Abstract
DNA recombination pathways are cell cycle regulated to coordinate with replication. Cyclin-dependent kinase (Cdk1) promotes efficient 5'-strand resection at DNA double strand breaks (DSBs), the initial step of homologous recombination and damage checkpoint activation. The Mre11–Rad50–Xrs2 complex with Sae2 initiates resection, whereas two nucleases, Exo1 and Dna2, and the DNA helicase/topoisomerase complex Sgs1–Top3–Rmi1 generate longer ssDNA at DSBs. Using Saccharomyces cerevisiae we provide evidence for Cdk1-dependent phosphorylation of the resection nuclease Dna2 at Thr4, Ser17 and Ser237 that stimulates its recruitment to DSBs, resection and subsequent Mec1-dependent phosphorylation. Poorly recruited dna2T4A S17A S237A and dna2ΔN248 mutant proteins promote resection only in the presence of Exo1, suggesting crosstalk between Dna2- and Exo1-dependent resection pathways.
Cyclin dependent kinases (Cdks) drive the cell cycle to coordinate processes such as DNA replication and chromosome segregation. Dysfunction of these kinases in mammals is associated with increased proliferation and genome instability of cancer cells1. Recently, several proteins involved in the DNA damage response were shown to be phosphorylated by Cdk1, revealing its role in co-ordinating DNA repair with replication2. The activities of the budding yeast DNA helicase Srs23, checkpoint proteins Rad53 and Rad9, and the Rad9 homologue Crb2 in fission yeast are regulated by Cdk-mediated phosphorylation4–7. In human cells, phosphorylation of the tumor suppressor protein BRCA2 by Cdk in M phase inhibits its interaction with RAD51, which likely minimizes unscheduled recombination when chromosomes segregate8.
Cdk1 in yeast controls the initial step of DSB-induced homologous recombination (HR), 5' strand resection. In G1 cells, DSB ends are poorly resected, thus enabling efficient repair by non-homologous DNA end-joining (NHEJ). In the S and G2 cells when sister chromatids are available, DSBs are resected promptly to generate a ssDNA substrate for HR9,10. Similarly in fission yeast, NHEJ and HR are cell cycle-regulated11 and Cdk activity is essential for the recruitment of the Rad51 recombinase to DSBs induced by ionizing radiation (IR)4. Finally, in human cells Cdk is also required for early steps of HR12. Consistent with decreased DSB resection, Cdk1-kinase deficient yeast cells also fail to activate the DNA damage checkpoint in response to a single DSB, even though the upstream checkpoint kinase, Mec1, remains at least partially active10,13,14. These results have stimulated a search for targets of Cdk1 that help control early HR steps. Sae2 protein and its vertebrate orthologue CtIP, both involved in the initiation of resection together with Mre11-Rad50-Xrs2 [MRX, (MRE11-RAD50-NBS1 or MRN in human)], were found to be substrates of Cdk1 and key regulators of DSB repair pathway choice15–17. The expression of the fission yeast Sae2 orthologue, Ctp1, is also regulated during the cell cycle18. Besides Sae2 there are likely additional targets of Cdk1 needed for resection because a SAE2 phospho-mimic allele does not efficiently bypass the need for Cdk1 in resection15,19. Evidence for the existence of additional targets comes from studies of resection in Cdk1 kinase deficient cells that lack also the Ku70–Ku80 complex, a central component of the NHEJ pathway. Several studies demonstrated that deletion of Ku proteins restores resection in Cdk1 deficient cells but extensive resection further from the break remains impaired13,20–22. Because Sae2 together with MRX likely act during the initial stages of resection, this result indicates that extensive resection is dependent on Cdk1 as well. We aimed to understand how Cdk1 controls extensive resection in budding yeast. Here we present our genetic and biochemical studies that reveal the role of Cdk1-mediated phosphorylation of Dna2, whose nuclease activity is important for extensive DSB resection in cells.
