Abstract
Currently, standard protocols for microbial DNA extraction from intestinal tissues do not exist. We assessed the efficiency of a commercial kit with and without mechanical disruption. Better quality DNA was obtained without mechanical disruption. Thus, it appears that bead-beating is not required for efficient microbial DNA extraction from intestinal biopsies.
Keywords: Intestinal mucosa, DNA extraction, Bead-beating, Quantitative PCR
Obtaining high yield of high quality microbial DNA from intestinal samples often proves to be a challenge. It has been extensively shown that the DNA extraction protocol is the most critical step to obtain a representative and unbiased picture of microbial diversity in environmental samples (de Lipthay et al., 2004). A positive effect of mechanical cell lysis has been demonstrated for stool samples, which are often used as a representation of intestinal tissue (Salonen et al., 2010). Consequently, mechanical disruption was also suggested for protocols of nucleic acid extraction from intestinal biopsies (Zoetendal et al., 2006). However, whereas the positive impact of mechanical disruption on stool samples is evident due to the often recalcitrant biostructure of stool (Swidsinski et al., 2008), mechanical disruption of mucosal biopsies might increase the concentration of eukaryotic DNA, which can bias PCR based methods by co-amplification of 16S rRNA gene sequences (Huys et al., 2008). Mechanical disruption also results in DNA shearing, and fragmented nucleic acids induce biases in subsequent PCR reactions (Liesack et al., 1991). Here, we compared DNA isolation from human ileal and colonic biopsies with a commercial extraction kit recommended but not yet tested on intestinal samples (Zoetendal, et al., 2006), with or without mechanical disruption by bead-beating.
A set of eight biopsies from two healthy individuals was used to assess the effect of bead-beating on DNA extraction efficiency. The manufacturer’s protocol (Qiagen DNA stool kit) was followed and the solution obtained after chemical lysis were aliquoted into two equal volumes. One aliquot for each sample was transferred into Lysing Matrix E tubes (MP Biomedicals, Ohio) (containing 1.4 mm ceramic spheres, 0.1mm silica spheres and one 4 mm glass sphere). These aliquots were then subjected to three cycles of bead-beating (15 seconds at 30 Hz) using a TissueLyser (Qiagen,). Subsequently, the protocol was carried out according to manufacturer’s instructions for both samples. DNA concentration were determined using the QubitR assay (Invitrogen) Real-time quantitative PCR (qPCR) was performed on all mucosal DNA extracts using the SYBRR Green PCR Master Mix (Applied Biosystems) with a particular emphasis on low abundant hydrogenotrophic microbes. Primer pair FTHFSf/FTHFSr (Leaphart and Lovell, 2001) targeting functional formyltetrahydrofolate synthetase (fhs) gene of reductive acetogens, ME1/ME2 (Hales et al., 1996) targeting the methyl coenzyme-M reductase (mcrA) gene of methanogenic archaea (MA) and DSR1fdeg/DSR4rdeg (Leloup et al., 2007) targeting the dissimilatory sulfate reductase (dsrA) gene of sulphate-reducing bacteria (SRB) were used. Primer pairs targeting 16S rRNA genes of Bacteria (8f/1541r (Embley, 1991)) and the Gram-positive genus Desulfotomaculum (DFM140/DFM842 (Daly et al., 2000)) were also used. Samples were amplified on a 7900HT Fast Real-Time PCR System (Applied Biosystems) using a dissociation curve. Standard curves were determined simultaneously using plasmids containing dsrA, mcrA and fhs or diluted PCR products from reference strains for the 16S rRNA genes. Gene copy numbers per ng of DNA were inferred from the standard curves.
