Abstract
Genetic variations in the CYP2A6 nicotine metabolic gene and the CHRNA5-CHRNA3-CHRNB4 (CHRNA5-A3-B4) nicotinic gene cluster have been independently associated with lung cancer. With genotype data from ever-smokers of European ancestry (417 lung cancer patients and 443 control subjects), we investigated the relative and combined associations of polymorphisms in these two genes with smoking behavior and lung cancer risk. Kruskal–Wallis tests were used to compare smoking variables among the different genotype groups, and odds ratios (ORs) for cancer risk were estimated using logistic regression analysis. All statistical tests were two-sided. Cigarette consumption (P < .001) and nicotine dependence (P = .036) were the highest in the combined CYP2A6 normal metabolizers and CHRNA5-A3-B4 AA (tag single-nucleotide polymorphism rs1051730 G>A) risk group. The combined risk group also exhibited the greatest lung cancer risk (OR = 2.03; 95% confidence interval [CI] = 1.21 to 3.40), which was even higher among those who smoked 20 or fewer cigarettes per day (OR = 3.03; 95% CI = 1.38 to 6.66). Variation in CYP2A6 and CHRNA5-A3-B4 was independently and additively associated with increased cigarette consumption, nicotine dependence, and lung cancer risk. CYP2A6 and CHRNA5-A3-B4 appear to be more strongly associated with smoking behaviors and lung cancer risk, respectively.
Context and Caveats
Prior knowledge
CYP2A6 and the nicotinic receptor subunit gene cluster CHRNA5-CHRNA3-CHRNB4 (CHRNA5-A3-B4) are involved in nicotine and tobacco-specific nitrosamine metabolism and signaling, respectively, and have been independently associated with cigarette consumption and lung cancer risk in previous studies, leading to the hypothesis that genetic variation in both CYP2A6 and CHRNA5-A3-B4 may influence lung cancer risk in smokers directly, as well as indirectly through altered cigarette exposure.
Study design
Ever-smokers of European ancestry selected from a published genome-wide association study (417 non-small cell lung cancer patients and 443 control subjects) were grouped by predicted CYP2A6 metabolic activity and according to genetic variation in the CHRNA5-A3-B4 cluster. Smoking variables among the genotype groups were compared, and the relationship between the genotype groups and lung cancer risk was examined.
Contribution
Combined genetic variation in CYP2A6 and CHRNA5-A3-B4 was associated with increased cigarette consumption and nicotine dependence. Variation in these genes was independently associated with an increased risk of lung cancer with an even higher relative increase in risk from these genes among the lighter-smoking stratum.
Implications
In addition to mediating lung cancer risk through smoking behavior, genetic variation in CYP2A6 and CHRNA5-A3-B4 may mediate carcinogenesis directly. Whereas variation in CYP2A6 may play a larger relative role in smoking behaviors, variation in CHRNA5-A3-B4 may play a larger relative role in lung cancer risk.
Limitations
The study population included ever-smokers of European ancestry and the application of the findings to other races/ethnicities is unclear. Also, cigarette consumption among the study participants was self-reported.
From the Editors
Genetic variation in CYP2A6 and in the nicotinic receptor subunit gene cluster, CHRNA5-CHRNA3-CHRNB4 (CHRNA5-A3-B4), has been modestly associated with lung cancer susceptibility in independent studies (1–5). CYP2A6 and CHRNA5-A3-B4 are both involved in the pharmacology of nicotine and nitrosamines, which are lung cancer procarcinogens (6). CYP2A6 inactivates nicotine and also activates tobacco-specific nitrosamines (1,7), whereas nicotinic receptors mediate nicotine-induced reward (8) and nitrosamine-induced carcinogenic signaling (9). Thus, genetic variations in CYP2A6 and CHRNA5-A3-B4 have the potential to influence lung cancer risk directly and indirectly through alteration of smoking behavior (10–13).
We investigated the associations between CYP2A6 and CHRNA5-A3-B4 polymorphisms, alone and in combination, with smoking behaviors and lung cancer risk. Ever-smoking non–small cell lung cancer patients (n = 417) and control subjects (n = 443) of European ancestry matched by age, sex, and smoking variables were selected from our previously published genome-wide association discovery set (4) (Supplementary Table 1, available online). Participants were genotyped for CYP2A6*2, *4, *9, and *12 (14,15), reduced enzymatic function alleles common in those of European ancestry, and grouped by predicted metabolic activity. CYP2A6 carriers were defined as either CYP2A6 normal metabolizers (participants who lacked variant alleles) or CYP2A6 reduced metabolizers (participants who carried at least one variant allele) (14,16).