Results
Dependence of Dna2-mediated long-range resection on Cdk1
First, we examined which of the protein components involved in the DNA motor driven path of resection23–25 - Exo1, Dna2, Sgs1 or MRX complex - remain active in yku70Δ Cdk1 kinase deficient cells. Resection was analyzed by Southern blots at an HO break located at MAT or 28 kb away from the break (FEN2 locus), to follow the initial removal of 5' strands and long-range resection, respectively26. We constructed exo1Δ, sgs1Δ and dna2Δ pif1-m2 derivatives of yku70Δ cdk1-as1 cells and tested resection in these cells either with or without the ATP analogue, 1-NMPP1, that inhibits cdk1-as1 kinase activity27. The pif1-m2 mutation suppresses the lethality of DNA2 deletion28. As previously noted22, in Cdk1 kinase-deficient cells, deletion of YKU70 restores resection only of sequences adjacent to the DSB, as evidenced by the lack of resection 28 kb away (Fig. 1a and Supplemental Fig. 1). We found that Exo1 but not Dna2 or Sgs1 was required for resection in yku70Δ cells where Cdk1 kinase is inactive. Importantly, in Cdk1 kinase deficient exo1Δ yku70Δ cells additional bands and smearing beneath the HO break intermediates that are typical for cells that lack both Exo1 and Dna2 accumulate. This confirms that the Dna2-dependent long range resection pathway is inactive in these conditions (Fig. 1b, Supplemental Fig. 1 and ref.26). As previously determined, MRX is responsible for the limited 5' strand cleavage (Fig. 1b). Similar to resection, DSB repair by ectopic recombination depends on Exo1 in Ku and Cdk1 kinase deficient cells (Supplemental Fig. 1). Consistent with this view, Ku blocks Exo1- and MRX-dependent resection in cycling cells29,30 but not as significantly as in Cdk1 kinase deficient cells. Our results suggest that Exo1 and MRX with Sae2 are active, at least partially so, in the absence of both Ku and Cdk1 kinase, and that the Dna2-dependent resection pathway is likely regulated by Cdk1.
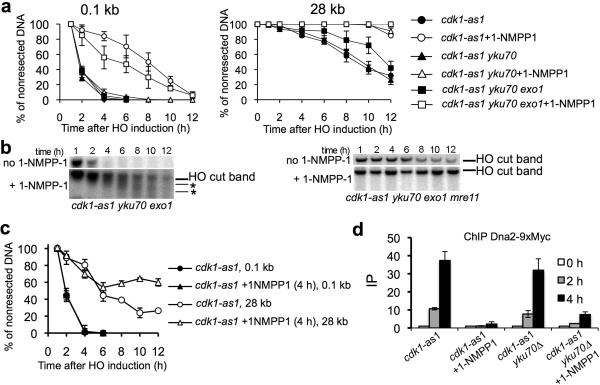
Figure 1.
Cdk1 regulates long range DSB resection by Dna2.
(a) Analysis of initial and extensive resection in mutants with either active or inactive Cdk1 kinase. The Southern blots corresponding to these experiments are presented in Supplemental Figure 1. Error bars correspond to s.d. (b) Southern blot analysis of initial resection in mutants. Smearing and additional bands below the HO cut band typical for mutants that lack extensive resection are indicated by asterisks. (c) Analysis of resection in cells where Cdk1 activity is blocked at 4 hours after break induction when all cells have initiated resection. (d) Recruitment of Dna2 to DSBs was monitored in indicated mutants by ChIP. Error bars correspond to s.d.
To directly test whether Cdk1 activity is needed to sustain normal resection rate further from the break, we measured resection in cells where for the first four hours after DSB induction Cdk1 is active and only then cdk-as1 kinase is blocked by 1-NMPP1. As shown in Figure 1c, long-range resection measured 28 kb from the DSB was still delayed when compared to cells that retained active Cdk1. Therefore Cdk1 controls both initial and long-range resection and, besides Sae2, additional targets of Cdk1 kinase likely exist.
One candidate protein is Dna2, previously shown to be a likely target of Cdk1 in genome wide screens31. First, we investigated whether Cdk1 activity is needed for Dna2 recruitment to DSBs. Recruitment of Dna2-9×Myc was followed using chromatin immunoprecipitation (ChIP) and qPCR using primers specific for sequences 1 kb upstream of the HO break. Recruitment of Dna2-9×Myc occurred efficiently in yku70Δ cdk-as1 cells only when Cdk1 remains active (Fig. 1d). Accordingly, fluorescence microscopy revealed that Dna2-GFP foci are formed in yku70Δ cdk-as1 cells only in presence of active Cdk1 (Supplemental Fig. 2). In contrast, the Sgs1 helicase, which works together with Dna2 in resection23,24,26, is recruited normally to the DSB in yku70Δ Cdk1-kinase deficient cells (Supplemental Fig. 2).
