the two strands with no intervening loop sequence), dT, dT2, dT3, dTTA, and dT4] were investigated, and for each sequence eight loop types (Ln, Lw, D, P3a, P3p, P2a, P4a, and P4p, Figure 2) were evaluated. Relative folding energies were also calculated for a subset of the single-loop models, and we found that generally short loops are less stable than longer loops. Finally, relative stabilities between anti-parallel, parallel, and parallel-dimer structures were calculated, and a hypothetical energy curve has been proposed for these structural forms.
Figure 2.
Schematic diagrams of the single-loop models investigated in this work: Ln, lateral narrow loop; Lw, lateral wide loop; D, diagonal loop; P3a/P3p, anti-parallel/parallel propeller loop spanning three quartet layers; P2a, anti-parallel propeller loop spanning two quartet layers; P4a/P4p, anti-parallel/parallel propeller loop spanning four quartet layers. For each of the eight models, six different loop sequences were investigated: d0, dT, dT2, dT3, dTTA, and dT4. Stems of the Ln, Lw, D, and P3a models share the same initial structure taken from the first model of the NMR structure 2GKU.(56)
Methods
Starting Structures
All of the Ln, Lw, D, and P3a models were built from the NMR structure 2GKU;(56) the P2a models were built from the 1L34 structure;(57) the P4a stems were built from the crystal structure 1JPQ with two loop K+ ions removed from the initial structure;(58) and the stem structures of the P3p/P4p models were obtained from the first three/four G-quartets of the 139D structure, respectively.(59) K+ ions(60) were added manually into the channel of the quadruplex stem between two adjacent quartets, except with the P4a models. The UCSF Chimera program(61) was used to manually superimpose different loops onto the stem structures. The initial structures of TTA-Ln, TTA-Lw, and TTA-P3a were obtained by keeping the third, second, and first loop of 2GKU, respectively; TTA-D was built by connecting the diagonal TTA loop from 143D(62) with the 2GKU stem; the TTA-P2a/P3p/P4p/P4a models were built by connecting the first loop of the 2GKU structure with the appropriate stems as stated above. T-Ln/Lw/D models were obtained by keeping the stem and the first loop residue of the described TTA-Ln/Lw/D models, respectively. The loop of the T-P3a model was built from the first loop of the 1XAV structure;(63) the T-P2a model was built by removing the first loop residue of the following T2-P3a model; and the T-P4p model was built by removing the second and the third loop residues of the T3-P4p model. The T2-Ln/Lw/D/P4p models were built by removing the third loop residue from the TTA-Ln/Lw/D/P4p models, respectively; an additional T2-Lw model (T2-Lw-2) was built from the third loop of the 1HUT(64) structure with the 2GKU stem. The loop of the T2-P3a model was mutated from the second loop of the 1XAV structure; the loop of T2-P2a model was mutated from the second loop of the 1L34 structure; and the T2-P4p model was obtained by removing the third loop residue of the T3-P4p model. The T3-Ln/Lw/D models were built by mutating the dA to dT from the TTA-Ln/Lw/D models; an additional T3-Lw model (T3-Lw-2) was built by connecting the loop of the 1A6H structure(65) with the 2GKU stem. T3-P2a and T3-P4p models were built by connecting the loop from the T3-P3a model with the stem of TTA-P2a and TTA-P4p, respectively. The T4- models were all built from the loop present in the 1JPQ structure.(58) Two additional loop geometries were used for the simulation of the T4-Lw model, one from the 1D59 structure(66) (T4-Lw-2) and one from the 1U64 structure(67) (T4-Lw-3). The building of the P4a models is analogous to the building of the P4p models. The d0 loop models were built by directly connecting the two ends of the two stem residues without any intervening residues (hence, the initial models were highly strained).
MD Simulation Protocol
All of the MD simulations and free energy analyses were carried out using AMBER 10 or AMBER 11(68) with the parmbsc0 modifications to the ff99 force field.69,70 Additional K+ ions(60) were added to net-neutralize each system, and then the model was solvated in an octahedral box of TIP3P(71) water with an extension of at least 10 Å from each side. Periodic boundary conditions were applied with the particle mesh Ewald method(72) to calculate the full electrostatic energy (charge grid spacing, ∼1 Å; cubic spline interpolation; and direct sum tolerance, 10–5 Å). The 10 Å non-bonded pair list was updated whenever any atom moved more than 0.5 Å since the last list update. SHAKE(73) constraints were applied on the bonds involving hydrogen (tolerance, 10–5 Å), and the time step was set to 2 fs. A cutoff of 9 Å was applied to the Lennard-Jones and direct space electrostatic interactions, with a uniform density approximation included to correct for the long-range van der Waals interactions.
The system was first minimized with a restraint of 100 kcal/(mol·Å2) applied on the DNA including the channel K+ ion(s) (500 steepest descent cycles followed by 500 conjugate gradient cycles). This minimization was followed by a 25 ps MD simulation with 100 kcal/(mol·Å2) positional restraints applied on the same solute atoms, and the temperature was slowly increased from 0 to 300 K. The temperature was regulated using Langevin dynamics(74) with a collision frequency of 0.2 ps–1. Then followed by another cycle of equivalent minimization and dynamics steps with a decreased restraint force constant of 25 kcal/(mol·Å2). Equilibration continued with five rounds of 1000 step minimizations, with solute restraint force constants of 20, 15, 10, 5, and 0 kcal/(mol·Å2) for each round, respectively. The equilibration was followed by a 250 ps unrestrained MD simulation during which the temperature was increased from 0 to 300 K with a collision frequency of 0.2 ps–1. The above equilibration steps were carried out at constant volume. Finally, a 250-ps unrestrained MD simulation at 300 K (Langevin dynamics; collision frequency, 1 ps–1) with 1 atm constant pressure was carried out to equilibrate the density, and this was followed by an at least 50 ns production MD simulation for each model under equivalent conditions.
Free Energy Calculations
The MM-PBSA Perl script(75) was used for free energy analyses. We followed established protocols to calculate the total free energy of the entire stem–loop system (Gtotal) and also calculated the free energy of the quadruplex stem (Gstem).(47) The free energy of the loop (Gloop) was calculated by subtracting Gstem from Gtotal.(47) In most cases, the final 10 ns of MD simulation was used for analysis, with sampling of configurations at 200 ps intervals. If the last 10 ns was not stable—as discussed and inferred from the all-atom root-mean-squared deviation (rmsd) to the initial structure—regions from the stable portions of the trajectories were chosen. For each specific loop sequence investigated in this work, the Gtotal values of the Ln, Lw, D, and P3a models were directly compared, as these models have the same number of atoms and composition. The P3a model was chosen, arbitrarily, as the reference to compare the relative stabilities. For the P3p, P2a, P4a, and P4p models, Gloop values were used to compare with the P3a model for relative stabilities. Solute entropy was estimated for a subset of the models through both standard normal-mode methods on 10 gas-phase minimized snapshots and also through quasi-harmonic methods(76) on 50 snapshots. However, the solute entropic results are not included in most analyses presented since they did not influence the relative stability ranking.
