Abstract
3D Magnetic Resonance (MR) and Diffusion Tensor Imaging (DTI) have become important noninvasive tools for the study of animal models of brain development and neuropathologies. Fully automated analysis methods adapted to rodent scale for these images will allow high-throughput studies. A fundamental first step for most quantitative analysis algorithms is skull-stripping, which refers to the segmentation of the image into two tissue categories, brain and non-brain. In this manuscript, we present a fully automatic skull-stripping algorithm in an atlas-based manner. We also demonstrate how to either modify an external atlas or to build an atlas from the population itself to present a self-contained approach. We applied our method to three datasets of rat brain scans, at different ages (PND5, PND14 and adult), different study groups (control, ethanol exposed), as well as different image acquisition parameters. We validated our method by comparing the automated skull-strip results to manual delineations performed by our expert, which showed a discrepancy of less than a single voxel on average. We thus demonstrate that our algorithm can robustly and accurately perform the skull-stripping within one voxel of the manual delineation, and in a fraction of the time it takes a human expert.
Keywords: MRI, DTI, small animal imaging, rat, segmentation, atlas building, alcohol, cocaine
1. AUTOMATIC SKULL-STRIPPING
The automatic skull-stripping method we propose is initialized using an atlas. In case an existing atlas appropriate for the population exists, this external atlas can be used for initializing automatic skull-stripping. If there is no external atlas that matches the population well enough (with respect to species, gender, age, etc.), manual skull-stripping can be used to initialize the process. The bias introduced by the initial skull-stripping, whether by a manual expert or via an external atlas, will be corrected for by repeating the process iteratively until convergence; the external skull-stripping will only be used in the first iteration of the process and the atlas built at the end of each iteration will be used in the subsequent iteration for improved fit. The atlas building process is discussed in Sec. 2.
The brain atlas can be used to create a probabilistic tissue segmentation atlas that can be subsequently applied for tissue segmentation via the tool itkEMS,1 which implements an Expectation-Maximization approach to segment the major brain tissue classes and correct for intensity homogeneities. The tissue classification can be performed either on structural MRI images (T1- or T2- weighted) or on the Fractional Anisotropy (FA), Isotropic Diffusion-Weighted Images (IDWI) and Mean Diffusivity (MD) maps computed from the diffusion-weighted MRI, depending on the available data. We will discuss DTI-based skull-stripping in the remainder of this manuscript; the use of a single structural MR image instead easily follows from a simplification of this algorithm.
If there is an ROI-segmentation of the brain atlas readily available, then these ROI's can be classified into white matter (WM), gray matter (GM) and intermediate categories. This intermediate category contains image regions with an appearance in between WM and GM, specifically, striatum, thalamus, superior and inferior colliculi, as suggested by Lee et al.2 The binary tissue segmentation thus obtained can be smoothed to obtain a pseudo-probabilistic atlas that can be used for itkEMS classification.
If an ROI-segmentation of the brain atlas is unavailable, a similar approach can be taken to create the probabilistic tissue atlas. In this case, the average MD atlas can be intensity-thresholded such that there will be three categories roughly corresponding to the WM, GM and intermediate categories outlined above. The particular values of the three threshold intensities will be dependent on the age group, as myelination and other developmental processes change the MD intensity ranges significantly during the early development. The main criterion for choosing the thresholds should be to create an outer class that mimics the GM, and two inner classes that divide the remainder of the brain roughly in two equal sized parts. The binary images thus created are next processed using the `close' operation from binary morphology to the segmentations to create more consistent clusters of “tissue”. The resulting binary maps are finally smoothed to obtain the “probabilistic” atlas.
The probabilistic tissue atlas thus prepared can be used to automatically skull-strip individual images. To this end, first, the diffusion tensors are reconstructed from the DWI image. Next, this DTI image is resampled to make it isotropic, and the FA, MD and IDWI maps are computed on it. The individual MD map is then rigidly aligned to the brain atlas. Next, the atlas is affinely aligned to the subject. This step is necessary because it is desirable to use a scale-matched atlas for each individual, however, it is undesirable to modify the scale of the individual scans as the volume of the brain structures is one of the measurements to be studied in subsequent analysis. Thus, each subject's image is processed using an individually-scaled atlas. The affine transformation is then propagated to the probabilistic tissue atlas, which is used as input to itkEMS for tissue classification.
The three tissue classes (white, gray, intermediate) computed by itkEMS are merged together for the skull-stripping step. The largest connected component of this combined segmentation is used to get rid of any extraneous voxels wrongly classified as brain tissue. Finally, the segmentation is “closed” using binary morphology to get rid of any voxels inside the brain wrongly classified as non-brain tissue. This final segmentation is used as a binary mask to skull-strip the individual brain.