Mec1- and Cdk1-dependent phosphorylation of Dna2
To verify whether Dna2 is phosphorylated, we examined the electrophoretic mobility of Dna2-9×Myc by Western blot. A mobility shift of Dna2-9×Myc was observed following DNA damage (Fig. 2a). The shift was due to phosphorylation as verified by treatment of immunoprecipitated Dna2 with lambda phosphatase (Supplemental Fig. 2). Dna2 phosphorylation is Cdk1-dependent, as it did not occur in kinase deficient cells (Fig. 2a). The maintenance of Dna2 phosphorylation also depends on Cdk1 activity, as revealed by adding 1-NMPP1 four hours after DSB induction (Fig. 2a). To investigate whether Dna2 phosphorylation requires damage checkpoint proteins we tested its phosphorylation in the mec1Δ sml1Δ mutant. Dna2 was not phosphorylated in mec1Δ sml1Δ mutant but remained phosphorylated in the absence of the signaling kinases Rad53, Chk1, or Dun1, suggesting that Dna2 is a direct target of Mec1 (Fig. 2a; data not shown). Immunoblotting of Dna2-TAP from cells 4 hours after HO break induction with a phospho-specific antibody confirmed that Dna2 is phosphorylated by Mec1 or its homolog Tel1 (Supplemental Fig. 2). Together, the results show that Cdk1 and Mec1 are required for Dna2 phosphorylation upon DNA damage.
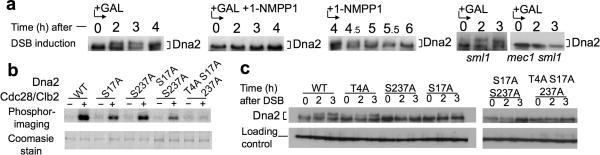
Figure 2.
Dna2 is phosphorylated by Cdk1 and Mec1.
(a) Western blot analysis of Dna2-9×Myc phosphorylation in cdk1-as1 with or without Cdk1 inhibitor and in checkpoint deficient cells in response to a single DSB. (b) In vitro phosphorylation of wild-type Dna2 or dna2 mutant proteins lacking single or multiple Cdk1 phosphorylation consensus sites. (c) Western blot analysis of Dna2 phosphorylation in wild type cells and indicated dna2 mutants cells.
Dna2 harbors 3 full S/T-P-x- K/R (Thr4, Ser17, Ser237) consensus sequences and 5 minimal S/T-P consensus sites for Cdk1 phosphorylation. The full consensus sites are conserved in related yeast species even though most of the N-terminal part of Dna2 is not (Supplemental Fig. 2). We purified wild-type Dna2 and dna2 mutants in which the serine or threonine within the full Cdk1 consensus sites had been replaced with alanine (Supplemental Fig. 2) and performed an in vitro phosphorylation assay with purified Cdk1–Clb2 as described31 (Fig. 2b). We found reduced phosphorylation of dna2S17A and dna2S237A, minimal phosphorylation of dna2S17A S237A, and absence of phosphorylation of dna2T4A S17A S237A (hereafter dna2-3A). These results demonstrate that Ser17, Ser237, and to a lesser degree Thr4, of Dna2 are phosphorylated by Cdk1. The five minimal S/T-P sequences are likely not targeted by Cdk1. The dna2-3A mutant retains biological activity, as it grows normally, whereas a complete deletion of dna2 is lethal28.
To examine whether the Cdk1 consensus sites in Dna2 are important for its phosphorylation in response to DSB induction, plasmid borne DNA2, dna2T4A, dna2S17A, dna2S237A and dna2-3A genes tagged with FLAG were introduced into dna2Δ pif1-m2 cells. The levels of wild-type and all the mutated proteins were similar, but a decrease in phosphorylation upon DSB induction was observed in dna2S17A and dna2S237A mutant cells and particularly in dna2S17A S237A double mutant and dna2-3A mutant cells (Fig. 2c). These results suggest an important role for Ser17 and Ser237 in Dna2 phosphorylation by Mec1 in response to DNA damage.