Results
MD Simulations—Summarizing the Observed Structural Features
An assessment of the structural behavior seen in the simulations can provide qualitative insight into the relative stability of the different G-DNA folds modeled. For example, the terminal quartets of the models T-Lw, T-D, T-P4p, and T2-D lost their characteristic quartet structure during the equilibration steps of the MD simulations; this suggests that these loops are not compatible with the G-quadruplex and are likely unstable. Therefore, these models were excluded from further discussion.
Visible buckle-like deformation of the terminal quartet was observed in four models: T-Ln (Figure 3a), T2-Lw, T3-D, and TTA-D. These deformations indicate strain in the structure despite “stable” MD trajectories as judged by plateaus in the rmsd values. With modern simulation protocols (including Ewald methods) and force fields, as applied here and in our many previous investigations of G-quadruplex structure, such buckle-like deformations are not normally observed in simulations of G-quadruplexes with Na+ or K+.
Figure 3.
Final 1 ns averaged loop structures of some models after 50 ns MD simulations. (a) T-Ln, (b) T3-Ln, (c) TTA-Lw, (d) T3-P3a, (e) T-P4a (the stem residue adjacent to the 3′-end of the loop is also shown in color), (f) 0-P3p (the two directly connected stem residues are shown in color), (g) T4-Lw-3, and (h) T4-Lw-2. The loops are shown as colored tubes, and the terminal quartets or stems are shown as black lines. The loop direction follows Figure 2, and the residue numbers are not labeled.
At the end of simulations, the final residue of all the Ln loops stacked on the terminal quartet (Figure 3b). Experimentally, Ln loops do not universally display this feature, and this is possibly due to the influence from terminal flanking sequences or other loops in the molecule. Common structural features were also observed with the T2-, T3-, and TTA-Lw loops: specifically, the first residue of the loop remained in the wider groove, and the second loop residue stacked above the terminal quartet (Figure 3c). For the diagonal loops, the last loop residue always stacked with the terminal quartet, similar to what was observed with the Ln loops. In some of the propeller loops, the final loop residue was also observed to fold back to stack with the bottom quartet; this was observed specifically with the T2-P2a, T3-P2a, TTA-P2a, and T3-P3a (Figure 3d) models. These observations with the propeller loops, together with the simulation results for all of the Ln and D loops investigated, suggest the importance of the final loop residue in stabilizing the loop–stem interactions. In parallel propeller loops, the folding back of the final loop residue to stack on the terminal quartet was not observed, is likely because the parallel propeller loops are flatter and therefore place their final loop residues farther away from the connecting stem residues (Figure S1). The single-nucleotide dT sequence is too short to form a P4p loop; however, it can be accommodated in the P4a model (Figure 3e). The sugar ring of the stem residue adjacent to the 3′-end of the T-P4a loop moved to the plane of its attached nucleobase ring (Figure 3e), and this may contribute to the accommodation of such a short loop spanning four quartet layers. The T2-P4a model was stable throughout the 50 ns simulation, whereas the terminal quartet of the T2-P4p loop lost its tetrad structure after 33 ns, indirectly suggesting that the T2-P4p loop is not as stable. In our simulations, in addition to the TTA-Ln and TTA-D loops, the dA residue of the TTA-Lw loop also stacked with the terminal quartet (Figure 3c), consistent with the fact that dA residues are good stacking partners due to their larger stacking surface and lower electrostatic polarity.
In anti-parallel propeller loops, the N3 atom of the stem residue adjacent to the 3′-end of the loop is close to its O4′ atom, and when the nearby phosphate group moves closer to the N3 atom, a stable cation binding pocket may form. Such stable binding sites were observed in the T-P3a, TTA-P3a, T4-P3a, T3-P3a, and T4-P2a models. In the simulations of the T3-P3a and TTA-P3a models, the K+ ion in this site had binding residency times of up to 20 ns. Figure S2 in the Supporting Information shows the correlation of the cation binding site with the loop geometry: when the loop adopted a conformation with closely spaced phosphates, the binding site formed and within ∼2 ns a K+ ion became trapped in this site; when the loop changed to a conformation with well-separated phosphate groups, the ion left quickly.
During revision, d0 models with loops that have no intervening residues (i.e., the two stem residues are directly connected without an intervening linker in a highly strained initial geometry; this topology is similar to the previously reported V-shaped loop(77)) were also built and tested. Among them, only the d0-P3p model maintained the quadruplex structure throughout the simulation (Figure 3f). In this structure, the sugar ring of the stem residue connecting to the 5′-end of the d0-P3p loop also moved to the plane of its attached nucleobase to accommodate this extremely short loop (Figure 3f).
Free Energy Estimation from MM-PBSA Calculations
In previous work, the loop free energies (Gloop) were typically estimated by subtracting the free energy of the stem (Gstem) from the total free energy of the entire stem–loop system (Gtotal).47,49 However, this approach has limitations. Although the stems tend to be very rigid, the stem structure and dynamics may be influenced by the loops attached to them and connecting their strands. For some short loops the strain within the loop potentially will bring distortion or instability to the stem; on the other hand, it is possible that with some other loops the stability of the stem (and of the whole structure) could be effectively increased by forming stacking interactions with the terminal quartets. Such effects are not fully included in the Gloop free energy values calculated as the difference between Gtotal and Gstem, and this may lead to errors when we assess the stabilities of different loop types using Gloop. In other words, when relying on the Gloop data, the loop–stem structural inter-dependence is not fully or properly included. To circumvent this, the present single-loop Ln, Lw, D, and P3a models were all built from a common stem structure (from the NMR structure 2GKU(56)). As these models have the same composition, they are directly comparable using the more complete and comparable Gtotal free energy estimates. This provides a consistent evaluation of the loop stabilities through direct comparison of the total free energies of the complete models, i.e., with full inclusion of the loop–stem interactions. However, due to different compositions, for the remaining systems we had to use the Gloop approach. Relative stabilities of different loop types are shown in Table 1, with more detailed data provided in the Supporting Information, Table S1. The P3a model was chosen to serve as the reference for all the other systems. As the P2a, P4a, and P4p models have different numbers of residues compared to the other models, their loop stabilities can only be assessed by comparing the estimated Gloop values with those calculated with the P3a model. Although the parallel P3p model has the same residue composition as the P3a model, since anti-parallel and parallel stems likely have different stabilities, both ΔGtotal and ΔGloop are shown in Table S1 as calculated by subtracting the P3pGtotal and Gloop values to get ΔGtotal and ΔGloop, respectively; in the analysis that follows, the ΔGloop energies are used for relative stability ranking with respect to the P3p models.