Participants were also grouped according to genetic variation in the CHRNA5-A3-B4 cluster, represented by the tag single-nucleotide polymorphism rs1051730 G>A (4). The AA genotype is the established risk genotype for smoking intensity and lung cancer risk (4,5,17), hence rs1051730 GG and GA individuals were grouped together. Individual allele and combined group genotype results are available in Supplementary Table 2 (available online). We used the Kruskal–Wallis test to compare smoking variables among genotype groups for all participants and separately in control subjects who were current smokers. Lung cancer odds ratios (ORs) were estimated using logistic regression analysis. This study was approved by Review Boards at the M.D. Anderson Cancer Center (Houston, TX), the Institute for Cancer Research Foundation (London, UK), and the University of Toronto (Toronto, ON, Canada).
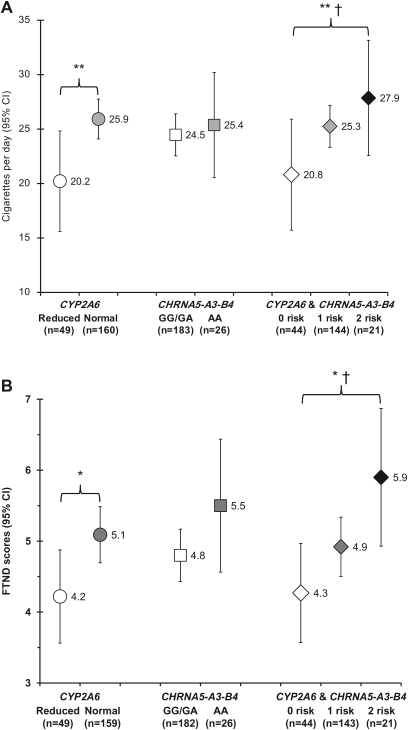
Daily cigarette consumption was statistically significantly associated with the CYP2A6 and CHRNA5-A3-B4 genotypes, alone and in combination, in the overall study population (Supplementary Table 3, available online). Here, we present the association in control subjects who were current smokers (n = 209) (Figure 1, A), because cancer diagnosis and recall bias among former smokers are minimized as confounders in this analysis. In control subjects who were current smokers, CYP2A6 normal metabolizers smoked statistically significantly more cigarettes per day (CPD) (mean = 25.9 CPD, 95% CI = 24.1 to 27.8 CPD) compared with reduced metabolizers (mean = 20.2 CPD, 95% CI = 15.6 to 24.8 CPD) (P < .001). The CHRNA5-A3-B4 AA genotype was associated with a statistically significant increase in CPD compared with the CHRNA5-A3-B4 GG and GA genotypes in the overall study population (Supplementary Table 3, available online) but did not reach statistical significance among control subjects who were current smokers (P = .416).
Figure 1.
Association of CYP2A6 and CHRNA5-A3-B4 genotype with smoking behaviors. A) Self-reported cigarettes smoked per day and B) Fagerström Test for Nicotine Dependence (FTND) scores are displayed for control subjects who were current smokers and are shown as the mean with 95% confidence intervals (CIs) by genotype group. The genotypes analyzed include the CYP2A6 genotype alone, the CHRNA5-A3-B4 genotype alone, and the combined CYP2A6 and CHRNA5-A3-B4 genotype groups according to the number of risk genotypes. The low-risk group (0 risk) included CYP2A6 reduced metabolizers with the CHRNA5-A3-B4 GG and GA genotypes. The intermediate-risk group (one risk) included participants with either the CYP2A6 normal metabolizer genotypes or the CHRNA5-A3-B4 AA genotype, and the high-risk group (two risk) included CYP2A6 normal metabolizers with the CHRNA5-A3-B4 AA genotype. P values were calculated by Kruskal–Wallis tests. *P < .05, **P < .001. Ptrend was calculated by a generalized linear model. †Ptrend < .05. All statistical tests were two-sided.
