Summary
The ClpS adaptor delivers N-end-rule substrates to ClpAP, an energy-dependent AAA+ protease, for degradation. How ClpS binds specific N-end residues is known in atomic detail and clarified here, but the delivery mechanism is poorly understood. We show that substrate binding is enhanced when ClpS binds hexameric ClpA. Reciprocally, N-end-rule substrates increase ClpS affinity for ClpA6. Enhanced binding requires the N-end residue and peptide bond of the substrate, as well as multiple aspects of ClpS, including, a side chain that contacts the substrate α-amino group and the flexible N-terminal extension (NTE). Finally, enhancement also needs the N domain and AAA+ rings of ClpA, connected by a long linker. The NTE can be engaged by the ClpA translocation pore, but ClpS resists unfolding/degradation. We propose a staged-delivery model that illustrates how intimate contacts between the substrate, adaptor, and protease reprogram specificity and coordinate handoff from the adaptor to the protease.
Keywords: ClpS adaptor, N degrons, ClpAP protease, N recognins
Introduction
The N-end rule relates degradation susceptibility to a protein’s N-terminal amino acid (Bachmair et al., 1986; Varshavsky, 2008). In bacteria, four N-terminal residues (Tyr, Phe, Trp, and Leu) serve as primary N-end degrons (Tobias et al., 1991). The ClpS adaptor binds these residues and delivers attached substrates to the AAA+ ClpAP protease for degradation (Erbse et al., 2006; Wang et al., 2007). In eukaryotes, a family of E3 ubiquitin ligases with a small region homologous to ClpS recognizes and covalently modifies N-end-rule substrates with polyubiquitin, targeting these modified proteins to the proteasome (Lupas et al., 2003; Tasaki and Kwon, 2007).
ClpAP, one of five degradation machines in Escherichia coli, consists of the ClpP14 protease and the ClpA6 unfoldase. ClpA6 is active as a hexamer composed of two AAA+ rings (D1 and D2) and also carries a family-specific N domain, which is flexibly attached to the D1 ring (Gottesman et al., 1992; Cranz-Milleva et al., 2008; Effantin et al., 2010). Using the energy of ATP binding and hydrolysis, machinery in the axial pore of ClpA6 unfolds and translocates protein substrates through this pore and into the ClpP14 chamber (Fig. 1A; Hinnerwisch et al., 2005; Kress et al., 2009).
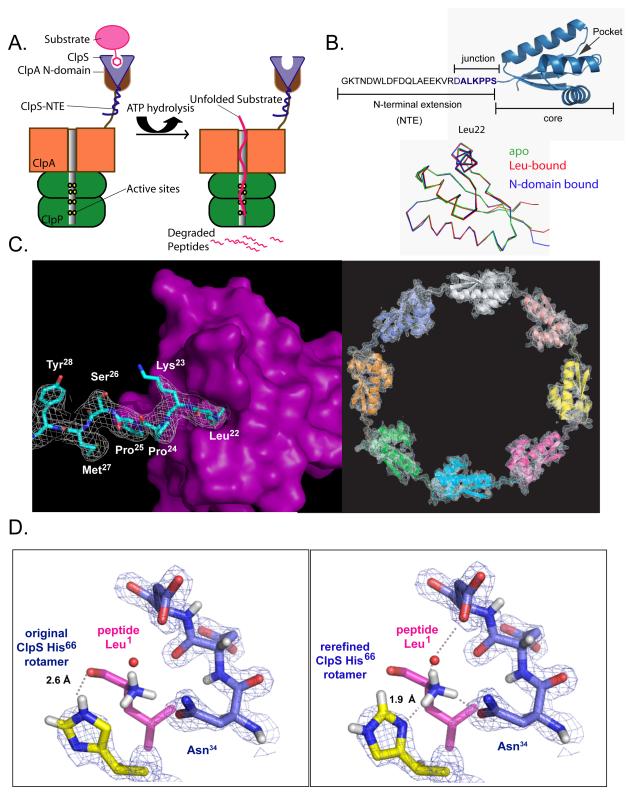
Figure 1. N-end-rule substrate recognition.
A) In bacteria, the ClpS adaptor (light blue) recognizes and binds N-end-rule substrates (pink) and delivers them for degradation by the ClpAP protease.
B) Top: ClpS has a flexible NTE required for N-end-rule substrate delivery and a folded ClpScore domain, which binds N-end-rule substrates. The ALKPPS sequence at the NTE-core junction is important for adaptor function. Bottom: Backbone Cα superposition (r.m.s.d. < 0.5 Å) of ClpScore (3O1F, green), a peptide-bound ClpS structure (2W9R, red), and ClpS from a complex with the ClpA N domain (1R6O, blue).
C) Left panel: In the rerefined 2WA9 structure, the side chain of Leu22 from an adjacent ClpS subunit was bound in the N-end-rule binding pocket, and density (1σ) for Leu22, Lys23, Pro24, and Pro25 was continuous with that for Ser26, Met27, Tyr28, and Lys29, which are part of ClpScore. Right panel: The rerefined map for the 2WA9 structure contained density (1.5 σ) for eight ClpS subunits in the asymmetric unit, arranged head to tail in a ring. The original 2WA9 structure (Schuenemann et al., 2009) had seven ClpS subunits, each with a bound N-end-rule peptide.
D) In the rerefined 2W9R structure (right panel), the correct rotamer of the His66 side chain make hydrogen bonds (dashed lines) with the α-NH3 group of the bound N-end peptide and fits nicely into the electron density. In the original 2W9R structure (Schuenemann et al., 2009; left panel), His66 rotamer chosen makes a poor hydrogen bond with the carbonyl oxygen of the first peptide residue and does not fit optimally into the electron density. In both panels, the electron density (1.25σ) is from the our rerefined map.
E. coli ClpS has a folded core domain (residues 26-106) and a poorly structured N-terminal extension (NTE; residues 1-25; Fig. 1B; Zeth et al., 2002; Guo et al., 2002). Importantly, the NTE is required for delivery of N-end-rule substrates, although it is not needed to bind substrates or ClpA, and shows little evolutionary sequence or length conservation (Hou et al., 2008) (Fig. S1). Crystal structures are known for E. coli ClpS bound to the N domain of E. coli ClpA, and for E. coli or Caulobacter crescentus ClpS bound to peptides beginning with Tyr, Phe, Trp, and Leu (Zeth et al., 2002; Guo et al., 2002; Xia et al., 2004; Wang et al., 2008a; Román-Hernández et al., 2009; Schuenemann et al., 2009). In each N-end-rule complex, the side chain of the N-end residue is completely buried in a deep hydrophobic pocket and the α-amino group and first peptide bond make additional contacts with ClpS. Differences in E. coli and C. crescentus ClpS binding to N-end-rule peptides have been proposed (Dougan et al., 2010), but we present evidence here for equivalent recognition by these highly homologous adaptors.
ClpS delivery of substrates to ClpAP must overcome several obstacles. For example, ClpS docks with the highly mobile N domain of ClpA, which could easily leave the substrate more than 80 Å from the axial pore of the D1 AAA+ ring, where unfolding/translocation initiates (Cranz-Mileva et al., 2008; Effantin et al. 2010). A similar issue occurs for the proteasome, where many substrates dock with receptors at sites far from the enzyme’s processing center (Striebel et al., 2009). Moreover, some experiments suggest that ClpS and ClpA both recognize the N-terminus of N-end rule substrates (Wang et al., 2007). Because the N-terminal side chain is buried in ClpS, substrate handoff to the ClpA pore would need to be actively promoted. However, little is known about the factors that control interactions between N-end-rule substrates, ClpS, and ClpA during substrate delivery.