Table 1. Approximate Stabilities of Various Loop Types Relative to P3a Loops (kcal/mol)a.
| Ln | Lw | D | P3a | P3p | P2a | P4a | P4p | parallel propensity | |
|---|---|---|---|---|---|---|---|---|---|
| T | 7 | –b | –b | 0 | –2 | 11 | –c | –b | –9 (P3p – Ln) |
| T2 | 2 | 4 | –b | 0 | 4 | 13 | –2 | 2 | 2 (P3p – Ln) |
| T3 | –5 | –9 | 0 | 0 | 1 | 13 | 1 | –1 | 10 (P3p – Lw) |
| TTA | –4 | –5 | 2 | 0 | 3 | 9 | –3 | 6 | 8 (P3p – Lw) |
| T4 | –7 | –19 | –10 | 0 | –3 | 2 | –7 | –4 | 16 (P3p – Lw) |
Gtotal is used to evaluate the relative stabilities for the Ln, Lw, D, and P3a loops with P3a as a reference. Gloop is used to evaluate the relative stabilities for the other loop types, still with P3a as the reference.
The quadruplex structure of these models was not maintained during the equilibration steps in the respective simulations.
The abnormal geometry of the sugar ring of the loop-connecting stem residue implies that the ΔGloop value is misleading; therefore, the data are not reported.
For some of the loops, several simulations were carried out if multiple experimental loop geometries are available. During one simulation, several geometries with different stabilities may also be sampled. In these cases, the most stable energy values were used for free energy comparisons. An exception was the sequentially over-stacked loop geometries (three sequentially continuous loop residues stack in a line, and together stack on the terminal quartet) that were observed in T2-Lw and T4-Lw-3 (Figure 3g). Free energy calculations indicate anomalously large stabilization of such structures (Table S1). For example, the T4-Lw-3 model with the over-stacked loop is ∼ –9 kcal/mol (Gtotal = −3308 vs −3299, Table S1) more stable than the T4-Lw-2 model (Figure 3h). The over-stacked loop geometries have also been observed in previous MD simulations but rarely in experimental structures. This specific geometry is possibly a force field artifact that needs further investigation and therefore was excluded from our energy ranking.
The estimated relative stabilities of different loop types compared to the P3a loop are shown in Table 1. Solute entropies were also calculated for a subset of the models (Table S1); however, these values do not significantly influence the trends and therefore are not included in Table 1. The data clearly show that the single-nucleotide dT sequence has a very strong preference to form a P3 loop, slightly favoring the P3p loop over the P3a loop (Table 1). This is fully consistent with the experiments reporting that single-nucleotide loops have a strong preference for parallel structures.32,36−38,78 In contrast, most of the dT2 (dinucleotide) loops have similar stabilities (Table 1), indicating that the dT2 loop does not have an obvious preference to fold into either parallel or anti-parallel structures. Both dT3 and dTTA favor lateral loops, and mutation of the last loop residue from dT to dA does not dramatically change the relative ranking of different loop types (Table 1). The dT4 sequence strongly favors the Lw or D loops (Table 1). These observations, or features of particular loop–sequence relationships, could aid in the design or engineering of particular folding topologies. Another observation is that most P2a loops (dT, dT2, and dT3) are significantly less stable than the alternative loop types (Table 1). This suggests that the propeller loops are extremely reluctant to fold within anti-parallel two-layer quadruplex structures.
The last column of Table 1 shows the “parallel propensity”, i.e., the energetic difference between the P3p loop and the most favorable anti-parallel loop (Ln, Lw, or D). This value provides a quantitative estimate of how likely a particular loop sequence will tend to fold into parallel over anti-parallel structures.
For those models with tilted terminal quartets or distorted stems (T-Ln, T2-Lw, T3-D, and TTA-D), the loops appear to be much more stable if judged by ΔGloop than if judged by ΔGtotal (Table S1), and this indicates that most of the strain resides in the quadruplex stems. For the T-P4a models, even though there are no apparent distortions in the stem quadruplex structure, the sugar ring of the loop-connecting stem residue adopts a rare puckering geometry (Figure 3e). This rare puckering geometry is unfavorable since the stem of T-P4a (Gstem= −3694 kcal/mol) is 10 kcal/mol less stable than that of the P4a model carrying longer loop sequences (e.g., Gstem= −3704 kcal/mol for T2-P4a) (Table S1). Thus, ΔGloop is not suitable to evaluate the loop stability for T-P4a, and separating the free energies into loop and stem contributions may be misleading, especially for short loops.
MD Simulations of the Left Propeller Loop
All the propeller (P) loop structures reported to date are right loops. Can left propeller loops form? We speculate that since all G-quadruplex stems display right-handed structures, if two parallel strands are connected with a left P loop, it would span a much longer distance compared to the right P loop (Figure 1a,b). Therefore, we hypothesized that a left P loop would be very unstable. To test this, we ran explicit MD simulations on a model built from the 2GKU stem structure with a left dT3 propeller loop (Figure 1b). Each residue of the T3 loop was positioned manually using UCSF Chimera,(61) with each nucleobase group initially pointing into the solvent. Significant stem distortion was observed soon after the positional restraints were removed during the equilibration process. The early instability in the restrained simulations suggests that the left P loop is unstable. Therefore, G-quadruplex topologies utilizing left P loops are unlikely to form. They are assumed to be out-competed by more suitable topologies and may be too unstable to form or be observed.