To assess the association of CYP2A6 and CHRNA5-A3-B4 genotypes in combination with smoking behavior, we separated participants into three groups as follows: CYP2A6 reduced metabolizers with the CHRNA5-A3-B4 GG and GA genotypes were identified as the low-risk group, CYP2A6 normal metabolizers with the CHRNA5-A3-B4 AA genotype were identified as the high-risk group, and control subjects with the either the CYP2A6 normal metabolizer genotypes or the CHRNA5-A3-B4 AA genotype were identified as the intermediate-risk group. Among control subjects who were current smokers, cigarette consumption was statistically significantly different (P < .001) between the low-risk group (mean = 20.8 CPD, 95% CI = 15.7 to 25.9 CPD), intermediate-risk group (mean = 25.3 CPD, 95% CI = 23.3 to 27.2 CPD), and high-risk group (mean = 27.9 CPD, 95% CI = 22.6 to 33.2 CPD) and increased linearly across these three genotype groups (Ptrend = .042). Similar results were observed for the overall study population (Supplementary Table 3, available online).
We also investigated the relationship between nicotine dependence and CYP2A6 and CHRNA3-A5-B4 genotypes, as assessed by the Fagerström Test for Nicotine Dependence (FTND). FTND scores were statistically significantly associated with the CYP2A6 and CHRNA5-A3-B4 genotypes, alone and in combination, in the overall study population (Supplementary Table 3, available online). In control subjects who were current smokers, statistically significantly higher FTND scores (P = .036) were observed for CYP2A6 normal metabolizers (mean = 5.1, 95% CI = 4.7 to 5.5) compared with reduced metabolizers (mean = 4.2, 95% CI = 3.6 to 4.9). The CHRNA5-A3-B4 AA genotype was associated with a statistically significant increase in FTND scores compared with the CHRNA5-A3-B4 GG and GA genotypes in the overall study population (Supplementary Table 3, available online) but did not reach statistical significance among control subjects who were current smokers (P = .137).
Nicotine dependence was also associated with the CYP2A6 and CHRNA5-A3-B4 genotypes in combination. Among control subjects who were current smokers, FTND scores were statistically significantly different (P = .036) between the low-risk group (mean = 4.3, 95% CI = 3.6 to 5.0), intermediate-risk group (mean = 4.9, 95% CI = 4.5 to 5.3), and high-risk group (mean = 5.9, 95% CI = 4.9 to 6.9) and increased linearly across these three genotype groups (Ptrend = .013). Similar results were observed for the overall study population (Supplementary Table 3, available online).
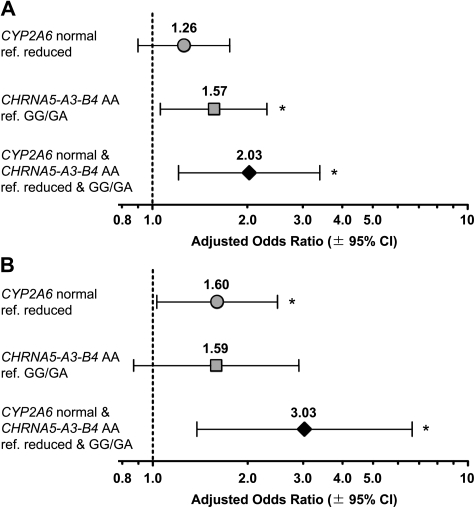
We then investigated the impact of CYP2A6 and CHRNA5-A3-B4 on lung cancer susceptibility (Figure 2, A). Because both CYP2A6 and CHRNA5-A3-B4 were associated with cigarette consumption, we adjusted odds ratios for cigarette pack–years. CYP2A6 normal metabolizers had a non-statistically significant increase in lung cancer risk compared with reduced metabolizers (OR = 1.26, 95% CI = 0.90 to 1.76; P = .180), whereas the CHRNA5-A3-B4 AA genotype was statistically significantly associated with increased lung cancer risk (OR = 1.57, 95% CI = 1.06 to 2.31; P = .024). Of note, the combination of both risk genotypes, CYP2A6 normal metabolizer and CHRNA5-A3-B4 AA, was statistically significantly associated with increased lung cancer risk (OR = 2.03, 95% CI = 1.21 to 3.40; P = .007).
Figure 2.
