Abstract
Messenger RNA surveillance, the selective and rapid degradation of mRNAs containing premature stop codons, occurs in all eukaryotes tested. The biological role of this decay pathway, however, is not well understood. To identify natural substrates of mRNA surveillance, we used a cDNA-based representational difference analysis to identify mRNAs whose abundance increases in Caenorhabditis elegans smg(−) mutants, which are deficient for mRNA surveillance. Alternatively spliced mRNAs of genes encoding ribosomal proteins L3, L7a, L10a, and L12 are abundant natural targets of mRNA surveillance. Each of these genes expresses two distinct mRNAs. A productively spliced mRNA, whose abundance does not change in smg(−) mutants, encodes a normal, full-length, ribosomal protein. An unproductively spliced mRNA, whose abundance increases dramatically in smg(−) mutants, contains premature stop codons because of incomplete removal of an alternatively spliced intron. In transgenic animals expressing elevated quantities of RPL-12, a greater proportion of endogenous rpl-12 transcript is spliced unproductively. Thus, RPL-12 appears to autoregulate its own splicing, with unproductively spliced mRNAs being degraded by mRNA surveillance. We demonstrate further that alternative splicing of rpl introns is conserved among widely diverged nematodes. Our results suggest that one important role of mRNA surveillance is to eliminate unproductive by-products of gene regulation.
Keywords: mRNA surveillance, ribosomal protein autoregulation, regulated alternative splicing
Eukaryotic mRNAs containing premature translation termination codons are almost always degraded more rapidly than their wild-type counterparts, a phenomenon known as nonsense-mediated mRNA decay (NMD) or mRNA surveillance (for review, see Czaplinski et al. 1999; Hilleren and Parker 1999). Cis-acting elements required for nonsense codon recognition include downstream exon junctions in mammalian mRNAs and sequences in yeast mRNAs that are found downstream of premature stop codons but upstream of normal termination codons (for review, see Hilleren and Parker 1999). Trans-acting factors required for mRNA surveillance have been identified genetically in both yeast and nematodes. Loss-of-function mutations affecting any of three Saccharomyces cerevisiae UPF/NMD genes (Leeds et al. 1992) or seven Caenorhabditis elegans smg genes (Pulak and Anderson 1993; Cali et al. 1999) eliminate mRNA surveillance, thereby causing nonsense mutant mRNAs to have normal, rather than reduced, half-lives. The molecular mechanisms of mRNA surveillance may be similar in all eukaryotes because yeast Upf1p (Leeds et al. 1992), nematode SMG-2 (Page et al. 1999), and Rent1/HUPF1, a human protein likely involved in mRNA surveillance (Perlick et al. 1996; Sun et al. 1998), are sequence homologs.
Although much is known about the sequences required in cis and the proteins required in trans for mRNA surveillance, we know relatively little about the substrates of mRNA surveillance in normal, wild-type organisms. Messenger RNA surveillance is not essential for viability of yeast or nematodes, as upf− and smg(−) mutants exhibit relatively mild phenotypes. For example, yeast upf− mutants have increased sensitivity to an inhibitor of translation (Leeds et al. 1992), impaired respiration (Altamura et al. 1992), and altered telomere length (Lewis and Fleming 1995), whereas nematode smg(−) mutants have modestly reduced brood sizes (Cali and Anderson 1998) and mild morphogenetic defects (Hodgkin et al. 1989). At least in yeast, such phenotypes may be an indirect consequence of eliminating mRNA surveillance. Transcriptional profiles of yeast demonstrate that the steady-state levels of mRNA for >8% of yeast genes are significantly increased or decreased in upf− mutants. Most of these effects, however, are indirect, as all affected mRNAs that have been tested have normal half-lives in upf− mutants (Lelivelt and Culbertson 1999).
One suggested role for mRNA surveillance is to act as a proofreading system to eliminate aberrant mRNAs arising from errors in gene expression (Pulak and Anderson 1993). Such errors might include germline or somatic mutations, transcriptional errors, inaccurate splicing, or inappropriate transport of pre-mRNAs to the cytoplasm. Aberrant mRNAs containing premature termination codons encode polypeptides truncated at their carboxyl termini, many of which may be deleterious. For example, a surprisingly large number of known or suspected C. elegans nonsense mutations are strongly dominant when present in a smg(−) genetic background but are recessive or only weakly dominant when in a smg(+) background (Cali and Anderson 1998; Pulak and Anderson 1993). Messenger RNA surveillance may thus protect cells from deleterious polypeptide fragments by rapidly eliminating the mRNAs that encode them.