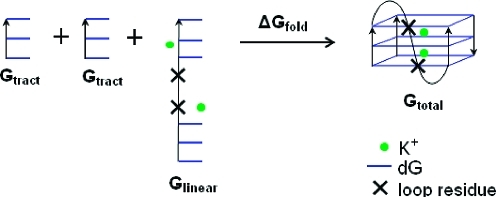
Effect of Loop Length on G-Quadruplex Stability
The results in Table 1 are useful to estimate the preferential loop type for a given sequence; however, the results do not directly provide insight into the influence of loop length on G-quadruplex stability. To understand the influence of different loop lengths on the stability of G-quadruplexes, we estimated relative folding energies for the different P3a models using a G-DNA formation process; this is schematically depicted in Figure 4, where ΔGfold = Gtotal – Glinear – 2Gtract. In this scheme, Glinear represents an idealized unfolded structure of the loop and its two connecting G-tracts, whereas 2Gtract represents the free energies of the two remaining unfolded G-tracts. This allows assessment of the relative folding energies via ΔΔGfold = Δ(Gtotal – Glinear), since the Gtract values will be equivalent for all of the P3a models. The Gtotal values for P3a models are shown in Table S1. To obtain the Glinear values, five linear sequences (dG3TG3, dG3T2G3, dG3T3G3, dG3TTAG3, and dG3T4G3) were built with the tleap module of AMBER into a fully extended initial structure. Since only crude estimates were needed, short explicit MD simulations (500 ps for each linear sequence) were run with the same equilibration strategy described earlier, and then MM-PBSA free energies were calculated and averaged at 6 ps intervals from the MD trajectories between 200 and 500 ps. Figure S3 shows a representative structure from the dG3T3G3 linear sequence at the end of a 500 ps simulation. Our calculations indicate, in general, that shorter loops are less favorable than longer loops (Table 2). This result seems contrary to previous experiments reporting that sequences with short loops usually show higher melting temperatures.36,43 However, it was also reported that sequences with short loops predominantly form dimers or trimers, even at very low concentrations,(38) and Tm is higher in multimers than in intramolecular G-quadruplexes.(79) This correlation suggests that the multimeric assemblies are parallel.(38) Parallel G-quadruplexes are flat, disk-like molecules and could easily stack upon each other through terminal quartets.80,81 In contrast, direct association of anti-parallel structures into higher order structures has not been observed experimentally, most probably due to the steric hindrance from lateral or diagonal loops. Therefore, we hypothesize that shorter loops themselves are not stabilizing factors per se, but they may bring more stability to G-quadruplex systems by favoring formation of parallel multimer.
Figure 4.
Illustration of folding energy calculation for the P3a models with varied loop lengths. For a particular model, ΔGfold = Gtotal – Glinear – 2Gtract, and therefore the relative folding energies between two models, ΔΔGfold = Δ(Gtotal – Glinear). Glinear was calculated with the two closest K+ cations included explicitly to make the free energies consistent.(46)
Table 2. Relative Folding Energies Calculated for the P3a Models (kcal/mol)a.
| folded model |
linear sequence |
without entropy |
with entropy |
|||||
|---|---|---|---|---|---|---|---|---|
| Gtotal | –TS | Glinear | –TS | (Gtotal – Glinear) | ΔΔGfold | (Gtotal – Glinear) | ΔΔGfold | |
| T-P3a | –2855.2 | –124.5 | –1633.2 | –130.7 | –1222.0 | 9 | –1215.8 | 6 |
| T2-P3a | –3004.6 | –124.1 | –1784.6 | –132.1 | –1220.0 | 11 | –1212.1 | 10 |
| T3-P3a | –3145.9 | –126.8 | –1914.9 | –136.0 | –1231.0 | 0 | –1221.7 | 0 |
| TTA-P3a | –3192.2 | –126.3 | –1946.1 | –141.0 | –1246.1 | –15 | –1231.5 | –10 |
| T4-P3a | –3283.9 | –131.5 | –2047.1 | –143.4 | –1236.8 | –6 | –1224.9 | –3 |
Relative folding energies of the P3a models were calculated on the basis of the scheme depicted in Figure 4. Entropy was calculated with a quasi-harmonic method on 50 snapshots. Glinear was estimated using the MM-PBSA method, with the two closest K+ cations included explicitly.
Potential Involvement of Multimeric G-Quadruplexes in Anti-parallel/Parallel Equilibrium
To test the plausibility of the above hypothesis, 50 ns MD simulations followed by free energy analyses were carried out on the anti-parallel monomer (2GKU), parallel monomer (1KF1), and parallel dimer (1KF1 dimer) with 5′-to-5′ arrangement obtained from the crystal structure of 1KF1.(80) Our free energy estimations indicate that the investigated unimolecular anti-parallel model structure with three dTTA loops is considerably more stable (∼30 kcal/mol) than its parallel counterpart. Considering the 1KF1 parallel dimer, the free energy gain upon association of the two parallel structures into a stacked dimer is ∼ –23.3 kcal/mol (neglecting rotational and translational entropy loss penalties, estimated from theory and experiment to be in the range of 3–10 kcal/mol,82−87 implying more accurate association free energies in the −13 to −20 kcal/mol range), which still favors the anti-parallel structure. Therefore, our calculations show the following relative stabilities for different forms of human telomeric quadruplexes—anti-parallel monomers > parallel dimers > parallel mononers—and a hypothetical energy curve is shown in Figure S4.
To get a brief idea of the influence of the loops on the dimer association, simulations were also run for the 1KF1 dimer with all the three dTTA loops changed to dT loops. The results show that the dimer association energy remains consistent (−24.5 kcal/mol) with shorter loops, while the anti-parallel structures become progressively less stable (Table 1). These computations suggest that the parallel multimer may become the predominant structural form in solution when shorter loops are present or under crowding conditions that favor multimer formation. This is consistent with very recent NMR work demonstrating, under crowding conditions, stabilization of the parallel form of human telomeric G-DNA and formation of higher order structure.(88) Our hypothesis and calculations on multimers could justify the discrepancy of the calculated instability of the short loops with the experimental results. However, because no consistent stacking mode has become available for multimeric G-quadruplexes,45,80,81,89,90 our calculations based solely on the dimer with simple 5′-to-5′ stacking of the terminal quartets are still quite preliminary and merit further investigation.