Lung cancer risk by CYP2A6 and CHRNA5-A3-B4 genotype. A) Overall risk of lung cancer and B) risk of lung cancer in the lighter-smoking stratum are shown as adjusted odds ratios with 95% confidence intervals (CIs). The lighter-smoking stratum was defined as individuals smoking 20 cigarettes or less per day on the basis of the median cigarette consumption in patients and control subjects. For each odds ratio, the lower-risk genotype group served as the reference (ref), ie, ref. CYP2A6 reduced metabolizers and/or CHRNA5-A3-B4 GG or GA. Odds ratios were adjusted by age (continuous), sex (male or female), and log pack–years (continuous). Lung cancer odds ratios and P values were estimated by logistic regression analysis with P < .05 denoted by an asterisk. All statistical tests were two-sided.
Adjusting the odds ratio for each genotype alone by variation in the other gene did not affect the association between genotype and lung cancer risk suggesting that variation in CYP2A6 and CHRNA5-A3-B4 independently affects lung cancer risk (for unadjusted, pack-year, and genotype-adjusted odds ratios, see Supplementary Table 4, available online). A statistically significant interaction between CYP2A6 and CHRNA5-A3-B4 was not found in a logistic regression model of lung cancer risk, suggesting that the combined genotype effects are additive.
In subgroup analyses, the CYP2A6 normal genotype was statistically significantly associated with lung cancer risk in the lighter-smoking stratum in which CPD was 20 or less, as defined by median CPD in patients and control subjects (OR = 1.60, 95% CI = 1.03 to 2.49; P = .036) (Figure 2, B). A statistically significant association with lung cancer risk was observed among those with both risk genotypes, CYP2A6 normal and CHRNA5-A3-B4 AA (OR = 3.03, 95% CI = 1.38 to 6.66; P = .006), whereas among the heavier-smoking stratum (CPD >20), no association with lung cancer risk was noted for either CYP2A6 or CHRNA5-A3-B4 alone or in combination (Supplementary Table 5, available online). This pattern of higher genetic risk in the lighter-smoking stratum supports the notion of a direct genotype contribution to lung carcinogenesis vs a sole contribution of genotype from altered smoking quantity and has been previously reported for the CHRNA5-A3-B4 genotype (5). Low-exposure gene effects (18) have also been observed for other polymorphic drug-metabolizing enzymes such as CYP1A1, NAT2, and MPO (19–22) and merit attention. Lighter smokers, a growing segment of the smoking population (23,24), tend to have reduced concerns about the negative health effects associated with smoking. Our findings, however, suggest that the genetic risk for lung cancer may remain high among lighter smokers. Furthermore, the levels of smoking may obscure genetic signals in association studies conducted in smoking populations. We also conducted a subgroup analysis by histology (Supplementary Table 6, available online) and found evidence of a stronger association of CYP2A6 and CHRNA5-A3-B4 with adenocarcinoma (OR = 2.09, 95% CI = 1.08 to 4.03, P = .029) vs squamous cell carcinoma (OR = 1.44, 95% CI = 0.65 to 3.20, P = .372).
The major limitation of the current study is the use of self-reported cigarette consumption without biochemical verification, such as plasma cotinine levels or total urinary nicotine equivalents. In addition, neither the smoking behavior analyses nor our lung cancer risk models were able to incorporate potential changes in smoking patterns of study participants over time.
This study demonstrates for the first time, to our knowledge, that genetic variation in CYP2A6 and CHRNA5-A3-B4 combines to increase cigarette consumption and nicotine dependence and independently and additively combines to increase lung cancer risk. Our results also suggest that variation in CYP2A6 appears to have a larger relative role in smoking behaviors, whereas variation in CHRNA5-A3-B4 may play a larger relative role in lung cancer risk. Given that metabolic activation of procarcinogens is an early step along the pathway to cancer (6), altered CYP2A6 nitrosamine activation may make a small contribution to lung cancer risk. The α5, α3, and β4 subunits are expressed in the respiratory tract (25) and have been implicated in the pathological effects of nitrosamines on epithelial cells (26). Thus, genetic variation in these subunits could influence nitrosamine carcinogenic signialing. These findings further our understanding of genetic risk factors for smoking and lung cancer and provide insight into mechanisms of lung cancer carcinogenesis.