Here, we dissect molecular interactions responsible for assembly of functional delivery complexes. We present evidence for complexes of ClpA6, ClpS, and substrate that differ markedly in stability and delivery activity. The most stable complex requires interactions mediated by the ClpS NTE, a ClpS residue that contacts the substrate N terminus, the substrate N-end residue and peptide bond, the AAA+ rings of ClpA, and a sufficiently long linker between the N and D1 domains of ClpA. Efficient substrate delivery also requires NTE residues, which appear to be engaged by the ClpA6 translocation machinery. Our results support a model in which formation of a high-affinity delivery complex reduces the mobility of the adaptor-bound substrate complex and positions the substrate’s N terminus close to the pore of the D1 AAA+ ring. This staged delivery mechanism illustrates an attractive general model to explain how substrates/adaptors that initially dock far from a AAA+ protease’s active center may be localized to the site where they are eventually processed.
Results
ClpS structures with N-end-rule peptides are highly conserved and unstrained
E. coli ClpS has been extensively used for studies of function (Dougan et al., 2002; Erbse et al., 2006; Wang et al., 2007a; 2008b; Hou et al., 2008; Román-Hernández et al., 2009; Schuenemann et al., 2009). However, a structure of the free protein had not been solved, leaving open the possibility that a conformational change occurs upon substrate and ClpA N-domain binding. Moreover, conflicting structures suggested that the details of N-end-rule recognition might differ in potentially important ways between the E. coli and C. crescentus adaptors (Dougan et al., 2010). We crystallized E. coli ClpScore (residues 26-106), and solved the structure at 1.4-Å resolution (Table 1). The backbone structure was similar to previously reported E. coli ClpS structures bound to the ClpA N domain or to N-end-rule peptides (Fig. 1B; Zeth et al., 2002; Guo et al., 2002; Xia et al., 2004; Schuenemann et al., 2009). Thus, major changes in the conformation of the ClpS core domain do not accompany N-domain or N-end-rule substrate binding.
Table 1.
Refinement statistics
| pdb code | 3O1F | 2W9R | 3O2H | 2WA8 | 3O2B | 2WA9 | 3O2O |
|---|---|---|---|---|---|---|---|
| E. coli ClpS residues | 26-106 | 2-108 ((b)) | 2-106 | 2-106 | 2-108 ((b)) | 2-106 ((c)) | |
| N-end peptide | none | LVKSKATNLLY | FRSKGEELFT | unclear ((d)) | none | ||
| space group | P1 | P1 | P1211 | C2 | |||
| unit cell a = | 28.0 Å | 28.1 Å | 32.2 Å | 171.9 Å | |||
| unit cell b = | 28.1 Å | 28.2 Å | 58.4 Å | 155.9 Å | |||
| unit cell c = | 51.6 Å | 38.9 Å | 56.4 Å | 71.2 Å | |||
| unit cell α = | 80.5° | 97.4° | 90.0° | 90.0° | |||
| unit cell β = | 77.9° | 106.5° | 101.9° | 114.6° | |||
| unit cell γ = | 72.3° | 92.4° | 90.0° | 90.0° | |||
| subunits/peptides per asu | 2/0 | 1/1 | 2/2 | 7/7 | 8/0 | ||
| refinement resolution | 22.5 - 1.4 Å | 25.0 - 1.7 Å | 30.2 - 2.15 Å | 30.2 - 2.05 Å | 25.0 - 2.9 Å | ||
| wavelength | 1.54 Å | 0.978 Å | 1.071 Å | 1.071 Å | |||
| Rsym (%) | 3.6 | 3.9 (20.9) | 11.3 (45.1) | 13.7 (73.6) | |||
| unique reflections | 23635 | 11540 | 9909 | 12292 | 19203 | ||
| completeness (%) | 82.4 (32) | 91.1 (86.1) | 94.0 (90.5) | 94.9 (96.0) | 99.3 (96.3) | ||
| data redundancy | 3.7 (3.1) | 2.2 (2.0) | 2.9 (2.7) | 7.5 (7.3) | |||
| average I/σI | 25.1 (5.2) | 12.4 (3.9) | 6.5 (2.3) | 11 (2.0) | |||
| Rwork (%) | 17.3 (21.7) | 22.6 (27.8) | 19.7 (25.6) | 22.5 (26.0) | 21.1 (27.1) | 23.7 (26.1) | 17.3 (29.4) |
| Rfree (%) | 19.7 (21.0) | 25.4 (32.8) | 22.7 (33.2) | 26.8 (33.5) | 24.8 (31.8) | 24.8 (28.5) | 20.1 (32.1) |
| TLS | no | yes ((e)) | no | no | no | yes | no |
| r.m.s.d. bond lengths (Å) | 0.007 | 0.009 | 0.009 | 0.010 | 0.006 | 0.015 | 0.005 |
| r.m.s.d. bond angles (°) | 1.19 | 1.19 | 1.17 | 1.54 | 1.02 | 1.57 | 0.79 |
| total protein atoms including H | 2637 | 861 (no H) | 1712 | 1660 (no H) | 3199 | 4880 (no H) | 10767 |
| solvent atoms | 402 | 68 | 128 | 91 | 140 | 0 | 0 |
| average B value | 17.9 | 22.1 | 24.1 | 33.2 | 33.6 | 50.0 | 73.7 |
| Ramachandran ((a)) favored/allowed/disallowed (%) |
100/0/0 | 100/0/0 | 100/0/0 | 93.5/3.0/3.5 | 100/0/0 | 94.4/2.0/3.6 | 99.5/0.5/0 |
| favorable rotamers (%) ((a)) | 100 | 93.6 | 100 | 89.9 | 100 | 89.7 | 100 |
| clash score ((a)) | 0.0 | 5.8 | 0.0 | 18.7 | 0 | 17.1 | 0 |
| Cβ deviations > 0.25 Å ((a)) | 0 | 0 | 0 | 3 | 0 | 4 | 0 |
| residues with bad angles ((a)) | 0 | 0 | 0 | 1 | 0 | 0 | |
| cis peptide bonds | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
Numbers in parenthesis represent values for the highest resolution bin. Unit cell and data collection statistics for the rerefined structures are from Schuenemann et al. (2009).
Favorable/allowed/disallowed Ramachandran angles, favorable rotamers, Cβ deviations, residues with bad angles, and the clash score (number of steric overlaps ≥ 0.4 Å per 1000 atoms) were calculated using MolProbity (Davis et al., 2007).
The original 2W9R and 2WA9 structures (Schuenemann et al., 2009) contain two additional C-terminal residues that are not present in the protein sequence. In our re-refined structures, there was no density for these “extra” residues and the chain terminated with Ala106 as expected.
As discussed in the text, it is possible that the polypeptide was cleaved between Ala21 and Leu22.
The peptide sequence is listed as LLT in the pdb file but as WRSKGEELFTGV in Schuenemann et al. (2009). In our rerefined structure, there was no peptide. Instead, an N-terminal segment of a neighboring ClpS molecule occupied the binding pocket.
ANISOU records are included in the 2W9R pdb file, although the header reports no TLS groups.
Rsym = ShSj ∣Ij(h) - <I(h)>∣ / ShSj <I(h)>, where Ij(h) is the jth reflection of index h and <I(h)> is the average intensity of all observations of I(h).
Rwork = Sh ∣Fobs(h) – Fcalc(h)∣ ∣ / Sh ∣Fobs(h)∣, calculated over the 93-95% of the data in the working set. Rfree equivalent to Rwork except calculated over 5-7% of the data assigned to the test set.
Validation of our ClpScore structure by MolProbity (Davis et al., 2007) revealed excellent geometry (Table 1). By contrast, analysis of the 2W9R, 2WA8, and 2WA9 complexes of E. coli ClpS with N-end-rule peptides (Schuenemann et al., 2009) revealed bad rotamers, poor bond angles, Ramachandran outliers, Cβ deviations, and unexpected cis peptide bonds (Table 1), which could arise if N-end peptide binding introduced strain or if these structures were incorrect. To resolve these issues and gain deeper insight into substrate recognition by ClpS, we re-refined these complexes, producing structures with no geometric anomalies and substantially improved refinement statistics (Table 1). Thus, binding of N-end-rule substrates does not introduce strain within ClpS.