Discussion
Purpose and Limitations of the Current Work
The main aim of this work is to provide qualitative insights into the relative stabilities of a wide range of different G-quadruplex loops and loop sequences when attached to different stem arrangements. The study is based on an extensive set of explicit solvent MD simulations of stems augmented by single loops followed by post-processing of the simulation trajectories via continuum-solvent-based MM-PBSA free energy computations. As computational studies have clear limitations, it is critical to understand the approximations and limitations to properly evaluate the results. Possible limitations include the following: (1) For the Ln, Lw, D, and P3a loops, we assessed loop stabilities by comparing ΔGtotal of the models instead of considering only the loop residues. As explained, this approach, in general, is more consistent as it properly includes the loop–stem interactions. However, it also has some disadvantages, namely that as larger systems are included in the MM-PBSA procedure, the free energy computations are likely associated with larger standard deviations or errors. (2) For longer loops, the energy surfaces are very complicated, resulting from a plethora of accessible conformations, and therefore the final conformations observed may still depend considerably on, or be biased by, the initial structure. Moreover, many of the studied loop models were not based on experimental structures and instead were built by hand. Fifty-nanosecond MD simulations of each model may be insufficient to fully sample the accessible conformational space of the loops.(91) Enhanced sampling methods were not applied, as these methods may introduce additional uncertainties and a priori do not guarantee that global minima are unambiguously located.(91) Therefore, our comparisons could be biased due to insufficient sampling. (3) With the single-loop models, although it is easier to get quantitative estimates of the importance of each loop in determining the G-quadruplex topology or stability, we did not include the possibility of loop–loop interactions that may be very important for determining G-quadruplex structure. (4) During our simulations, stable ions or very high ion densities were observed for some loops, but to simplify our comparisons we did not include these ions in our free energy calculations, which could lead to errors.(46) (5) The MM-PBSA method is based on several approximations.(92) (6) The present nucleic acid force fields still show deficiencies in the representation of the exact structures of single-stranded G-DNA loops.47,91 Due to all these limitations, careful comparison to experimental observation is crucial.
Comparison of Simulation and Experimental Observations
dT Loops
MD simulation and free energy analyses suggest that the most stable single-nucleotide loop is the propeller loop spanning three quartets (P3 loops). Moreover, the significant difference between the T-P3a/T-P3p and T-Ln stabilities indicates that single-nucleotide sequences strongly prefer the formation of parallel G-quadruplex structures. This result is consistent with a number of previous experiments on G3-tract sequences with varied loop lengths.32,36−38,43,78 For example, the sequence in the c-Myc promoter with two dT loops and one dTA loop forms a parallel structure,(63) as does the guanine-rich sequence in the c-Kit oncogene promoter, which forms a parallel quadruplex with one dC loop, one dA loop, and a long CGCGA propeller loop.(93)
dT2 Loops
The results suggest that dT2 sequences favor the Ln loop over the Lw loop by a small margin, which is consistent with previous NMR structures: both T2-Ln and T2-Lw loops exist in each of the two inter-converting bimolecular anti-parallel G-quadruplex structures formed from d(TG4T2G4T).(94) If, in fact, T2-Ln and T2-Lw loops actually had significantly different relative stabilities, strands in this molecule would easily rearrange to form the more stable loop structures, i.e., either two Ln loops or two Lw loops. Both loop forms are observed since the loops are nearly isoenergetic and since different anti-parallel G4 strand arrangements tend to have similar stem stabilities.(30) Unlike the two-nucleotide repeat sequences discussed above, the four-repeat sequence d(T2G4)4 forms an unexpected three-layer structure,(95) indicating that dinucleotide loops are slightly detrimental to the stability of G-quadruplexes; this suggests that three dinucleotide loops cannot easily be accommodated in a single molecule. When two dinucleotide loops combine with a trinucleotide, such as for the TBA sequence d(G2T2G2TGTG2T2G2),(96) stable chair-type structures can form. This observation is consistent with our energetic estimates that suggest that short loops are less stable than longer loops (Table 2).
dT3 Loops
Lateral dT3 loops are favored over diagonal loops (Table 1), and this result is consistent with experiments and previous simulations by Hazel et al.(48) We also found that dT3 sequences favor Lw loops over Ln loops. A good experimental example to validate this result is the bimolecular anti-parallel quadruplex formed from d(GCGGTTTGCGG)2, with two Lw loops.(65) Another example is the crystal structure of dGGGGBrUTTGGGG,(97) where the single head-to-tail quadruplex has two Lw loops. The other two quadruplex molecules in the crystal unit cell stack together, and each molecule has one Lw loop and one Ln loop instead of forming two Lw loops. This may indicate that the two blunt ends with syn-syn-anti-anti glycosidic bond orientation patterns are more beneficial during quadruplex aggregation than two blunt ends with syn-anti-syn-anti conformations.
dT4 Loops
The simulation results further suggest the preferential formation of Lw loops with the dT4 sequence. Although this is consistent with the observation of dT4-Lw loops in bimolecular anti-parallel structures,98,99 diagonal loops are more common,58,100−103 which suggests that the diagonal loop is more stable than the Lw loop for dT4 sequences. This discrepancy may reflect known inaccuracies of the force field. In high-resolution X-ray structures, the diagonal dT4 loops are often associated with an ion binding site at the stem–loop junction.(58) Compared to the equilibrium exchange observed on a time scale of seconds in previous NMR studies,104,105 this ion usually leaves the junction site in the first few nanoseconds of the MD simulations and is therefore thought to be a force field artifact.47,91 If the ion binding site is important for the energetics of the diagonal loop, then it is not surprising that our results for the dT4 loop are not fully satisfactory. The failure to get fully consistent ranking likely implies the importance of including the ions explicitly for ranking the various T4 loops, consistent with our previous work where explicit ions were included and shown to be critical for the MM-PBSA analysis of G-quadruplex stems.(46)
Parallel versus Anti-parallel
The last column of Table 1 shows a “parallel preference” for each loop sequence. This quantity reflects the relative preference to form a parallel-stranded structure based on the estimated relative free energies. For unimolecular quadruplexes possessing three loops, we suggest that the relative preference for forming parallel structures can be estimated by summing the parallel preferences of the three loops. Table 3 shows six d(G3WiG3WkG3WkG3) sequences with varied loop lengths ordered according to decreased parallel structure signals in CD spectra(38) (see Figure 2 of the cited paper). As shown in the table, the suggested experimental propensity to form parallel structure is entirely consistent with the trends predicted by the computations.
Table 3. Consistency of the Calculated Parallel Preferences with the Parallel Signals in CD Spectraa,(38).
| sequence | parallel signal in CD spectra (260 nm) | calculated parallel preference (kcal/mol) |
|---|---|---|
| 1-1-2 | ↓ decreasing | –16 |
| 1-3-1 | –8 |
|
| 2-2-1 | –5 |
|
| 3-2-1 | 3 |
|
| 3-3-1 | 11 |
|
| 3-3-3 | 30 |
Parallel preference of a sequence was calculated by summing the parallel preferences of the three loops. Parallel preferences for the various single loops are shown in Table 1.