Note that once all the images in the population were automatically skull-stripped using the first version of the atlas, it is beneficial to repeat the atlas building step (Sec. 2) and use this new atlas for skull-stripping the individual images. As noted above, this will ensure that any bias introduced by using an external atlas or an initial manual skull-stripping is minimized.
2. ATLAS BUILDING
Once the images are successfully skull-stripped, a new unbiased atlas is built from the skull-stripped images. In the current data sets, we have used MD maps for this step to match the modality of our initial template; FA or IDWI maps can also be used. In any case, the images to be used should be affinely registered as well as intensity-matched (via histogram matching) to the current atlas before the unbiased atlas building step to facilitate the deformable registration process. We follow an unbiased atlas building approach3, 4 based on a fluid, diffeomorphic deformation model using an iterative continuous joint registration. The resulting unbiased atlas image, which is the Frechet mean anatomical image of the population, serves as a natural average representation for the anatomy within the images and provides a coordinate system for the analysis of local measurements.
The deformation fields computed in the atlas building stage provide a means of bringing each image to the common coordinate frame defined by the atlas. These deformation fields can be applied to the MD, FA and IDWI maps of each subject, and these warped images can be averaged to create mean DTI maps for the population.
Other than being used to iteratively improve the skull-stripping quality as described above, the atlases thus built are valuable for a range of applications from automatic segmentation and cortical thickness studies to statistical analysis of the deformation fields.
3. RESULTS AND DISCUSSION
We applied our algorithm to three datasets of rat DTI's and compared results with our expert's manual delineations. All scans were acquired at UNC Small Animal MRI facilities with a 9.4T Bruker scanner. To compare the automatic and manual segmentations for each subject, we have computed four different evaluation metrics:
Volume difference: Difference between the volume of the automated and manual segmentations, in mm3;
Volume percentage difference: The volume difference expressed as a percentage of the manual segmentation;
Average surface distance: We extracted surfaces from the binary segmentations and computed the closest point from each point on the automated-mesh to each point on the manual-mesh; we are reporting the average of the error measurements between these points, in mm;
Tanimoto error, which is a measure of overlap (where 0 means complete agreement).
In this manuscript, we are reporting the mean and standard deviations of these 4 metrics for each dataset.
The first dataset was a population of 6 adult male Wistar rats that have been treated with ethanol vapor for 14 hours/day for 5 weeks starting from PND35 with matched controls. A 13-hour protocol with 12 diffusion gradient directions and 2 baseline images with (0.16mm)3 resolution was used. To initialize our algorithm, we used the Brookhaven C57 mouse brain atlas5 that was scaled up to match the rat size. The average skull-stripped brain from the population itself was used for subsequent iterations. Separately, our expert has manually skull-stripped all 12 images. The converged results from our automatic algorithm are compared to the expert in Table 1. Note that the average surface distance between the automatic and manual results is 0.15mm, which is less than a single voxel. This demonstrates the robustness of our method even when initialized with a relatively inappropriate external atlas (mismatched species, size and acquisition parameters).
Table 1.
Manual and automated skull-strip comparison on adult male Wistar rats. Half of the population was intermittently exposed to ethanol. Note that on average, the two surfaces are less than one voxel away from each other.
| n=12 | Auto Vol. | Manual Vol. | Volume Diff. | % Abs. Volume Diff. | Avg. Distance | %Tanimoto Error |
|---|---|---|---|---|---|---|
| Mean | 2.11 | 2.04 | −0.07 | 3.38 | 0.15 | 8.47 |
| StdDev | 0.09 | 0.07 | 0.08 | 3.80 | 0.03 | 1.33 |
The second dataset included 20 PND14 healthy Sprague-Dawley rats that were scanned with a 15-hour protocol that captured 21 diffusion gradient directions and 3 baseline images with (0.12mm)3 resolution. To initialize our algorithm, we used the Brookhaven C57 atlas as it is since the PND14 rat brains are approximately the size of adult mouse brains. As before, the subsequent iterations used the average brain from the population itself. The results from our automatic algorithm are compared to the expert results in Table 2. Note again that the average distance is less than a single voxel.
Table 2.
Manual and automated skull-strip comparison on PND 14 Sprague-Dawley rats. Note that on average, the two surfaces are less than one voxel away from each other.
| n=20 | Auto Vol. | Manual Vol. | Volume Diff. | % Abs. Volume Diff. | Avg. Distance | %Tanimoto Error |
|---|---|---|---|---|---|---|
| Mean | 1.12 | 1.08 | −0.04 | 3.86 | 0.10 | 6.82 |
| StdDev | 0.08 | 0.07 | 0.02 | 1.83 | 0.02 | 1.12 |
The third dataset consisted of 20 PND5 healthy Sprague-Dawley rats that were scanned with 21 diffusion gradient directions and 3 baseline images with (0.12mm)3 resolution. 3 scans were excluded from this study for various reasons (perfusion problems, etc). To initialize the process, we manually skull-stripped 5 of the images and used their average as the initial atlas, as the Brookhaven C57 atlas would have been highly inappropriate for this age group where intense neurodevelopment is ongoing. As before, the subsequent iterations used the average brain from the population itself. The results from our automatic algorithm are compared to the expert results in Table 3. Note that the average distances in this group are somewhat higher than the other datasets we presented (slightly over one voxel); this is due to the highly variable nature of the anatomy at this young age.