Role of Dna2 phosphorylation in the DNA damage response
To test the role of phosphorylation of the Cdk1 target residues in resection, we compared the rates of 5' resection of the HO break in dna2Δ cells harboring a centromeric plasmid carrying either wild-type DNA2 or the dna2-3A mutant allele. As control, dna2Δ pif1-m2 cells that show a dramatic defect in resection were included. Initiation of resection was identical in all the mutants, but the rate of resection further from the break site was decreased in the dna2-3A mutant strain (Fig. 3a, Supplemental Fig. 3). Similar resection rates were observed in the dna2Δ pif1-m2 strain background complemented with plasmids carrying the dna2 mutant alleles (Supplemental Fig. 3). A short delay in resection observed in dna2-3A cells suggests that phosphorylation of Ser17 and Ser237 is important for timely resection by Dna2. A similar partial defect was observed in mutants bearing a truncation of the N-terminal 248 residues of Dna2 (dna2ΔN248) where all three full Cdk1 consensus sites are eliminated. The growth rate of dna2-3A and N-terminal truncation mutants is comparable to that of the wild type strain. Importantly, the resection defect exhibited by the dna2 mutant harboring 8 point mutations eliminating all possible Cdk1 consensus sites was no more severe than that in the dna2-3A strain (data not shown).
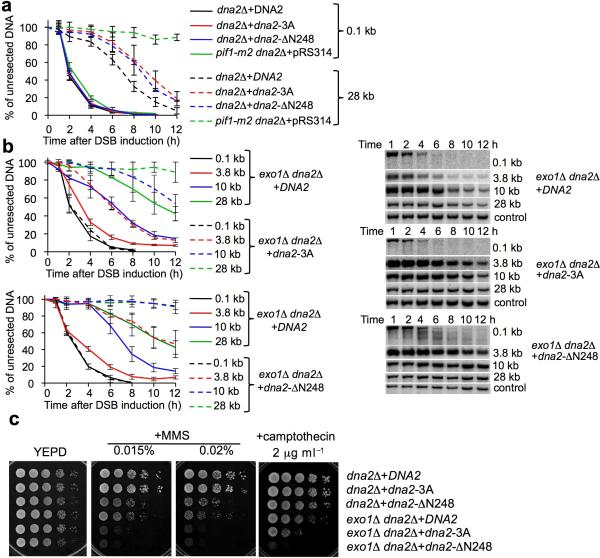
Figure 3.
Dna2 phosphorylation by Cdk1 stimulates resection.
(a) Analysis of 5' strand resection in dna2Δ cells complemented with plasmids carrying either wild-type or a mutant DNA2 allele. Error bars correspond to s.d. The Southern blots corresponding to these experiments are presented in Supplemental Figure 3. (b) Analysis of resection in exo1Δ dna2Δ cells complemented with plasmids carrying either wild-type or a mutant DNA2 allele. (c) Analysis of sensitivity to MMS and camptothecin in the indicated mutants.
Exol defines a second pathway of long range resection, thus it is possible that the dna2 mutants are compensated for by Exo1. To test this possibility we constructed an exo1Δ dna2Δ strain expressing either wild-type DNA2, dna2-3A or dna2ΔN248 and tested resection rates. The growth rate of exo1Δ exo1Δ dna2-3A and exo1Δ dna2ΔN248 is comparable. As shown in Figure 3b, the exo1Δ dna2-3A strain exhibited a very significant defect in extensive resection. An even more severe defect was observed in exo1Δ dna2Δ cells harboring dna2ΔN248. Accordingly, exo1Δ mutants bearing Dna2 lacking the Cdk1 consensus sites or with the N248 deletion are much more sensitive to DNA damage than exo1Δ cells (Fig. 3c). Overexpression of dna2-3A is able to slightly restore resistance to DNA damage indicating that a higher level of mutant protein can partially bypass the need for Cdk1 dependent phosphorylation. Importantly, the dna2S17D S237D mutant that mimics phosphorylation showed normal resection and resistance to DNA damage identical to wild type Dna2 in presence of Cdk1 (Supplemental Fig. 3). We concluded that first, Dna2 phosphorylation by Cdk1 stimulates its role in resection and DNA damage resistance and second, the dna2-3A or dna2ΔN248 mutant can contribute to extensive resection mostly in an Exo1-dependent manner, suggesting crosstalk between the Exo1- and Dna2-dependent resection pathways.