Predominant Geometries of Human Telomeric G-Quadruplexes
Our previous work has shown that the most stable geometry of G-quadruplex stems involving G3-tracts is the (3+1) scaffold: three parallel strands each occupying the most stable 5′-syn-anti-anti-3′ glycosidic bond orientation pattern and the fourth strand oriented anti-parallel, adopting a less stable 5′-syn-syn-anti-3′ pattern.(30) The (3+1) scaffolds are often also referred to as a “hybrid” type quadruplex topology. There are six possible arrangements that have three strands in one direction within anti-parallel unimolecular G-quadruplexes: ↓L↑P↑P↑, ↑P↑P↑L↓, ↑P↑L↓L↑, ↑L↓L↑P↑, ↑L↓D↑P↑, or ↑P↑D↓L↑, where the strand orientation is denoted with an arrow and intervening loops with the previously defined nomenclature (i.e., L is a lateral loop, D diagonal, and P is a propeller loop). Based on our calculations, the P and D loops are unfavorable for the dTTA sequence (Table 1). Thus, the ↑P↑L↓L↑ and ↑L↓L↑P↑ structures should be the most stable geometries for the human telomeric sequences, and indeed these correspond to the hybrid-I and hybrid-II structures observed in K+ solution.106−108 These two specific geometries have three syn-anti-anti strands, one syn-syn-anti strand, two lateral loops, and one propeller loop (just arranged in different orders), so their free energies should be similar. Very subtle differences such as the 5′- or 3′-flanking sequences would affect the final selection of the two conformations.(109) As left propeller loops are unstable according to our simulations, the presence of the propeller loop in the first or third position implies that these two geometries can only progress in the anti-clockwise direction following de Silva’s definition,(55) and this is consistent with experimental observations.106−108 In the asymmetric bimolecular G-quadruplex with a (3+1) scaffold arranged as ↑ + ↑L↓L↑, the less stable propeller loop is avoided.(110) In this case, the absence of the propeller loops suggests that both anti-clockwise and clockwise structures could form, and also that terminal flanking sequences may play an important role in determining the progression direction.
G-Quadruplex Multimers
If the trends are correct, the hypothetical energy curve shown in Figure S4 suggests that factors disfavoring anti-parallel structure may quite easily shift the equilibrium in favor of the parallel multimers, even when the parallel structure per se, as estimated with the current force field, remains less stable. Similarly, factors affecting free energies of formation of multimers of parallel structures would affect the anti-parallel vs parallel structure equilibrium. Although speculative (due to the approximations of the computational methods), this hypothesis could be helpful to qualitatively explain observations from G-quadruplex experiments. For example, molecular crowding agents like polyethylene glycol effectively increase local quadruplex concentrations and therefore may promote multimer formation, shifting the parallel/anti-parallel equilibrium toward parallel structures.108,109 Crystallization conditions have a similar effect that favors multimer formation, and therefore parallel structures of human telomeric sequences are observed in crystals,(80) while anti-parallel structures are the predominant geometries observed in K+ solution. Very recent NMR work supports these observations showing preferential stabilization of parallel G-DNA structures, and formation of higher order structure, in crowded solution.(88)
Conclusions
Biomolecular simulations of a wide variety of single-loop G-quadruplex models confirm and extend previous findings regarding the influence of G-quadruplex loops, specifically: (1) The single-nucleotide dT loop has a strong preference to form parallel structures, dinucleotide dT2 loops are compatible with both parallel and antiparallel structures, and trinucleotide or longer loop sequences prefer the formation of anti-parallel structures. (2) Short loops by themselves are less stable than longer loops. (3) Single-nucleotide loop sequences are able to form lateral loops over narrow grooves. (4) The left propeller loops are extremely unstable. (5) The final loop residues frequently stack with nearby terminal quartets in MD simulations. Preliminary work also suggests that multimers might affect anti-parallel/parallel equilibrium and justify the discrepancies between the calculated and experimental results with respect to the influence of short loops on G-quadruplexes stability, even though further work is necessary before conclusive arguments can be made. Overall, despite the known limitations of the computational methods, approach, and nucleic acid force fields, the encouraging consistency between the computational results and most experiments suggest that the computations are sufficiently robust to correctly dissect and reflect stability trends associated with different combinations of stem and loop topologies and sequences. Additionally, the results provide predictions testable by future experiments, although further simulation work is necessary to dissect the influences of other factors such as multimerization, different salts, salt concentration, and capping sequence effects.
Acknowledgments
T.E.C. acknowledges support from the NIH R01-GM59306890, and J.S. acknowledges Ministry of Education funding from the Czech Republic (AVOZ50040507, AVOZ50040702, and LC06030), from the Grant Agency of the Academy of Sciences of the Czech Republic (IAA40040802), and from the Grant Agency of the Czech Republic (203/09/1476 and P208/11/1822). Supporting computer time comes from the Center for High Performance Computing at the University of Utah and also substantial computer allocations and support from the NSF TeraGrid MCA01S027.
Supporting Information Available
Detailed analysis of the estimated free energies and molecular images of backbone differences in parallel and anti-parallel loops and unfolded G-tracts. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Sen D.; Gilbert W. Nature 1988, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- Laughlan G.; Murchie A. I.; Norman D. G.; Moore M. H.; Moody P. C.; Lilley D. M.; Luisi B. Science 1994, 265, 520–524. [DOI] [PubMed] [Google Scholar]