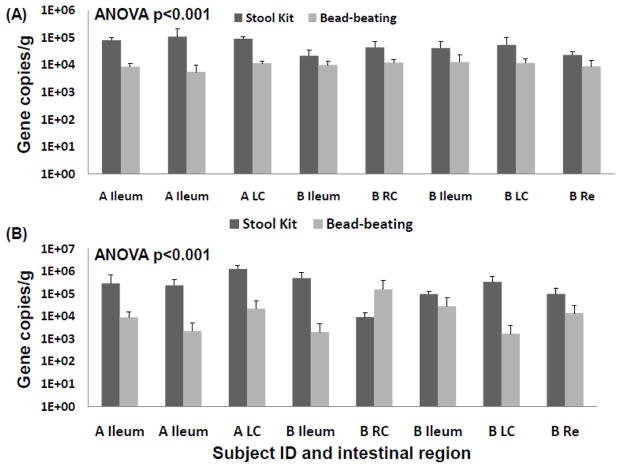
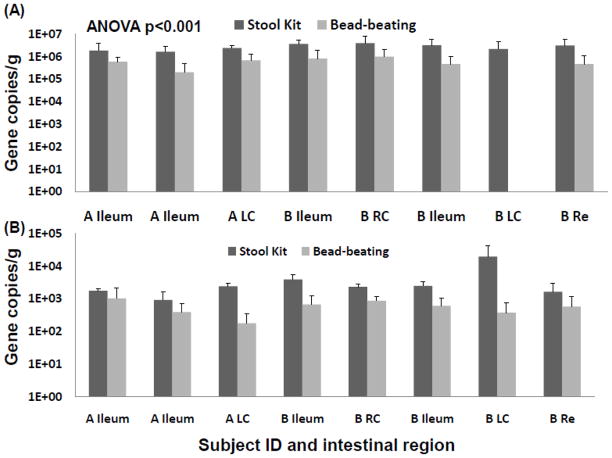
The addition of a bead-beating step clearly increased the DNA yield, which was generally two to four times higher (1.46-2.08 ng/μL for the DNA stool kit alone, 2.73-5.86 ng/μL with bead-beating) than with the kit protocol alone. The quality of the DNA in terms of detection of microbial target genes was assessed by calculation of the number of gene copies per ng of DNA. The quantity of Bacteria 16S rRNA gene copies was similar in DNA extracted with and without bead-beating (Figure 1). Detection of low abundant hydrogenotrophic microbiota in intestinal mucosa was of particular interest. Methanogenic archaea have also been reported to be refractory to chemical lysis (Jarrell et al., 1992, Kandler and Hippe, 1977). A greater relative abundance of mcrA and dsrAB was detected in extracts that were not subjected to mechanical cell disruption (Figure 2). Gram-positive bacteria are generally more recalcitrant to chemical and enzymatic lysis, and thus it is assumed that their effective lysis requires mechanical disruption (von Wintzingerode et al., 1997). Most reductive acetogens harbouring the fhs gene and all Desulfotomaculum spp. are Gram-positive (Drake et al., 2002, Junier et al., 2010) and their corresponding genes were also detected at greater relative abundance in extracts that were not subjected to mechanical cell disruption (Figure 3).
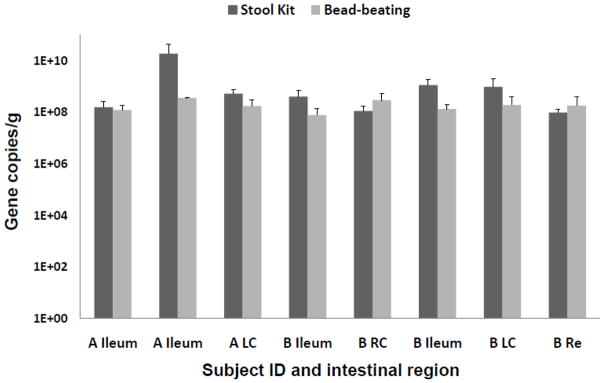
Figure 1.
Bacteria 16S rRNA gene copy numbers per ng of DNA extract obtained from the DNA stool kit with and without bead-beating for two individuals (A and B). Error bars indicate the Standard Error to the Mean (SEM) based on three technical replicates. RC: Right Colon, LC: Left Colon, Re: Rectum.
Figure 2.
Bacteria 16S rRNA gene copy numbers per ng of DNA extract obtained from the DNA stool kit with and without bead-beating for two individuals (A and B). Error bars indicate the SEM based on three technical replicates. RC: Right Colon, LC: Left Colon, Re: Rectum.
Figure 3.
(A) fhs and (B) Desulfotomaculum 16S rRNA gene copy numbers per ng of DNA extract obtained from the DNA stool kit with and without bead-beating for two individuals (A and B). Error bars indicate the SEM based on three technical replicates. RC: Right Colon, LC: Left Colon, Re: Rectum.