Table 3.
Manual and automated skull-strip comparison on PND 5 Sprague-Dawley rats. There is slightly more error in this dataset due to the variation in these young pups.
| n=17 | Auto Vol. | Manual Vol. | Volume Diff. | % Abs. Volume Diff. | Avg. Distance | %Tanimoto Error |
|---|---|---|---|---|---|---|
| Mean | 0.42 | 0.41 | 0.00 | 5.93 | 0.14 | 12.51 |
| StdDev | 0.03 | 0.00 | 0.03 | 3.13 | 0.04 | 3.77 |
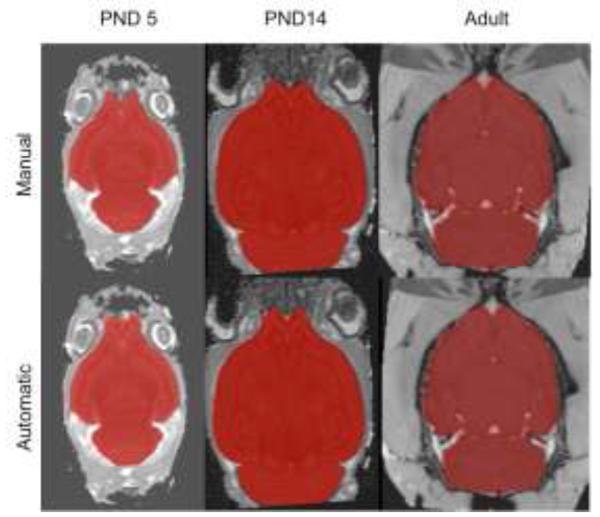
Figure 1 illustrates the manual and automatic skull-strip results for 3 different cases, one from each age group. Note that the automatic skull-strip is practically indistinguishable from the manual expert delineation throughout most of the image. The only noticeable differences are near the olfactory bulb region, where the signal to noise ratio is typically low and even the expert delineation can be inconsistent. Furthermore, note that the automatic segmentations have smoother boundaries than the manual ones. This is an advantage for subsequent processing steps as the jagged boundaries of the manual segmentation can cause numeric unstableness in deformable registration and other similar algorithms. Finally, note that the automatic segmentation has worked well even in the case where part of the brain is cut off due to poor animal positioning (the cerebellum region of the PND14 case in Figure 1). This further demonstrates the robustness of our algorithm.
Figure 1.
Manual and automated skull-strip comparison on typical subjects of each dataset. Note that the automated results are smoother; however, there are slight discrepancies near the olfactory bulbs, where the SNR is typically lower.
In summary, we propose a robust automated skull-stripping method that we have demonstrated to be reliable for usage on MR/DTI images of rats of different strain, age, gender and experimental group, acquired with a variety of acquisition protocols.
ACKNOWLEDGEMENTS
This research was funded by the NIH Program Project IP01DA022446-02, the UNC Neurodevelopmental Disorders Research Center HD 03110, the NIH STTR grant R41 NS059095, the NIH grant RC1AA019211, the UNC Bowles Center for Alcohol Studies as well as the NIH grants AA06059 and AA019969.
REFERENCES
- 1.Prastawa M, Gilmore J, L. W, Gerig G. Automatic segmentation of MR images of the developing newborn brain. Medical Image Analysis. 2005:457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Lee J, Jomier J, Aylward S, Tyszka M, Moy S, Lauder J, Styner M. Evaluation of atlas based mouse brain segmentation. Proceedings of SPIE. 2009 Jan;7259:725943–725943–9. doi: 10.1117/12.812762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004 Jan;23(Suppl 1):S151–60. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 4.Davis B, Lorenzen P, Joshi S. Large deformation minimum mean squared error template estimation for computational anatomy. Proceedings of the IEEE Symposium on Biomedical Imaging. 2004:173–176. [Google Scholar]
- 5.Ma Y, Hof P, Grant S, Blackband S, Bennett R, Slatest L, McGuigan M, Benveniste H. A 3d digital atlas database of the adult c57bl/6j mouse brain by mr microscopy. Neuroscience. 2005;135(4):1203–15. doi: 10.1016/j.neuroscience.2005.07.014. [DOI] [PubMed] [Google Scholar]