Dna2 phosphorylation stimulates its recruitment to DSBs
To understand the role of Cdk1-dependent Dna2 phosphorylation we compared nuclease activity and protein-protein interactions of wild-type Dna2 and mutant dna2-3A proteins purified from yeast. We showed that dna2-3A possesses normal nuclease activity and interacts with other resection proteins as well as wild type Dna2 (Supplemental Fig. 4). We therefore looked for clues about the function of Dna2 phosphorylation in cells. Since Dna2 is not recruited to DSBs in Cdk1-kinase deficient cells (Fig. 1d), we asked whether Cdk1-dependent phosphorylation at Ser17 and Ser237 is required for Dna2 recruitment. To compare recruitment of wild-type and mutants, we constructed dna2Δ strains carrying GFP-tagged DNA2 or mutant dna2S17A, dna2S237A, dna2S17A S237A, dna2-3A and dna2ΔN248 on a centromeric plasmid. We tested the recruitment of the GFP-tagged proteins to an HO-induced DSB 4 hours after break induction using fluorescence microscopy. The dna2S237A-GFP and dna2S17A-GFP mutant protein showed decreased intensity of foci compared to wild type, and the dna2S17A S237A-GFP and dna2-3A-GFP mutants formed only barely visible foci. An even greater defect in focus formation was observed for dna2ΔN248-GFP (Fig. 4a and Supplemental Fig. 5). As noted previously32, nuclear localization was diminished in the dna2S17A-GFP mutant. These data suggest that serines 17 and 237 of Dna2 are important for its nuclear localization and recruitment to DSBs. Consistent with the microscopy data, ChIP analysis to examine for protein association 1 kb from the DSB ends showed decreased recruitment of the single mutants dna2S17A-FLAG and dna2S237A-FLAG, and severely impaired recruitment of dna2S17A S237A-FLAG, dna2-3A-FLAG, and dna2ΔN248-FLAG (Fig. 4b).
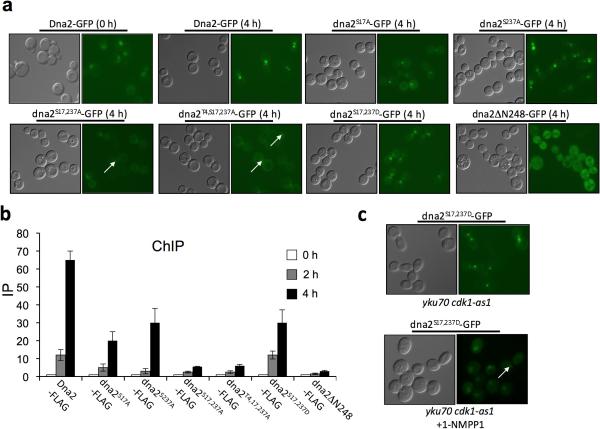
Figure 4.
Dna2 phosphorylation by Cdk1 is needed for its recruitment to DSBs.
(a) Analysis of DSB recruitment of GFP-tagged wild-type Dna2 and indicated mutant dna2 proteins. (b) Analysis of recruitment of FLAG-tagged wild-type Dna2 and indicated mutant dna2 proteins to DSB ends by ChIP using primers specific for sequences located 1 kb upstream of the DSB. Error bars correspond to s.d. (c) Analysis of recruitment of GFP-tagged phosphomimic dna2S17D S237D protein to DSB ends in Cdk1 and Ku deficient cells.