Funding
This work was supported by the Canadian Institutes of Health Research (MOP86471 to RFT); the Centre for Addiction and Mental Health; a Canada Research Chair in Pharmacogenetics (RFT); the National Institutes of Health (U01 DA020830 to RFT; CA55769 and CA127219 to MRS, CA133996 and CA121197 to CIA); the Kleberg Center for Molecular Markers at the M.D. Anderson Cancer Center; and the Flight Attendant Medical Research Institutes.
Supplementary Material
Footnotes
The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the article; or the decision to submit the article for publication.
References
- 1.Rossini A, de Almeida Simao T, Albano RM, Pinto LF. CYP2A6 polymorphisms and risk for tobacco-related cancers. Pharmacogenomics. 2008;9(11):1737–1752. doi: 10.2217/14622416.9.11.1737. [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 4.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100(21):1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Toxicol. 2008;21(1):160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8(10):1385–1402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 8.Changeux JP. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat Rev Neurosci. 2010;11(6):389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 9.Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer. 2009;9(3):195–205. doi: 10.1038/nrc2590. [DOI] [PubMed] [Google Scholar]
- 10.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin Pharmacol Ther. 2005;77(3):145–158. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 11.TAG. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum L, Lerer B. Differential contribution of genetic variation in multiple brain nicotinic cholinergic receptors to nicotine dependence: recent progress and emerging open questions. Mol Psychiatry. 2009;14(10):912–945. doi: 10.1038/mp.2009.59. [DOI] [PubMed] [Google Scholar]
- 14.Lerman C, Jepson C, Wileyto EP, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther. 2010;87(5):553–557. doi: 10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mwenifumbo JC, Zhou Q, Benowitz NL, Sellers EM, Tyndale RF. New CYP2A6 gene deletion and conversion variants in a population of Black African descent. Pharmacogenomics. 2010;11(2):189–198. doi: 10.2217/pgs.09.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benowitz NL, Swan GE, Jacob P, III, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80(5):457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100(18):1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taioli E, Zocchetti C, Garte S. Models of interaction between metabolic genes and environmental exposure in cancer susceptibility. Environ Health Perspect. 1998;106(2):67–70. doi: 10.1289/ehp.9810667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrie AK, Nyberg F, Warholm M, Rannug A. Influence of CYP1A1, GSTM1, GSTT1, and NQO1 genotypes and cumulative smoking dose on lung cancer risk in a Swedish population. Cancer Epidemiol Biomarkers Prev. 2004;13(6):908–914. [PubMed] [Google Scholar]
- 20.Ishibe N, Wiencke JK, Zuo ZF, McMillan A, Spitz M, Kelsey KT. Susceptibility to lung cancer in light smokers associated with CYP1A1 polymorphisms in Mexican- and African-Americans. Cancer Epidemiol Biomarkers Prev. 1997;6(12):1075–1080. [PubMed] [Google Scholar]
- 21.Sorensen M, Autrup H, Tjonneland A, Overvad K, Raaschou-Nielsen O. Genetic polymorphisms in CYP1B1, GSTA1, NQO1 and NAT2 and the risk of lung cancer. Cancer Lett. 2005;221(2):185–190. doi: 10.1016/j.canlet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Schabath MB, Spitz MR, Hong WK, et al. A myeloperoxidase polymorphism associated with reduced risk of lung cancer. Lung Cancer. 2002;37(1):35–40. doi: 10.1016/s0169-5002(02)00034-x. [DOI] [PubMed] [Google Scholar]
- 23.Duval S, Jacobs DR, Jr., Barber C, et al. Trends in cigarette smoking: the Minnesota Heart Survey, 1980-1982 through 2000-2002. Nicotine Tob Res. 2008;10(5):827–832. doi: 10.1080/14622200802029517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiffman S. Light and intermittent smokers: background and perspective. Nicotine Tob Res. 2009;11(2):122–125. doi: 10.1093/ntr/ntn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam DC, Girard L, Ramirez R, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67(10):4638–4647. doi: 10.1158/0008-5472.CAN-06-4628. [DOI] [PubMed] [Google Scholar]
- 26.Arredondo J, Chernyavsky AI, Grando SA. Nicotinic receptors mediate tumorigenic action of tobacco-derived nitrosamines on immortalized oral epithelial cells. Cancer Biol Ther. 2006;5(5):511–517. doi: 10.4161/cbt.5.5.2601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.