Thus, in the case of intestinal biopsies, the benefit of bead-beating is not supported and the present data indicate that better DNA quality is obtained without mechanical cell disruption. Overall, the bead-beating step increases the quantity of microbial DNA, but it also most likely increases the presence of eukaryotic DNA from epithelial cells. Eukaryotic DNA can be co-amplified by bacterial 16S primers (Huys, et al., 2008), which might explain the observation Bacteria 16S rRNA gene copy numbers were more similar in reactions using primers targeting this domain Figure 1). Increased eukaryotic DNA can also have inhibitory effects on microbial targeted PCR by physical obstruction (Cogswell et al., 1996). Importantly, low abundant microbes such as MA and SRB were detected in higher relative abundance in DNA extracted without mechanical disruption. Substantial MA and SRB diversity was described using this method on another set of colonic biopsies (Nava et al., 2011), and these observations motivated the present comparative study. Even more unexpected was the observation that Gram-positive bacteria were also detected in greater abundance. T he mucosa-associated microbiota represents only a thin layer of cells in biopsy tissue and it is likely that gentler mechanical disruption (vortexing) coupled with chemical and enzymatic lyses is sufficient to dislodge and break microbial cells from intestinal tissues.
Highlights.
Standard microbial DNA extraction protocols for intestinal biopsies are needed
DNA quality is improved when cells are not mechanically disrupted
A commercial DNA extraction kit is more efficient for such delicate samples
Acknowledgments
This work was supported by a grant from the Carle Foundation as well as a Carle Hospital-University of Illinois Translational Research Program grant. The subjects are thanked for their participation in the study and Carle Foundation Hospital staff for cooperation in obtaining specimens and subject data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cogswell FB, Bantar CE, Hughes TG, Gu Y, Philipp MT. Host DNA can interfere with detection of Borrelia burgdorferi in skin biopsy specimens by PCR. Journal of clinical microbiology. 1996;34:980–982. doi: 10.1128/jcm.34.4.980-982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly K, Sharp RJ, McCarthy AJ. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology. 2000;146:1693–1705. doi: 10.1099/00221287-146-7-1693. [DOI] [PubMed] [Google Scholar]
- de Lipthay JR, Enzinger C, Johnsen K, Aamand J, Sorensen SJ. Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol Biochem. 2004;36:1607–1614. [Google Scholar]
- Drake HL, Kusel K, Matthies C. Ecological consequences of the phylogenetic and physiological diversities of acetogens. Antonie Van Leeuwenhoek. 2002;81:203–213. doi: 10.1023/a:1020514617738. [DOI] [PubMed] [Google Scholar]
- Embley TM. The Linear PCR Reaction - a simple androbust method for sequencing mmplified ribosomal-RNAgenes. Lett Appl Microbiol. 1991;13:171–174. doi: 10.1111/j.1472-765x.1991.tb00600.x. [DOI] [PubMed] [Google Scholar]
- Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huys G, Vanhoutte T, Joossens M, Mahious AS, De Brandt E, Vermeire S, Swings J. Coamplification of eukaryotic DNA with 16S rRNA gene-based PCR primers: Possible consequences for population fingerprinting of complex microbial communities. Curr Microbiol. 2008;56:553–557. doi: 10.1007/s00284-008-9122-z. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Faguy D, Hebert AM, Kalmokoff ML. A general-method of isolating high molecular weight DNA from methanogenic archaea (archaebacteria) Can J Microbiol. 1992;38:65–68. doi: 10.1139/m92-010. [DOI] [PubMed] [Google Scholar]
- Junier P, Junier T, Podell S, Sims DR, Detter JC, Lykidis A, Han CS, Wigginton NS, Gaasterland T, Bernier-Latmani R. The genome of the Gram-positive metal- and sulfate-reducing bacterium Desulfotomaculum reducens strain MI-1. Environ Microbiol. 2010;12:2738–2754. doi: 10.1111/j.1462-2920.2010.02242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O, Hippe H. Lack of peptidoglycan in cell walls of Methanosarcina barkeri. Arch Microbiol. 1977;113:57–60. doi: 10.1007/BF00428580. [DOI] [PubMed] [Google Scholar]
- Leaphart AB, Lovell CR. Recovery and analysis of formyltetrahydrofolate synthetase gene sequences from natural populations of acetogenic bacteria. Appl environ microbiol. 2001;67:1392–1395. doi: 10.1128/AEM.67.3.1392-1395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup J, Loy A, Knab NJ, Borowski C, Wagner M, Jorgensen BB. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol. 2007;9:131–142. doi: 10.1111/j.1462-2920.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16s rDNA analysis of a mixed culture of strict barophilic Bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- Nava GM, Carbonero F, Croi JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2011 doi: 10.1038/ismej.2011.90. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, Palva A, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008;135:568–579. doi: 10.1053/j.gastro.2008.04.017. [DOI] [PubMed] [Google Scholar]
- von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Heilig HGHJ, Klaassens ES, Booijink CCGM, Kleerebezem M, Smidt H, de Vos WM. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1:870–873. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]