When Ser17 and Ser237 were replaced with aspartic acid to mimic phosphorylation, Dna2 nuclear localization and DSB recruitment were largely restored (Fig. 4a–b, Supplemental Fig. 5). We therefore tested whether dna2S17D S237D-GFP is normally recruited to DSBs in yku70Δ Cdk1-kinase deficient cells where resection is close to DSB ends is normal. However, even nuclear localization occurred normally, DNA damage focus formation of dna2S17D S237D-GFP was impaired when Cdk1 kinase activity was blocked (Fig. 4c). Consistent with the poor DSB recruitment, the phosphomimetic dna2S17D S237D mutation does not restore long range resection in Cdk1 and Ku deficient cells (data not shown). These results suggest that another Cdk1-controlled event is needed for efficient Dna2 recruitment to DSBs. An upstream protein that may be important for Dna2 recruitment is the trimeric replication protein A (RPA), the Rfa2 subunit of which is phosphorylated in a cell cycle-dependent manner33. As described above we have also demonstrated that Dna2 is phosphorylated by the checkpoint kinase Mec1. This phosphorylation is not needed for Dna2 phoshorylation by Cdk1, Dna2 nuclear localization, recruitment to DSBs, or Dna2-dependent resection as analyzed in Exo1 and Mec1 deficient cells (Supplemental Fig. 6). Once the Mec1-specific phosphorylation sites in Dna2 are identified it will be possible to define their significance.
Discussion
Cdk1 kinase is needed for proper 5' strands resection. Previously, Sae2 was reported to be a target of Cdk115 but the molecular function of Sae2 phosphorylation was not determined. Here we have provided evidence that in addition to Sae2, Dna2 is also phosphorylated by Cdk1. Two serines within Dna2 are targeted by Cdk1, an important event for the DSB recruitment of Dna2. A decrease in Dna2 recruitment results in impaired resection that is further aggravated by the deletion of EXO1. Clearly, the cell cycle control of resection by Cdk1 is more complex than previously anticipated, and additional Cdk1 target(s) relevant for DSB resection must exist, as Dna2 and Sae2 pseudo-phosphorylation is insufficient for normal resection in Cdk1 deficient cells.
The budding yeast S. cerevisiae has two resection pathways, controlled by the Exo1 and Sgs1 with Dna2, that can generate long ssDNA at DSBs23,24,26,34. These two pathways appear to be conserved in human cells35. Interestingly, BLM, the Sgs1 orthologue in humans, seems to stimulate both nucleases36. The precise reason for the existence of two resection pathways is not yet known. The Exo1-dependent pathway plays the major role in the resection of Spo11 induced DSB's during meiosis37–39, in processing UV damage40 or within the context of stalled replication forks in checkpoint deficient cells41. Why the Sgs1 with Dna2 pathway does not contribute in these circumstances remains to be established. In mitotic cells, elimination of Exo1 and particularly Dna2 slows down the rate of resection, suggesting that one pathway cannot completely substitute for the other, whereas elimination of both Dna2 and Exo1 eliminates long range resection26. It is possible that, while the two pathways can act independently, they normally collaborate during resection. Indeed, Exo1 and Dna2 may be present simultaneously at DSB ends, since a Dna2-GFP focus is present in all cells with an induced DSB and we have been able to show by ChIP that both Exo1 and Dna2 are associated with regions proximal to and farther away from the DSB site26,30. We have shown here that the dna2-3A and dna2ΔN248 mutants exhibit a modest defect in resection when compared to complete elimination of Dna2. However, Exo1 becomes indispensible for resection in these mutants, suggesting that dna2 mutant proteins that are recruited poorly to DSBs contribute to resection in conjunction with Exo1. One possibility is that the two nucleases are needed for resection because each may be impeded by unique DNA sequences or structures that are less inhibitory to the other.
Based on our results, we propose that Cdk1-mediated phosphorylation of Dna2 ensures an optimal response to DNA damage in S-phase when most of the spontaneous DNA damage occurs. However, it is likely that Cdk1-dependent phosphorylation of Dna2 is also important for other well documented functions of Dna2, most notably in Okazaki fragment processing42. Our study makes a significant contribution toward delineating the cell cycle dependence and regulation of DNA damage repair and replication.
Methods
Strains and plasmids
All strains used in this study are derivates of JKM139 (hoΔ MATa hml∷ADE1 hmr∷ADE1 ade1-100 leu2-3,112 trp1∷hisG' lys5 ura3-52 ade3∷GAL∷HO). Strains used are listed in Supplemental Table 1. All plasmids were constructed using standard PCR and cloning techniques as described in Supplementary Methods.