Re-refinement allowed us to identify and correct additional errors. For example, the 2WA9 structure purportedly contained a peptide with an N-terminal Trp side chain, which was interpreted to be poorly ordered although it should have been snugly bound in the ClpS hydrophobic pocket (Schuenemann et al., 2009). By contrast, in our re-refined structure, a well-ordered Leu22 side chain from the NTE of a neighboring molecule occupied this pocket, with unambiguous electron density connecting the intervening residues to the adjacent ClpScore domain (Fig. 1C). This same head-to-tail interaction was observed in eight ClpS molecules, which formed a closed ring in the asymmetric unit (Fig. 1C). The original 2WA9 structure contained seven subunits in the asymmetric unit, and the NTE density was incorrectly interpreted as an N-end Trp peptide included during crystallization.
In the apo structure, the His66 side chain occupied part of the N-end binding pocket. In the re-refined complexes, the His66 ND1 nitrogen hydrogen-bonded to the α-amino group of the N-end residue, which required a 180° side-chain flip from the original structures, but the new position fit the electron-density well and made better chemical sense (Fig. 1D, Fig. S2). In the rerefined 2WR9 structure, for example, the unprotonated ND1 nitrogen of His66 accepts a hydrogen bond (1.9 Å; 170°) from a peptide −NH3 proton, whereas the proton on the His66 NE2 nitrogen donates a hydrogen bond (2.2 Å; 166°) to a side-chain oxygen from Glu94 in a neighboring molecule (Fig. S2). By contrast, when we added hydrogens to the original 2WR9 structure using REDUCE (Word et al., 1999), the nonpolar HD2 hydrogen of the His66 ring clashed with a peptide NH3 proton, a hydrogen bond between the peptide carbonyl oxygen and the proton on the NE2 nitrogen had poor geometry (2.6 Å; 117°), and the close interaction with the Glu94 carboxylate involved a non-polar hydrogen on the His66 ring. Importantly, in the rerefined complexes, contacts between E. coli ClpS and the N-terminal substrate residue were essentially indistinguishable from those observed in complexes of N-end-rule peptides with C. crescentus ClpS, including hydrogen bonds with the side chains of Asn34 (Fig. 1D) and a water molecule that bridges the α-amino group and Asp35 side chain (Wang et al., 2008a; Román-Hernández et al., 2009). We conclude that the mechanism of recognition of N-end-rule peptides by ClpS is highly conserved.
Enhanced N-end-rule affinity requires the ClpS NTE and ClpA6
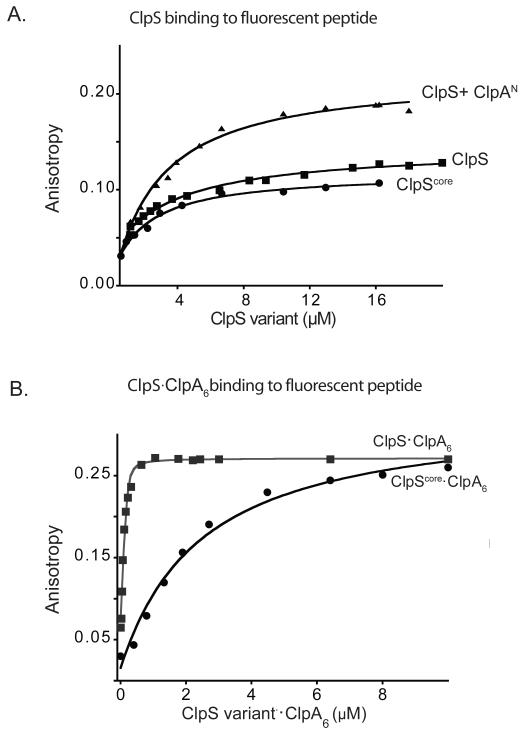
The NTE is required for substrate delivery (Hou et al., 2008), but its functional role is obscure. We established that the NTE does not affect binary binding to N-end-rule substrates, as intact ClpS and ClpScore bound to a fluorescent N-end-rule peptide (LLYVQRDSKEC-fl) with comparable affinities (~3 μM) in assays monitored by fluorescence anisotropy (Fig. 2A). Strikingly, ClpS binding to this peptide was much tighter (~40 nM) in the presence of ClpA and ATPγS, which stabilizes ClpA hexamers (Fig. 2B) The final anisotropy value was also higher with ClpA present, as expected for slower tumbling of the larger ClpA-ClpS-peptide complex compared to a ClpS-peptide complex. Importantly, tighter binding was not observed when ClpScore was used in place of intact ClpS, when the N domain of ClpA or ΔNClpA were used in place of ClpA, or when ATPγS was omitted (Fig. 2A, 2B; not shown). Thus, N-end-rule binding by ClpS is substantially strengthened in an NTE-dependent manner by interactions with the N domain and hexameric ring of ClpA6.
Figure 2. N-end-rule degrons bind more tightly to the ClpS-ClpA6 complex.
A) A fluorescent N-end-rule peptide (LLYVQRDSKEC-fl; 200 nM) was bound with similar affinities (KD ~3 μM) by ClpS, by ClpScore, and by ClpS in complex with the ClpA N domain, as assayed by changes in anisotropy. The molecular weights and maximum anisotropies of each complex differ.
B) Increasing concentrations of 1:1 molar mixtures of ClpA6 and ClpS or ClpScore were titrated against the LLYVQRDSKEC-fl peptide (100 nM). The ClpS-ClpA6 complex bound more tightly (Kapp = 42 ± 6 nM) than the ClpScore-ClpA6 complex (Kapp = 1.5 ± 0.25 μM), demonstrating that the ClpS NTE is required for affinity enhancement. Assays contained 4 mM ATPγS to promote ClpA hexamer formation.
ClpS binds ClpA6 more tightly in the presence of N-end rule peptides
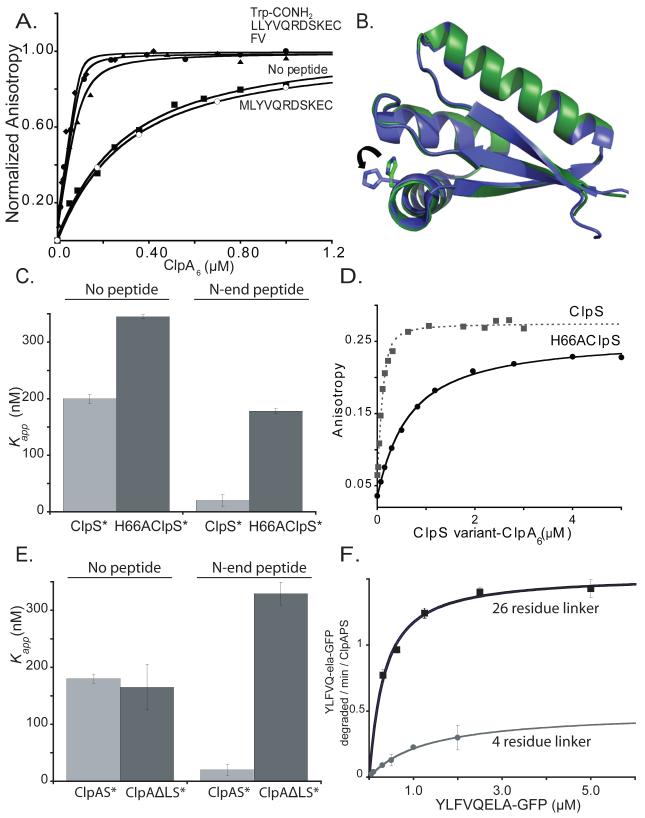
We constructed ClpS* (C73V; C101S; E96C), which contains one surface-exposed cysteine and was fully active in multiple functional assays (not shown), labeled it with fluorescein (ClpS*F), and measured fluorescence anisotropy in the presence of increasing concentrations of ClpA6 (stabilized by ATPγS). The fitted Kapp value was ~180 nM (Fig. 3A). Next, we assayed binding in the presence of excess LLYVQRDSKEC, a Phe-Val dipeptide, or Trp-conh2, all of which have good N-end-rule side chains. In each case, ClpA6 bound ClpS*F at least 9-fold more tightly than observed with a MLYVQRDSKEC peptide or in the absence of ligand (Fig. 3A). Although binding was too tight to calculate accurate Kapp values, Trp-conh2 enhanced ClpA6 affinity for ClpS*F as well as the longer N-end-rule peptides, indicating that the N-terminal residue and a few nearby atoms play the dominant role in enhanced binding.