- Lane A. N.; Chaires J. B.; Gray R. D.; Trent J. O. Nucleic Acids Res. 2008, 36, 5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R. K.; Buckingham J. M.; Cram L. S.; Dani M.; Deaven L. L.; Jones M. D.; Meyne J.; Ratliff R. L.; Wu J. R. Proc. Natl. Acad. Sci. U.S.A. 1988, 85, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J. R. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 703–730. [DOI] [PubMed] [Google Scholar]
- Huppert J. L.; Balasubramanian S. Nucleic Acids Res. 2007, 35, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T. FEBS J. 2010, 277, 1107–1117. [DOI] [PubMed] [Google Scholar]
- Kim N. W.; Piatyszek M. A.; Prowse K. R.; Harley C. B.; West M. D.; Ho P. L.; Coviello G. M.; Wright W. E.; Weinrich S. L.; Shay J. W. Science 1994, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- Han H.; Hurley L. H. Trends Pharmacol. Sci. 2000, 21, 136–142. [DOI] [PubMed] [Google Scholar]
- Raymond E.; Soria J. C.; Izbicka E.; Boussin F.; Hurley L.; Von Hoff D. D. Invest. New Drugs 2000, 18, 123–137. [DOI] [PubMed] [Google Scholar]
- Hurley L. H. Biochem. Soc. Trans. 2001, 29, 692–696. [DOI] [PubMed] [Google Scholar]
- Doherty K. M.; Sharma S.; Gupta R.; Brosh R. M. Jr. Recent Pat. Anti-Cancer Drug Discovery 2006, 1, 185–200. [DOI] [PubMed] [Google Scholar]
- Huppert J. L. Philos. Trans. 2007, 365, 2969–2984. [DOI] [PubMed] [Google Scholar]
- Patel D. J.; Phan A. T.; Kuryavyi V. Nucleic Acids Res. 2007, 35, 7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L. Chem. Soc. Rev. 2008, 37, 1375–1384. [DOI] [PubMed] [Google Scholar]
- Ou T. M.; Lu Y. J.; Tan J. H.; Huang Z. S.; Wong K. Y.; Gu L. Q. ChemMedChem 2008, 3, 690–713. [DOI] [PubMed] [Google Scholar]
- Tan J. H.; Gu L. Q.; Wu J. Y. Mini Rev. Med. Chem. 2008, 8, 1163–1178. [DOI] [PubMed] [Google Scholar]
- Neidle S. FEBS J. 2010, 277, 1118–1125. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S.; Bugaut A.; Huppert J. L.; Balasubramanian S. Nat. Chem. Biol. 2007, 3, 218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps H. J.; Rhodes D. Trends Cell Biol. 2009, 19, 414–422. [DOI] [PubMed] [Google Scholar]
- Hagihara M.; Yamauchi L.; Seo A.; Yoneda K.; Senda M.; Nakatani K. J. Am. Chem. Soc. 2010, 132, 11171–11178. [DOI] [PubMed] [Google Scholar]
- Wanrooij P. H.; Uhler J. P.; Simonsson T.; Falkenberg M.; Gustafsson C. M. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 16072–16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M.; Lipsett M. N.; Davies D. R. Proc. Natl. Acad. Sci. U.S.A. 1962, 48, 2013–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S.; Parkinson G. N.; Hazel P.; Todd A. K.; Neidle S. Nucleic Acids Res. 2006, 34, 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M. A. Biopolymers 2000, 56, 123–146. [DOI] [PubMed] [Google Scholar]
- Simonsson T. Biol Chem 2001, 382, 621–628. [DOI] [PubMed] [Google Scholar]
- Gaynutdinov T. I.; Neumann R. D.; Panyutin I. G. Nucleic Acids Res. 2008, 36, 4079–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires J. B. FEBS J. 2010, 277, 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang X.; Sponer J.; Cheatham T. E. III Nucleic Acids Res. 2011, 39, 4499–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov I.; Shafer R. H. Biochemistry 2000, 39, 1462–1468. [DOI] [PubMed] [Google Scholar]
- Hazel P.; Huppert J.; Balasubramanian S.; Neidle S. J. Am. Chem. Soc. 2004, 126, 16405–16415. [DOI] [PubMed] [Google Scholar]
- Risitano A.; Fox K. R. Nucleic Acids Res. 2004, 32, 2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevec M.; Plavec J. Biochemistry 2005, 44, 15238–15246. [DOI] [PubMed] [Google Scholar]
- Rachwal P. A.; Findlow I. S.; Werner J. M.; Brown T.; Fox K. R. Nucleic Acids Res. 2007, 35, 4214–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaut A.; Balasubramanian S. Biochemistry 2008, 47, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.; Maiti S. Nucleic Acids Res. 2008, 36, 5610–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargiasso N.; Rosu F.; Hsia W.; Colson P.; Baker E. S.; Bowers M. T.; De Pauw E.; Gabelica V. J. Am. Chem. Soc. 2008, 130, 10208–10216. [DOI] [PubMed] [Google Scholar]
- Arora A.; Maiti S. J. Phys. Chem. B 2009, 113, 8784–8792. [DOI] [PubMed] [Google Scholar]
- Balkwill G. D.; Garner T. P.; Searle M. S. Mol. Biosyst. 2009, 5, 542–547. [DOI] [PubMed] [Google Scholar]
- Balkwill G. D.; Garner T. P.; Williams H. E.; Searle M. S. J. Mol. Biol. 2009, 385, 1600–1615. [DOI] [PubMed] [Google Scholar]
- Hu L.; Lim K. W.; Bouaziz S.; Phan A. T. J. Am. Chem. Soc. 2009, 131, 16824–16831. [DOI] [PubMed] [Google Scholar]
- Guedin A.; Gros J.; Alberti P.; Mergny J. L. Nucleic Acids Res. 2010, 38, 7858–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. W.; Lacroix L.; Yue D. J.; Lim J. K.; Lim J. M.; Phan A. T. J. Am. Chem. Soc. 2010, 132, 12331–12342. [DOI] [PubMed] [Google Scholar]
- Kuryavyi V.; Phan A. T.; Patel D. J. Nucleic Acids Res. 2010, 38, 6757–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R.; Cheatham T. E. III; Spackova N.; Fadrna E.; Berger I.; Koca J.; Sponer J. Biophys. J. 2003, 85, 1787–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrna E.; Spackova N.; Stefl R.; Koca J.; Cheatham T. E. III; Sponer J. Biophys. J. 2004, 87, 227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel P.; Parkinson G. N.; Neidle S. Nucleic Acids Res. 2006, 34, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel P.; Parkinson G. N.; Neidle S. J. Am. Chem. Soc. 2006, 128, 5480–5487. [DOI] [PubMed] [Google Scholar]
- Sponer J.; Spackova N. Methods 2007, 43, 278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnikov R. V.; Golovin A. V.; Kopylov A. M. Biochemistry (Moscow) 2010, 75, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Rosu F.; Gabelica V.; Poncelet H.; De Pauw E. Nucleic Acids Res. 2010, 38, 5217–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J.; Rosu F.; Amrane S.; De Cian A.; Gabelica V.; Lacroix L.; Mergny J. L. Nucleic Acids Res. 2007, 35, 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin C.; Leroy J. L. Nucleic Acids Res. 2008, 36, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webba da Silva M. Chemistry (Weinheim an der Bergstrasse, Germany) 2007, 13, 9738–9745. [DOI] [PubMed] [Google Scholar]