Analysis of resection at DSB ends
Resection was analyzed at an HO endonuclease-induced DSB at the MAT locus on chromosome III using Southern blots as previously described26. Briefly, Genomic DNA was isolated using a standard phenol extraction method. Purified DNA was digested with EcoRI and separated on a 0.8% (w/v) agarose gel. Southern blotting and hybridization were performed as described previously43. Intensities of target bands were analyzed with ImageQuant TL (Amersham Biosciences) and normalized to the TRA1 probe. Resection efficiency was measured with probes located 0.1 kb (MAT), 3.8 kb (BUD5), 10 kb (SNT1) or 28 kb (FEN2) upstream of the DSB. Analysis was performed at least three times in each mutant strain.
Ectopic recombination assay
DSB repair kinetics and efficiency of DSB repair 8 hr after HO endonuclease induction was measure using ectopic recombination between chromosome III and chromosome V as described previously44.
Fluorescence microscopy
Dna2-GFP foci were photographed using an EM-CCD digital camera (Hamamatsu) connected to an Axiovert 200M microscope (Zeiss). The images were analyzed using the AxioVision Software (Zeiss). To measure Dna2-GFP foci intensity eleven pictures were taken along the Z-axis at 0.3 mm intervals with an acquisition time of 750 ms. Foci intensity was measured in 20 cells in each strain. For DAPI staining we used mounting medium with DAPI H-1200 from Vector Laboratories, Inc. Burlingame. Nuclear localization of Dna2-GFP was verified in at least 100 cells per strain.
Chromatin immunoprecipitation
ChIP assays were performed as described by Sugawara et al. (2003)45. Briefly, cultures were grown to a density of ~1 × 107 cells per ml in pre-induction medium YEP-Raffinose (1% yeast extract, 2% peptone, 2% (w/v) raffinose) and the HO endonuclease was induced by adding 2% (w/v) galactose. Proteins were crosslinked for 10 min at room temperature by the addition of 1% (w/v) (final concentration) formaldehyde, followed by quenching with glycine (125 mM final concentration) for 5 min. Cells were lysed with glass beads, and the extracts were sonicated to shear the DNA to an average size of 0.5 kb. IP samples were incubated with affinity-purified anti-Myc (Sigma M4439) or anti-FLAG (Sigma M2) antibody for overnight at 4°C and bound to protein-G agarose beads for 4 hr at 4°C. The protein bound beads were carried through a series of washes, followed by elution of the proteins and reversal of crosslinking (overnight at 65°C). Samples were treated with proteinase K followed by phenol extraction and ethanol precipitation. Purified DNA was analyzed by real-time quantitative PCR using primers that anneal ~1.1 kb from the DSB.
Whole cell extract preparation
Yeast cells were grown overnight in YEP raffinose medium to a density of ~1 × 107 cells per ml. The HO break was induced by adding 2% (w/v) galactose. Cells from 5 ml of culture were washed with water and resuspended in 10% trichloroacetic acid. The cells were lysed by vortexing with glass beads and the protein lysates were pelleted by centrifugation at 20,000 × g for 15 min. The pellet was washed with 80% acetone and proteins were dissolved in 2× SDS sample loading buffer by boiling samples for 5 min. Samples were centrifuged for 5 min at top speed in a microcentrifuge and the supernatant was retained as the protein extract.
Protein phosphatase treatment
To examine that Dna2 is phosphorylated, yeast cells carrying chromosomal tagged DNA2-TAP were cultured in YEPD to early log phase. Dna2 phosphorylation was induced by adding DSB-inducing agent phleomycin to culture. The TAP tagged Dna2 was immunoprecipitated and treated with Lambda protein phosphatase (New England Biolabs, P0753S). Drug treatment, cell lysis and protein phosphatase treatment were performed as described in Supplementary Methods.
Western blot analysis
Proteins samples were resolved on a 6.5% SDS-PAGE gel and transferred onto a PVDF membrane (Immobilon-P; Millipore) using semi-dry method following manufacturer's protocol. Mouse monoclonal antibodies anti-Myc (M4439) and anti-FLAG (M2) were bought from Sigma, anti-TAP antibody was obtained from GenScript, and the antibodies against phospho-(Ser/Thr) ATM and ATR substrates and phospho-Ser Cdk1 substrate were purchased from Cell Signaling Technology. The blots were developed using the ECL Western Blotting substrate (GE Healthcare).