Figure 3. ClpS binds ClpA6 more tightly in the presence of N-end-rule peptides.
A) As assayed by anisotropy, ClpA6 bound 200 nM fluorescent ClpS*F tightly in the presence 20 μM Trp-conh2, LLYVQRDSKEC, or FV N-end-rule peptides (Kapp < 20 nM) and more weakly in the absence of peptide or with 20 μM MLYVQRDSKEC peptide (Kapp ~ 180 nM).
B) The H66 residue of ClpS is one of the side chains involved in the formation of one of the three hydrogen bonds that the adaptor forms with the α-amino group of the N-end degron. Overlay of the apo (green, PDB 3O1F) and the peptide-bound (blue, 2W9R) crystal structures of ClpS reveal no major global changes occur upon peptide binding. The most substantial change is the rotation of the H66 side chain, which appear to need to move away from the pocket in the apo form to accommodate the N-degron in the peptide binding site.
C) ClpA6 bound ClpS*F (KD = 200 ± 6 nM) and H66AClpS*F (KD = 345 ± 3 nM) with similar affinities. Addition of 20 μM LLYVQRDSKEC N-end-rule peptide enhanced ClpA6 affinity for ClpS*F substantially (Kapp = 20 ± 10 nM) but increased affinity for H66AClpS*F only modestly (Kapp = 178 ± 4 nM).
D) H66AClpS-ClpA6 complex binds more weakly to an N-end-rule fluorescent peptide (LLYVQRDSKEC-fl) when compared to ClpS-ClpA6 (Kapp = 560 nM vs Kapp = 42 ± 6 nM for wild type).
E) An N-end-rule peptide (LLYVQRDSKEC; 20 μM) enhanced binding of ClpS*F to ClpA6 but not to ΔLClpA6, which has shorter linker between the N and D1 domains (Cranz-Mileva et al., 2008).
F) Michaelis-Menten plots showed that substituting ΔLClpA6 (4-residue linker) for ClpA6 (26-residue linker) decreased KM and Vmax for ClpAPS degradation (100 nM ClpA6 or ΔLClpA6; 200 nm ClpP14; 600 nm ClpS) of the N-end-rule substrate ylfvqela-GFP.
If there are no ligand-induced conformational changes in ClpS, as the structures argue, then how does Trp-conh2 enhance ClpA6 binding to ClpS? Because the side chain of Trp-conh2 would be buried in ClpS, binding stimulation might involve contacts between ClpA6 and exposed main-chain atoms of Trp-conh2, or contacts between ClpA6 and ClpS side chains whose conformations are stabilized by contacts with Trp-conh2. Therefore, we tested if His66 in ClpS might participate in binding enhancement, as this residue contacts the substrate N terminus (Fig. 1D), the H66A mutation increased KM and reduced Vmax for ClpAP degradation of N-end-rule substrates (Wang et al., 2008a), and His66 adopted different conformations in the apo and peptide-bound structures of E. coli ClpS (Fig. 3B). Importantly, we found that peptide-free H66AClpS*F bound ClpA6 nearly as well as ClpS*F, but peptide-bound H66AClpS*F bound ClpA6 ~9-fold more weakly than the parent (Fig. 3C). Thus, this mutant does not form the high-affinity complex. Although the H66A mutation reduces N-end substrate affinity (Fig. 3D), the experiment in Fig. 3C was performed using a peptide concentration 35-fold higher than Kapp for H66AClpS•ClpA6 binding, ensuring that most mutant ClpS molecules were peptide bound. We conclude that the His66 side chain stabilizes a high-affinity complex of ClpA6, ClpS, and substrate.
De Donatis et al. (2010) reported that NTE deletion reduced binary ClpA6 affinity. We found that NTE deletion in a ClpS*F variant reduced binary ClpA6 affinity ~10-fold and also reduced the anisotropy observed at binding saturation (Fig. S3). These results show that the NTE helps stabilize the ClpS•ClpA6 complex, both in the presence and absence of substrate, and suggest that the NTE-mediated interaction reduces the segmental mobility of ClpA6-bound ClpS.
ClpS-ClpA6 collaboration requires mobility of the ClpA N domain
ClpScore docks with the ClpA N domain, which is linked to the D1 ring by a 26-residue tether. Using a ClpA variant with a 4-residue tether (ΔLClpA; Cranz-Mileva et al., 2008) allowed us to probe the importance of linker length. In the absence of substrate, ClpS*F bound ClpA6 and ΔLClpA6 equally well (Fig. 3E), suggesting that the shorter linker does not prevent formation of the NTE-mediated contacts with ClpA6. Importantly, however, when binding was measured in the presence of N-end-rule peptide, ΔLClpA6 showed much weaker binding (Fig. 3E). Thus, an N-D1 linker of sufficient length is important in forming stable ternary complexes with ClpS and N-end-rule substrates. Analysis of the steady-state kinetics of ClpAPS degradation of ylfvqela-GFP revealed a 7-fold weaker KM and 3-fold lower Vmax when ΔLClpA6 was substituted for wild-type ClpA6 (Fig. 3F). Thus, the longer linker is also required for efficient substrate delivery to ClpAP. Using a different substrate, Cranz-Mileva et al. (2008) also found defects in degradation using ΔLClpA6, albeit significantly smaller than those shown in Fig. 3F.
Defining NTE lengths required for efficient ClpS delivery and formation of high-affinity complexes
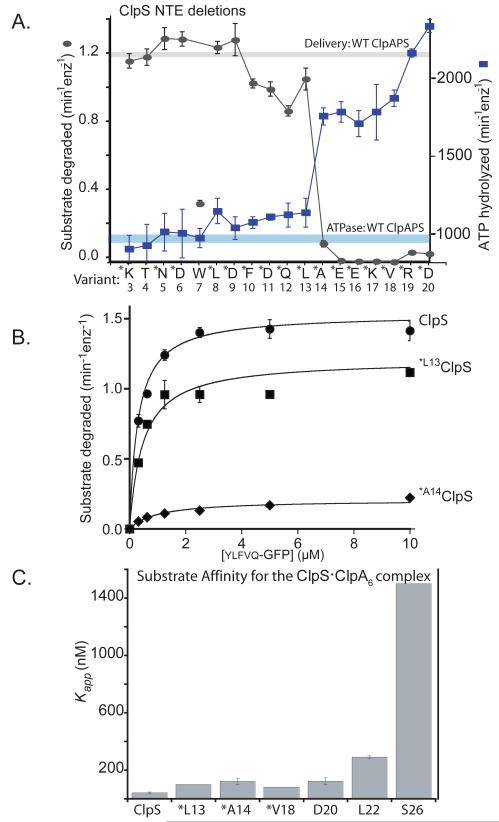
To probe the importance of NTE length, we purified ClpS mutants with N-terminal truncations from 3 to 20 amino acids and assayed their ability to slow ATP hydrolysis by ClpA and to deliver ylfvqela-GFP for ClpAP degradation (Fig. 4A). Strikingly, a precipitous decline in delivery and loss of ability to suppress ATPase rates occurred over a very narrow truncation range defined by the Met-Leu13 and Met-Ala14 variants. To investigate this delivery defect in greater detail, we determined the steady-state kinetics of ClpAP degradation of different concentrations of ylfvqela-GFP in the presence of ClpS or the variants starting with Met-Leu13 and Met-Ala14 (Fig. 4B). The Met-Leu13 mutant delivered the substrate with a 1.3-fold reduction in Vmax compared to ClpS. By contrast, delivery by the Met-Ala14 variant displayed a 7.5-fold decrease in Vmax. These results demonstrate that the ClpS NTE must have a critical minimum length to promote efficient delivery to ClpAP.