- Luu K. N.; Phan A. T.; Kuryavyi V.; Lacroix L.; Patel D. J. J. Am. Chem. Soc. 2006, 128, 9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryavyi V.; Majumdar A.; Shallop A.; Chernichenko N.; Skripkin E.; Jones R.; Patel D. J. J. Mol. Biol. 2001, 310, 181–194. [DOI] [PubMed] [Google Scholar]
- Haider S.; Parkinson G. N.; Neidle S. J. Mol. Biol. 2002, 320, 189–200. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Patel D. J. J. Mol. Biol. 1993, 234, 1171–1183. [DOI] [PubMed] [Google Scholar]
- Joung I. S.; Cheatham T. E. III J. Phys. Chem. B 2008, 112, 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. J. Comput. Chem. 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Patel D. J. Structure 1993, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- Ambrus A.; Chen D.; Dai J.; Jones R. A.; Yang D. Biochemistry 2005, 44, 2048–2058. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K.; Padmanabhan K. P.; Ferrara J. D.; Sadler J. E.; Tulinsky A. J. Biol. Chem. 1993, 268, 17651–17654. [DOI] [PubMed] [Google Scholar]
- Kettani A.; Kumar R. A.; Patel D. J. J. Mol. Biol. 1995, 254, 638–656. [DOI] [PubMed] [Google Scholar]
- Kang C.; Zhang X.; Ratliff R.; Moyzis R.; Rich A. Nature 1992, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- Crnugelj M.; Sket P.; Plavec J. J. Am. Chem. Soc. 2003, 125, 7866–7871. [DOI] [PubMed] [Google Scholar]
- Case D. A.; Cheatham T. E. III; Darden T. A.; Gohlker H.; Luo R.; Merz K. M. Jr.; Onufriev A. V.; Simmerling C.; Wang B.; Woods R. J. Comput. Chem. 2005, 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Cieplak P.; Kollman P. A. J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar]
- Perez A.; Marchan I.; Svozil D.; Sponer J.; Cheatham T. E. III; Laughton C. A.; Orozco M. Biophys. J. 2007, 11, 3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J.; Klein M. L. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar]
- Pastor R. W.; Brooks B. R.; Szabo A. Mol. Phys. 1988, 65, 1409–1419. [Google Scholar]
- Jayaram B.; Sprous D.; Beveridge D. L. J. Phys. Chem. B 1998, 102, 9571–9576. [Google Scholar]
- Schlitter J. Chem. Phys. Lett. 1993, 215, 617–621. [Google Scholar]
- Kuryavyi V.; Patel D. J. Structure 2010, 18, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.; Sahoo B.; Varun K. A.; Maiti S.; Maiti S. Nucleic Acids Res. 2008, 36, 4433–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z.; Chen Y.; Li X.; Fang W. Analyst 2011, 136, 2367–2372. [DOI] [PubMed] [Google Scholar]
- Parkinson G. N.; Lee M. P.; Neidle S. Nature 2002, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- Kato Y.; Ohyama T.; Mita H.; Yamamoto Y. J. Am. Chem. Soc. 2005, 127, 9980–9981. [DOI] [PubMed] [Google Scholar]
- Page M. I.; Jencks W. P. Proc. Natl. Acad. Sci. U.S.A. 1971, 68, 1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. K.; Given J. A.; Bush B. L.; McCammon J. A. Biophys. J. 1997, 72, 1047–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans J.; Wang L. J. Am. Chem. Soc. 1997, 119, 2707–2714. [Google Scholar]
- Luo R.; Gilson M. K. J. Am. Chem. Soc. 2000, 122, 2934–2937. [Google Scholar]
- Yu Y. B.; Privalov P. L.; Hodges R. S. Biophys. J. 2001, 81, 1632–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T.; Masunov A.; Gandolfo F. Proteins 2002, 47, 194–208. [DOI] [PubMed] [Google Scholar]
- Heddi B.; Phan A. T. J. Am. Chem. Soc. 2011, 133, 9824–9833. [DOI] [PubMed] [Google Scholar]
- Renciuk D.; Kejnovska I.; Skolakova P.; Bednarova K.; Motlova J.; Vorlickova M. Nucleic Acids Res. 2009, 37, 6625–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraccone L.; Garbett N. C.; Chaires J. B.; Trent J. O. Biopolymers 2010, 93, 533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrna E.; Spackova N. a.; Sarzynska J.; Koca J.; Orozco M.; C. T. E. III; Kulinski T.; Sponer J. J. Chem. Theory Comput. 2009, 5, 2514–2530. [DOI] [PubMed] [Google Scholar]
- Srinivasan J.; Cheatham T. E. III; Cieplak P.; Kollman P. A.; Case D. A. J. Am. Chem. Soc. 1998, 120, 9401–9409. [Google Scholar]
- Hsu S. T.; Varnai P.; Bugaut A.; Reszka A. P.; Neidle S.; Balasubramanian S. J. Am. Chem. Soc. 2009, 131, 13399–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Modi Y. S.; Patel D. J. J. Mol. Biol. 2004, 338, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Patel D. J. Structure 1994, 2, 1141–1156. [DOI] [PubMed] [Google Scholar]
- Schultze P.; Macaya R. F.; Feigon J. J. Mol. Biol. 1994, 235, 1532–1547. [DOI] [PubMed] [Google Scholar]
- Esposito V.; Randazzo A.; Piccialli G.; Petraccone L.; Giancola C.; Mayol L. Org. Biomol. Chem. 2004, 2, 313–318. [DOI] [PubMed] [Google Scholar]
- Bouaziz S.; Kettani A.; Patel D. J. J. Mol. Biol. 1998, 282, 637–652. [DOI] [PubMed] [Google Scholar]
- Kettani A.; Bouaziz S.; Gorin A.; Zhao H.; Jones R. A.; Patel D. J. J. Mol. Biol. 1998, 282, 619–636. [DOI] [PubMed] [Google Scholar]
- Schultze P.; Smith F. W.; Feigon J. Structure 1994, 2, 221–233. [DOI] [PubMed] [Google Scholar]
- Keniry M. A.; Strahan G. D.; Owen E. A.; Shafer R. H. Eur. J. Biochem. 1995, 233, 631–643. [DOI] [PubMed] [Google Scholar]
- Schultze P.; Hud N. V.; Smith F. W.; Feigon J. Nucleic Acids Res. 1999, 27, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. L.; Strobel S. A.; Loria J. P. Nucleic Acids Res. 2006, 34, 4506–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbevsek P.; Sket P.; Plavec J. J. Am. Chem. Soc. 2008, 130, 14287–14293. [DOI] [PubMed] [Google Scholar]
- Podbevsek P.; Hud N. V.; Plavec J. Nucleic Acids Res. 2007, 35, 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Punchihewa C.; Ambrus A.; Chen D.; Jones R. A.; Yang D. Nucleic Acids Res. 2007, 35, 2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Carver M.; Punchihewa C.; Jones R. A.; Yang D. Nucleic Acids Res. 2007, 35, 4927–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Luu K. N.; Patel D. J. Nucleic Acids Res. 2007, 35, 6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Carver M.; Yang D. Biochimie 2008, 90, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.; Phan A. T.; Patel D. J. J. Am. Chem. Soc. 2005, 127, 17277–17285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.