Protein isolation and in vitro phosphorylation analysis
For in vitro kinase reaction, 500 ng of Dna2 or dna2 mutant was incubated with 4 ng of purified Cdk1-Clb2 in 10 μl of kinase reaction buffer (40 mM Tris-HCl pH 7.4, 2 mM MgCl2, 2 mM ATP, 100 μg ml−1 BSA, 1 mM DTT, 50 mM KCl and 5 μCi [γ-32P]-ATP) at 30°C for 20 min. The reaction was stopped by the addition of 2× SDS loading buffer and heating at 95°C for 3 min. The samples were split and fractionated by 7.5 % SDS-PAGE. The gels were dried and analyzed by phosphorimaging or stained with Coomassie blue.
Protein purification, Dna2 nuclease assay and Affinity pulldown assay
Dna2, dna2 mutants, the Mre11-Rad50-Xrs2 (MRX) complex, the Top3-Rmi1 (TR) complex and yeast RPA were purified to near homogeneity form yeast or E. coli cells tailored to express them, as previously described24. The Cdk1-Clb2 complex was purified from yeast cells, as described before46. Dna2 nuclease activity assay was conducted using a 32P-labeled partial duplex harboring a 5' single-strand overhang with or without RPA exactly as described24. Affinity pulldown to check for interaction of the dna2-3A mutant with the MRX or TR complex was conducted exactly as described24.
Supplementary Material
Acknowledgements
We thank Nick Lyons and David Morgan (University of California, San Francisco) for providing Cdk1 kinase and valuable advice. This work was supported by the National Institutes of Health (NIH) grants GM080600 and 3R01GM080600 to G.I., RO1GM57814 and RO1ES07061 to P.S and GM071011 and 3R01 GM071011 to S.E.L.
References
- 1.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 2.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saponaro M, et al. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 2010;6:e1000858. doi: 10.1371/journal.pgen.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granata M, et al. Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diani L, et al. Saccharomyces CDK1 phosphorylates Rad53 kinase in metaphase, influencing cellular morphogenesis. J Biol Chem. 2009;284:32627–34. doi: 10.1074/jbc.M109.048157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schleker T, Shimada K, Sack R, Pike BL, Gasser SM. Cell cycle-dependent phosphorylation of Rad53 kinase by Cdc5 and Cdc28 modulates checkpoint adaptation. Cell Cycle. 2010;9:350–63. doi: 10.4161/cc.9.2.10448. [DOI] [PubMed] [Google Scholar]
- 8.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 9.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. Embo J. 2004;23:4868–75. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–7. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18:2249–54. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 13.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janke R, et al. A truncated DNA-damage-signaling response is activated after DSB formation in the G1 phase of Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:2302–13. doi: 10.1093/nar/gkp1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–92. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–65. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–3. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–46. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3' overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810–8. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–46. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 22.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. Embo J. 2008;27:1875–85. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–6. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–11. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–85. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases dna2 and exo1 resect DNA double-strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 28.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–69. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim EY, et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–80. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 32.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–6. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–77. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 34.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–4. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–72. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimonkar A, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes and Development. 2011;25:350–62. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgson A, et al. Mre11 and Exo1 contribute to the initiation and processivity of resection at meiotic double-strand breaks made independently of Spo11. DNA Repair (Amst) 2011;10:138–48. doi: 10.1016/j.dnarep.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Keelagher RE, Cotton VE, Goldman AS, Borts RH. Separable roles for Exonuclease I in meiotic DNA double-strand break repair. DNA Repair (Amst) 2011;10:126–37. doi: 10.1016/j.dnarep.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 39.Zakharyevich K, et al. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol Cell. 2010;40:1001–15. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giannattasio M, et al. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Cotta-Ramusino C, et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–9. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Budd ME, et al. A Network of Multi-Tasking Proteins at the DNA Replication Fork Preserves Genome Stability. PLoS Genet. 2005;1:e61. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984;81:1991–5. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–11. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–19. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 46.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.