Figure 4. A minimal NTE length is required for ClpS function.
A) ClpS variants (1 μM) with N-terminal truncations were assayed for delivery of ylfvqela-GFP (1 μM) for ClpAP degradation (gray curve) and for effects on ClpAP ATP hydrolysis (blue curve) using 100 nM ClpA6 and 270 nM ClpP14 for both assays. Data points represent averages (n=3) ± 1 SD. Each ClpS variant is named by the first wild-type residue in the construct. Those marked with an asterisk contain an additional N-terminal methionine and are therefore one residue longer than the labels indicate; these mutants were expressed as SUMO-fusion proteins and cleaved in vitro (see Experimental Procedures) or were expressed as standard non-fusion proteins but retained the initiator Met (verified by mass spectrometry). The T4 and W7 variants were also expressed as standard non-fusion proteins but mass spectrometry and/or N-terminal sequencing showed that the initiator Met was removed from both of these proteins. Note the sharp activity transitions between *L13 ClpS (starting Met12Leu13) and *A14 ClpS (starting Met13Ala14). The processing of the W7 variant is inconsistent with canonical methionine aminopeptidase activity and generates a good N-end-rule residue, which may be responsible for the poor activity of this ClpS variant in delivering other N-end-rule substrates.
B) Michaelis-Menten plots of ylfvqela-GFP degradation by ClpAP and ClpS or variants (100 nM ClpA6; 200 nm ClpP14; 600 nm ClpS or variants). Wild-type ClpS and *L13 ClpS (beginning Met12Leu13Ala14) supported roughly similar steady-state degradation kinetics, but delivery by *A14 ClpS (beginning Met13Ala14Glu15) resulted in a substantial decrease in Vmax. Thus, the NTE must have a critical minimal length to support efficient substrate delivery. The solid lines are a global fit to a model in which the ClpS-substrate complex binds ClpA in an initial bimolecular step (K1 = 1.1 μM) and then is engaged for degradation in a second unimolecular step (K2), which depends on NTE length. In this model, apparent Vmax = Etotal•kdeg/(1+K2) and apparent KM = K1•K2/(1+K2). For the fits shown, the kdeg value was 2.1 min−1 and the K2 values were 0.37 (wild-type ClpS), 0.74 (*L13 ClpS), and 9.2 (*A14 ClpS).
C) Binding to an N-end-rule peptide (LLYVQRDSKEC-fl; 150 nM) by complexes of ClpA6 with ClpS variants (1 ClpS per ClpA6) showed that ClpS junction residues are important for formation of the high-affinity delivery complex. Variants marked * have an additional N-terminal methionine. Apparent affinity constants were 43 nM (wild-type ClpS), 100 nM (*L13), 130 nM (*A14), 83 nM (*V18), 130 nM (D20), 290 nM (L22), and 1500 nM (S26).
We also determined Kapp values for binding of several ClpS-NTE variants to a fluorescent N-end-rule peptide in the presence of ClpA6/ATPγS (Fig. 4C). Mutants beginning at Asp20 or earlier formed relatively stable substrate complexes with ClpA6 (Kapp 80-120 nM), albeit less stable than the wild-type ClpS complex. Thus, an NTE extending past residue 14 is not required for relatively stable ternary-complex formation but is needed for efficient substrate delivery. Affinity was weakened further when the truncated ClpS variant began with Leu22 (~300 nM) and substantially more when it started at Ser26 (ClpScore; ~1500 nM; Fig 2C; Fig. 4C). These results establish that 4-5 residues at the junction between the NTE and ClpScore are important for stabilizing high-affinity complexes. This junction sequence was substantially conserved among orthologs, whereas the rest of the NTE showed almost no sequence conservation (Fig. S1, Zeth et al., 2002; Guo et al., 2002), supporting a model in which the NTE-junction residues form specific docking contacts with ClpA.
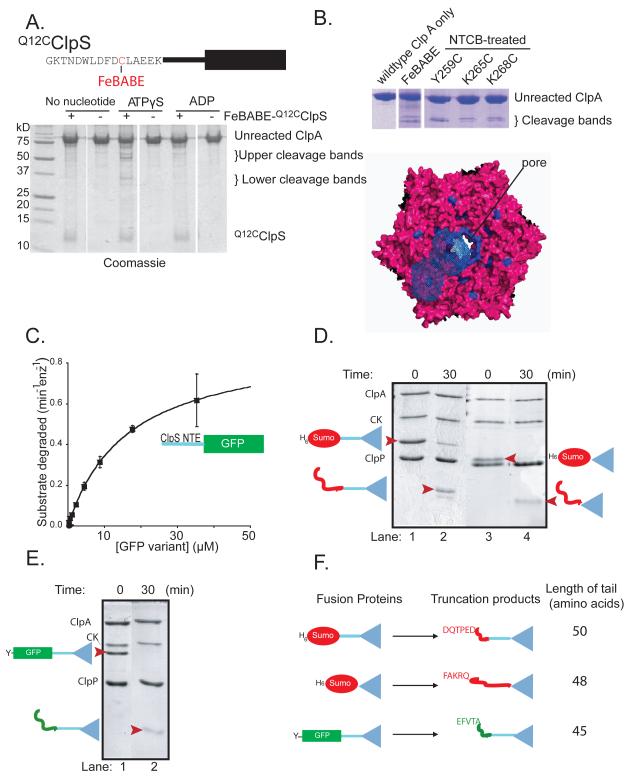
FeBABE mapping places the ClpS NTE near the ClpA pore
To probe regions of contact between the NTE and ClpA, we attached FeBABE to residue 12 of Q12CClpS (Fig. 5A). In the presence of ascorbic acid and hydrogen peroxide, the Fe3+ atom in the NTE-bound FeBABE generate free radicals which can cleave regions of ClpA in close proximity. ClpS was mixed with ClpA containing a C-terminal FLAG tag in the presence of ATPγS, ADP, or no nucleotide. After 30 min, cleavage was initiated, allowed to proceed for 30 s, quenched, and the products where analyzed by SDS-PAGE. In presence of ATPγS, FeBABE cleavage resulted in two fragments of ~50 kDa and two of ~29 kDa either in the absence (Fig. 5A) or presence (not shown) of N-end-rule peptides. No specific cleavage products were observed without nucleotide or with ADP, suggesting that cleavage requires ATPγS-dependent formation of ClpA6•ClpS complexes (Fig. 5A).
Figure 5. The ClpS NTE contacts ClpA near the axial pore.
A) Top: FeBABE was attached to residue 12 of Q12CClpS variant for cleavage studies. Bottom: As assayed by SDS-PAGE, cleavage of ClpA required FeBABE-modified Q12CClpS and ATPγS.
B) ClpA residues 259-268 are highlighted in blue in a top view of a model of the hexameric D1 ring (Guo et al., 2002). In one ClpA subunit, blue-wire shading shows regions within 12 Å of residues 259-268, which represents the approximate reach of the tethered FeBABE.
C) A substrate consisting of residues 2-26 of ClpS fused to GFP was efficiently degraded by ClpAP, as shown by Michaelis-Menten analysis (KM = 16.4 μM; Vmax = 0.62 min−1 enz−1).
D) Assays monitored by SDS-PAGE showed that ClpAP only partially degraded the H6-Sumo-ClpS and H6-Sumo-ClpScore fusion proteins, resulting in truncated products of a lower molecular weight (marked by red arrowheads in lanes 2 & 4).
E) ClpAP partially degraded the ylfvqela-GFP-ClpS fusion protein, resulting in a lower molecular weight truncation product (marked by a red arrowhead in lane 2).
F) Depiction of the ClpS fusion proteins used to test degradation by ClpAP (left) and the corresponding truncation products produced by degradation (right). N-terminal sequencing of the truncation products revealed that the new N-termini corresponded to an internal sequence in the protein fused to ClpS (either Sumo or GFP). The truncation products consisted of the ClpS core and an additional N-terminal tail of 45-50 amino acids.
Western blots using anti-FLAG antibody indicated that the larger FeBABE cleavage products corresponded to the C-terminal portion of ClpA and the smaller bands were N-terminal segments (not shown). Edman sequencing of these products was unsuccessful. However, based on molecular-weight standards and fragments produced by cleavage before cysteines in the Y259C, K265C, and K268C variants of ClpA, FeBABE cleavage appeared to occur near ClpA residue 260 (Fig. S4). Residues 259-268 are located near the axial pore of the D1 AAA+ ring of ClpA. The FeBABE-ClpS linkage would allow the reagent to reach regions of ClpA within ~12 Å of the site of NTE attachment. Mapping these potential contacts on a model of the ClpA hexamer suggested that the ClpS NTE could physically contact the central pore of ClpA6 and/or the top of the D1 ring (Fig. 5B).
The NTE is a degradation signal but ClpS resists ClpAP proteolysis
ClpAP does not degrade ClpS (Dougan et al., 2002). Nevertheless, because the NTE makes contacts near the ClpA pore, we hypothesized that it might be engaged by the translocation/unfolding machinery. To test this model, we appended the mature NTE of E. coli ClpS (residues 2-26) to the N-terminus of GFP and assayed degradation. Untagged GFP is not degraded by ClpAP (Weber-Ban et al., 1999), but the NTE-fusion protein was efficiently degraded (Fig. 5C), with a KM of 16 μM and Vmax similar to other GFP substrates. These results support a model in which the ClpA pore can engage the ClpS NTE, but the ClpS core domain resists proteolysis. To confirm that ClpScore is refractory to degradation, we purified H6-SUMO-NTE-ClpScore, H6-SUMO-ClpScore, and ylfvqela-GFP-NTE-ClpScore fusion proteins, and assayed ClpAP proteolysis. In each case, partial proteolysis was observed (Fig. 5D, 5E), but Edman degradation of the resistant fragment demonstrated that ClpScore remained intact as did an N-terminal tail of 45-50 amino acids before the core domain (Fig. 5F). Tails of this length result when AAA+ proteases are unable to unfold a very stable domain in the midst of a multi-domain substrate (Lee et al., 2001; Koodathingal et al., 2009), strongly supporting a model in which ClpAP cannot unfold or degrade the ClpScore domain. As discussed below, this degradation-resistant character of ClpS is likely to be critical for its function as an efficient adaptor.
Discussion
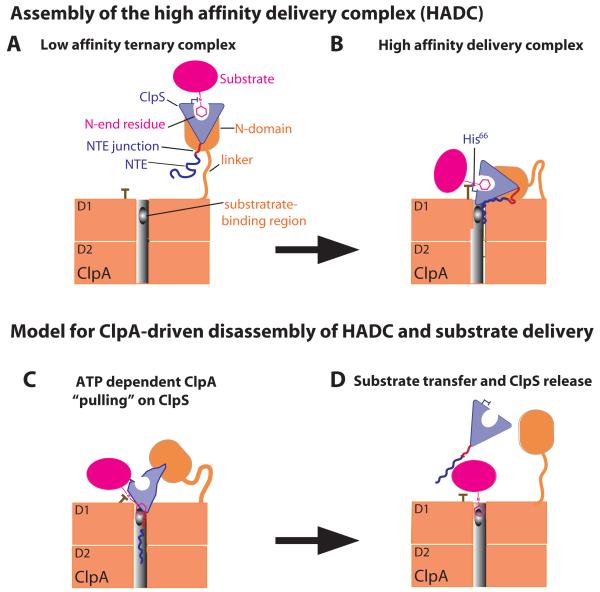
The work presented here elucidates important new aspects of the molecular mechanism of ClpS delivery and ClpAP degradation of N-end-rule substrates. Our current view of these processes is shown in the model of Fig. 6, which begins with formation of a low-affinity ternary complex (LATC; Fig. 6A), proceeds to a high-affinity delivery complex (HADC; Fig. 6B), and ends with active substrate handoff from ClpS to ClpA (Figs. 6C and 6D). As illustrated in Fig. 6A, uncoupled sets of binary contacts between ClpS and the N-end-rule substrate and between ClpScore and the ClpA N domain stabilize the LATC. Formation of the HADC involves additional interactions mediated by junction residues of the NTE of ClpS, by His66 of ClpS, by the N-end residue of the substrate, and by the D1 ring of ClpA (Fig. 6B). The ClpS•substrate portion of the complex is highly mobile in the LATC, because of the flexible tethering of the N domain to the D1 ring of ClpA, but is constrained in the HADC by additional contacts with the D1 ring.
Figure 6. Model for staged delivery of N-end-rule substrates.
A) Independent binding of ClpS to the ClpA N domain and of the substrate N-end residue in the ClpS pocket results in a low-affinity ternary complex.
B) A high-affinity delivery complex is stabilized by additional interactions between the D1 ring of ClpA and NTE-junction residues and between the D1 ring, the His66 side chain of ClpS, and the N-end residue of the substrate.
C) Translocation-mediated ClpA tugging on the NTE distorts the ClpScore structure, weakens ClpS interactions with the N-end residue, and facilitates transfer of the N-degron of the substrate to a site in the ClpA pore.
D) ClpS slips from the grasp of ClpA, clearing the pore and allowing subsequent degradation of the N-end-rule substrate.
The properties of the LATC are based on previous studies of the interaction of ClpS with substrates or the N domain of ClpA. Our current work supports the existence of the HADC and defines many of its properties. For example, we find that degrons containing just the N-end residue and peptide bond enhance ClpS affinity for ClpA6 and are bound far more tightly by ClpS and ClpA6 together than by either individual protein. Moreover, high-affinity binding requires multiple regions of ClpS (including the junction region of the NTE and His66, which contacts the N-end residue of the substrate), as well as the AAA+ body of ClpA and a suitably long linker between the ClpA D1 ring and N domain. Consistent with the Fig. 6A and 6B models, we find that the mobility of the ClpS portion of complexes with ClpA is higher when the NTE is absent. Work presented here and previously (Hou et al., 2008; Wang et al., 2008a; Schuenemann et al., 2009) shows that mutation of His66 or deletion of the NTE severely compromises ClpS delivery of N-degron substrates to ClpAP, indicating that formation of the HADC is a critical step in substrate delivery.
It is not currently known what parts the AAA+ body of ClpA make contacts with the junction residues of the NTE or with His66 and the N-end residue in the HADC, but our studies set the stage for future experiments to define these interactions in greater molecular detail. FeBABE-cleavage experiments do show that residues near the center of the NTE can contact the D1 AAA+ ring of ClpA6, and given the size of ClpS, other contacts with the AAA+ body of ClpA6 would likely also be restricted to the D1 ring. For example, a residue in the D1 ring of ClpA could contact the His66 of ClpS and stabilize its interaction with the α-amino group of the N-degron, explaining the importance of all of these elements in stabilizing the HADC. Alternatively, the conformation of the His66 side chain could change in the HADC, allowing one set of D1 interactions with His66 and another set of interactions with the N-degron. Interestingly, a sufficiently long linker between the ClpA N domain and AAA+ ring is needed to allow formation of the ClpA contacts mediated by the N-end degron and ClpS binding pocket.
In addition to its role in delivery of N-degron substrates, ClpS binding prevents recognition and degradation of other types of substrates by ClpAP (Dougan et al., 2002). In the absence of N-end-rule substrates, it would be counterproductive if ClpS bound ClpA too tightly as this would preclude degradation of other substrates. However, we find that ClpS binds ClpA6 ~10-fold more tightly when N-degron substrates are present, providing an elegant solution to this problem. Substrate-dependent affinity enhancement would help to ensure the formation of a ClpAPS complex when N-end-rule substrates were available but also keep ClpAP largely free to perform other functions when these substrates were absent.
Our working model for substrate delivery culminates with engagement of the substrate N-degron by the ClpA pore (Fig. 6). A key feature of this model is binding of a portion of the ClpS NTE in the ClpA pore (Fig. 6B), allowing the translocation/unfolding machinery to pull on ClpS and facilitate transfer of the N-degron from ClpS to ClpA (Fig. 6C and 6D). Although aspects of the transfer model are speculative, it accounts for many experimental observations. For example, we found that a truncated ClpS variant beginning at NTE-residue 13 mediated efficient substrate degradation, whereas deleting one additional NTE residue dramatically reduced delivery. The presence of the extra residue could allow the NTE to reach a binding site in the ClpA6 pore that was critical for initiating substrate delivery. Indeed, our FeBABE-cleavage results suggest that this central region of the NTE could contact the pore of the ClpA D1 ring. Engagement of the NTE by the ClpA pore is also supported by our finding that appending the ClpS NTE to GFP, a protein which is not normally degraded, results in efficient ClpAP degradation. Despite NTE engagement, our results also show that the folded portion of ClpS resists ClpAP degradation. In combination, these results account for our observation that delivery-competent NTE truncations result in lower ClpA ATPase rates than delivery-incompetent truncations. For example, AAA+ unfoldases hydrolyze ATP more slowly during attempts to unfold a protein (Kenniston et al., 2003; Wolfgang et al., 2009), and the lower ATPase rates seen using delivery-competent NTE truncations are therefore consistent with failed ClpA attempts to unfold ClpS.
How could ClpA tugging on ClpS facilitate handoff of N-end rule substrates? Given that the NTE is distant from the ClpS substrate-binding pocket, an attractive model is that translocation-mediated pulling on the NTE deforms ClpScore, facilitating transfer of the N-end degron to a site in the ClpA pore (Fig. 6C). This model requires independent recognition of the N-degron by ClpA, which is supported by the observation that ClpAP alone can recognize and degrade N-end-rule substrates, albeit with relatively low KM’s compared to values obtained with ClpS (Wang et al. 2007). Moreover, in resisting unfolding, ClpS could slip from the grasp of ClpA, as observed for other difficult-to-unfold proteins (Kenniston et al., 2005), clearing the pore as a prelude to substrate degradation (Fig. 6D). Experiments with the related ClpXP enzyme also reveal that multiple polypeptide chains can simultaneously occupy the pore (Burton et al., 2001; Bolon et al., 2004).
There are parallels between the Fig. 6 model and the delivery of ssrA-tagged substrates to the ClpXP protease by the SspB adaptor. For example, one region of SspB binds the N domain of ClpX, another part of SspB binds to a segment of the ssrA degron, a different part of this degron binds to the ClpX pore, and each binary interaction is substantially weaker than the overall ternary interaction (Levchenko et al., 2000; 2003; Wah et al. 2003; Bolon et al., 2004; Martin et al., 2008). Because the ssrA tag of the substrate is positioned in the pore of the ClpX AAA+ ring in the high-affinity complex, ATP-fueled translocation allows tag contacts with the adaptor to be broken at the same time that degradation is initiated.
Assembly of increasingly stable macromolecular complexes frequently drives biological recognition. This mechanism provides directionality by proceeding downhill to a thermodynamic minimum but also results in an energy well from which spontaneous escape is difficult, creating a problem if the high-affinity complex is not the final product. For example, recombination catalyzed by MuA transposase is driven by increasingly stable protein-DNA complexes, which eventually must be disassembled in an ATP-dependent process by ClpX (Burton and Baker, 2005). As shown here and previously, adaptor-mediated delivery of substrates to AAA+ proteases also involves a progression from low-affinity to high-affinity complexes. This type of assembly has several advantages. From a kinetic perspective, splitting the overall pathway into discrete bimolecular and unimolecular steps speeds assembly. For example, ClpS with bound N-degron substrate could initially dock with any of the six N domains of ClpA6. Moreover, these N domains are highly mobile, further increasing the chances for productive collisions. Subsequent assembly steps would then be unimolecular, allowing the use of relatively weak interactions to position the substrate/adaptor near the translocation machinery of ClpA6.
We propose that adaptors for AAA+ proteases will fall into two general categories. In one category, exemplified by SspB, enzymatic pulling on the substrate disrupts the HADC and initiates degradation. In the second category, exemplified by ClpS, enzymatic tugging on the adaptor destabilizes the HADC, allowing substrate transfer and degradation. Many adaptors that function by a ClpS-type mechanism are likely to be degradation resistant. For example, Rad23 facilitates interactions between ubiquitinated substrates and the proteasome and is refractory to degradation (Heessen et al., 2005; Fishbain et al., 2010). However, a ClpS-type mechanism could also work if the adaptor were degraded. Indeed, the MecA adaptor is degraded by ClpCP during substrate delivery (Turgay et al., 1998).
Experimental Procedures
Proteins and Peptides
Mutants were generated by the QuikChange method (Stratagene) or PCR. ClpS, ClpS mutants, and substrates were initially fused to the C terminus of H6-Sumo in pET23b (Novagen). Following expression, fusion proteins were purified by Ni-NTA chromatography (Qiagen) and cleaved with Ulp1 protease. The cleaved H6-Sumo fragment was removed by passage through Ni-NTA, and the protein of interest was purified by gel filtration on Superdex-75 (GE Healthsciences) and/or ion-exchange chromatography on MonoQ. ClpS variants were concentrated and stored in 20 mM HEPES (pH 7.5), 150 mM KCl, 1 mM DTT, and 10% glycerol. ClpA and ClpP were purified as described (Hou et al., 2008). Trp-conh2 and Phe-Val (FV) were purchased (Sigma). All remaining peptides were synthesized by standard FMOC techniques using an Apex 396 solid-phase instrument.
Crystallography
Crystals of E. coli ClpS26-106 were obtained after 3 weeks at 20 °C in hanging drops containing 0.5 μL of protein solution (7.5 mg/ml) and 1 μL of reservoir solution (0.2 M ammonium formate, 20% PEG 3350). Crystals were frozen without additional cryoprotection and X-ray diffraction data were collected on a Rigaku Micromax 007-HF rotating anode equipped with Varimax-HR mirrors, an RAXIS-IV detector, and an Oxford cryo-system. Data were processed using HKL-2000 (Otwinowski and Minor, 1997). Initial phases were obtained by molecular replacement using PHASER (Storoni et al., 2004) with E. coli ClpS 2W9R as a search model. The final structure was obtained by iterative model building using COOT (Emsley and Cowtan, 2004), refinement using PHENIX (Adams et al., 2002), and had excellent geometry and refinement statistics (Table 1). Re-refinement of the 2W9R, 2WA8, and 2WA9 structures, using COOT and PHENIX, also produced structures with excellent geometry and substantially improved refinement statistics (Table 1).
Fluorescent labeling
Peptides were labeled with fluorescein maleimide as described (Wang et al., 2008). ClpS* variants (50 μM) containing a single cysteine were incubated with 50 mM DTT in 100 mM TrisCl (pH 8) for 1.5 hours at 4 °C, buffer exchanged into 100 mM Na2PO4 (pH 8) and 1 mM EDTA, and labeled by addition of 0.3 mg/mL of fluorescein maleimide (Thermo Scientific) for 2 h at room temperature in the dark. Excess reagent was removed by size-exclusion chromatography, and the modified protein was stored in 10 mM HEPES (pH 7.5), 200 mM KCl, and 1 mM DTT. Binding assays monitored by fluorescence anisotropy were performed using a Photon Technology International Fluorimeter. Data were fitted using a non-linear squares algorithm to a hyperbolic binding isotherm or to a quadratic equation for tight binding. Reported KD and Kapp values are averages (n ≥ 2) with errors calculated as SQRT((K-Kavg)2/n).
FeBABE cleavage
For FeBABE labeling, Q12CClpS was incubated in 30 mM MOPS (pH 8.1), 4 mM EDTA at 4 °C overnight, desalted into 30 mM MOPS (pH 8.1), 100 mM NaCl, 1 mM EDTA, 5% glycerol, and incubated with 5 μg/μL of Fe(III) (s)-1-(p-bromoacetamidobenzyl)-EDTA (Pierce) for 1 h at 37 °C. FeBABE-ClpS and ClpA were buffer exchanged into 50 mM MOPS (pH 8.1), 300 mM NaCl, 10 mM MgCl2, 10% glycerol, mixed together with no nucleotide or 1 mM ATPγS/ADP, and incubated at room temperature for 30 min. Cleavage was initiated by the addition of 40 mM ascorbate, 10 mM EDTA, followed immediately by 40 mM hydrogen peroxide, 10 mM EDTA. The reaction was quenched by adding SDS-PAGE sample buffer with 40% glycerol and analyzed by SDS-PAGE.
To generate size standards for FeBABE cleavage, Y259C, K265C, and K268C ClpA variants were constructed, purified, incubated with 50 mM DTT at 37 °C for ~10 min, and exchanged into 200 mM Tris acetate (pH 8), 1 mM EDTA, 5 M urea, 0.1% SDS. This sample was incubated with 2 mM 2-nitro-5-thiocyanobenzoate at 37 °C for 20 min to modify the cysteines, and then buffer exchanged into 200 mM Tris acetate (pH 9) and incubated at 45 °C for 2 h to allow protein cleavage.
Degradation and ATPase Assays
ClpAPS degradation assays were performed as described (Wang et al., 2008). Briefly, ClpA6 (100 nM), ClpP14 (200 nM), and ClpS or variants (600 nM) were preincubated in reaction buffer (50 mM HEPES (pH 7.5), 300 mM NaCl, 20 mM MgCl2, 0.5 mM DTT, and 10% glycerol) with substrate for 3 min at 30 °C before adding ATP regeneration mix (4 mM ATP, 50 mg/ml creatine kinase, 5 mM creatine phosphate) to initiate degradation. GFP degradation was assayed by loss of fluorescence and the data were fitted by a non-linear least-squares algorithm to a quadratic version of the Michaelis-Menten equation to obtain KM and Vmax. Reported values of kinetic parameters were averages (n=3) ± 1 SD. ATPase rates were monitored under similar conditions but used a coupled ATP-hydrolysis assay (Kim et al,. 2001).
Supplementary Material
Highlights.
Atomic details of ClpS, adaptor for N-end rule substrates, are clarified
Multiple contacts between ClpS and AAA+ enzyme ClpA contribute to substrate docking
The N terminal region of ClpS is engaged by the ClpAP pore for substrate delivery
ClpS is un-degradable by ClpAP, a feature that contributes to delivery mechanisms
Acknowledgements
We thank S. Bissonnette and P. Chien for helpful discussions and advice, A. de Regt for assistance, and E. Weber-Ban for providing the ΔLClpA construct. This work was supported by the Howard Hughes Medical Institute (HHMI) and the National Institutes of Health (grants AI-15706 and GM-049224). T.A.B. is an employee of HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers Coordinates for the apo E. coli ClpS crystal structure (3O1F) and the re-refined co-crystal structures (3O2B, 3O2H, and 3O2O) have been deposited with the Protein Data Bank.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BM, Baker TA. Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase. Protein Sci. 2005;14:1945–1954. doi: 10.1110/ps.051417505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranz-Mileva S, Imkamp F, Kolygo K, Maglica Z, Kress W, Weber-Ban E. The flexible attachment of the N-domains to the ClpA ring body allows their use on demand. J Mol Biol. 2008;378:412–424. doi: 10.1016/j.jmb.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Donatis GM, Singh SK, Viswanathan S, Maurizi MR. A single ClpS monomer is sufficient to direct the activity of the ClpA hexamer. J Biol Chem. 2010;285:8771–8781. doi: 10.1074/jbc.M109.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- Effantin G, Ishikawa T, De Donatis GM, Maurizi MR, Steven AC. Local and global mobility in the ClpA AAA+ chaperone detected by cryo-electron microscopy: functional connotations. Structure. 2010;18:553–562. doi: 10.1016/j.str.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- Fishbain S, Prakash S, Herrig A, Elasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Maurizi MR. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–29. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Hou JY, Sauer RT, Baker TA. Distinct structural elements of the adaptor ClpS are required for regulating degradation by ClpAP. Nat Struct Mol Biol. 2008;15:288–294. doi: 10.1038/nsmb.1392. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Wah DA, Sauer RT, Baker TA. Structure of a delivery protein for a AAA+ protease in complex with a peptide degradation tag. Mol Cell. 2003;12:365–372. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Koretke KK. Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J Struct Biol. 2003;141:77–83. doi: 10.1016/s1047-8477(02)00582-8. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of Substrate Denaturation and Translocation by the ClpXP Degradation Machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Kress W, Mutschler H, Weber-Ban E. Both ATPase domains of ClpA are critical for processing of stable protein structures. J Biol Chem. 2009;284:31441–31452. doi: 10.1074/jbc.M109.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr., Sweet RM, editors. Methods in Enzymology. Volume 276: Macromolecular Crystallography, part A. Academic Press; New York, USA: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Román-Hernández G, Grant RA, Sauer RT, Baker TA. Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc Natl Acad Sci USA. 2009;106:8888–8893. doi: 10.1073/pnas.0903614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueneman VJ, Kralik SM, Albrecht R, Spall SK, Truscott KN, Dougan DA, Zeth K. Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS. EMBO Rep. 2009;10:508–514. doi: 10.1038/embor.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. Discovery of cellular regulation by protein degradation. J. Biol. Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Flexible linkers leash the substrate-binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 2007;21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Oakes ESC, Sauer RT, Baker TA. Tuning the strength of a bacterial N-end rule degradation signal. J Biol Chem. 2008b;283:24600–24607. doi: 10.1074/jbc.M802213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Román-Hernández G, Grant RA, Sauer RT, Baker TA. The molecular basis of N-end rule recognition. Mol Cell. 2008a;32:406–414. doi: 10.1016/j.molcel.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Wolfgang K, Weber-Ban E. The alternating power stroke of a 6-cylinder AAA protease chaperone engine. Mol Cell. 2009;35:574–85. doi: 10.1016/j.molcel.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- Xia D, Esser L, Singh SK, Guo F, Maurizi MR. Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J Struct Biol. 2004;146:166–179. doi: 10.1016/j.jsb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Zeth K, Ravelli RB, Paal K, Cusack S, Bukau B, Dougan DA. Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat Struct Biol. 2002;9:906–911. doi: 10.1038/nsb869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.