What are the sources of mRNAs containing premature stop codons in normal cells? Only a handful of natural targets of mRNA surveillance have been identified to date. In yeast, unspliced cytoplasmic pre-mRNAs of CYH2, RP51B, MER2, and CRY2 contain stop codons within the retained introns and are efficiently eliminated by mRNA surveillance (He et al. 1993; Li et al. 1995). Certain mRNAs that undergo leaky scanning for translation initiation sites are also subject to mRNA surveillance in yeast (Welch and Jacobson 1999). In nematodes, certain SR protein mRNAs contain early termination codons in alternatively spliced exons and are eliminated by mRNA surveillance (Morrison et al. 1997). In mammals, mRNA for selenium-dependent glutathione peroxidase, in which a UGA codon encodes selenocysteine, is down-regulated by mRNA surveillance under conditions of dietary selenium deficiency (Moriarty et al. 1998). In cells of the mammalian immune system, gene rearrangements of immunoglobulin and T-cell receptor β genes often result in out-of-frame mRNAs that are down-regulated relative to in-frame transcripts (Baumann et al. 1985; Carter et al. 1995). Such effects are consistent with an important proofreading role for mRNA surveillance. The variety of such substrates of surveillance suggests that many natural targets exist, but they have not been systematically studied or characterized.
To better understand the role of mRNA surveillance in wild-type organisms, we have undertaken a general screen for natural targets of surveillance in C. elegans. Our approach is based on the assumption that the abundance of such mRNAs will be increased in smg(−) mutants relative to smg(+) controls. We identified alternatively spliced mRNAs of four ribosomal protein genes as abundant natural targets of mRNA surveillance and have investigated the regulatory circuitry responsible for their production.
Results
RDA identifies messages subjected to mRNA surveillance
We performed a representational difference analysis (RDA; Hubank and Schatz 1994) of smg(+) and smg-2(−) cDNAs to identify mRNAs whose abundance increases in smg-2 mutants. We first prepared cDNA populations from poly(A)+ mRNA of each strain, and subsequently subtracted smg(+) cDNAs from smg-2(−) cDNAs (see Materials and Methods). In principle, such methods should identify mRNAs whose abundance is increased or which contain novel sequences in smg-2 mutants compared to wild type. Following two to three rounds of subtraction and enrichment, we generated cDNA clones of candidate mRNAs and characterized them by direct sequencing. To confirm that cDNA candidate clones represented mRNAs subjected to mRNA surveillance, we measured their levels of expression in wild-type and smg(−) mutants by Northern blot hybridization.
As a control to demonstrate that the methods of RDA were successful, both the smg(+) and smg-2(−) strains that we used also contained unc-54(r315), a nonsense allele of a myosin heavy chain gene (Bejsovec and Anderson 1990). Previous work demonstrates that the abundance of unc-54(r315) mRNA is approximately 20-fold higher in smg(−) mutants than in smg(+) controls (Pulak and Anderson 1993). Of 104 tested cDNA clones derived from RDA enrichment, 17 were clones of unc-54, as determined either by sequencing or hybridization (data not shown). This demonstrated that the RDA technique could successfully identify mRNAs whose abundance is increased in smg(−) mutants.
Alternatively spliced mRNAs of four ribosomal protein genes are natural targets of mRNA surveillance
Of 87 non-unc-54 cDNA clones described above, 14 represented clones of one of four ribosomal protein genes (three clones of rpl-3, three of rpl-7a, two of rpl-10a, and six of rpl-12) that were spliced in an unexpected manner. In all cases, comparing the sequences of cDNA clones obtained by RDA to those of genomic DNA (Ainscough et al. 1998) and a database of C. elegans expressed sequence tags (see http://www.ddbj.nig.ac.jp) identified an alternative splice site located within an intron of the relevant rpl gene as shown, for example, in Figure 1. rpl-7a, rpl-10a, and rpl-12 contain alternative 5′ splice sites within introns 3, 1, and 2, respectively, whereas rpl-3 contains an alternative 3′ splice site within intron 1. The sequences of all splice sites thus defined (yielding both conventional and alternative mRNAs) conform well to C. elegans 5′ and 3′ splice consensus sequences (Blumenthal and Steward 1997). In each case, the conventional mRNA encodes a full-length RPL protein, whereas the alternative mRNA retains a portion of the affected intron. The retained intron sequences vary in size from 85 nucleotides (rpl-7a) to 125 nucleotides (rpl-10a), and in all cases, contain in-frame translation termination codons that presumably cause the mRNAs to be substrates of mRNA surveillance (see below). The alternative mRNAs are predicted to encode truncated proteins ranging in size from 25% (rpl-10a) to 72% (rpl-12) of the corresponding full-length RPL protein.
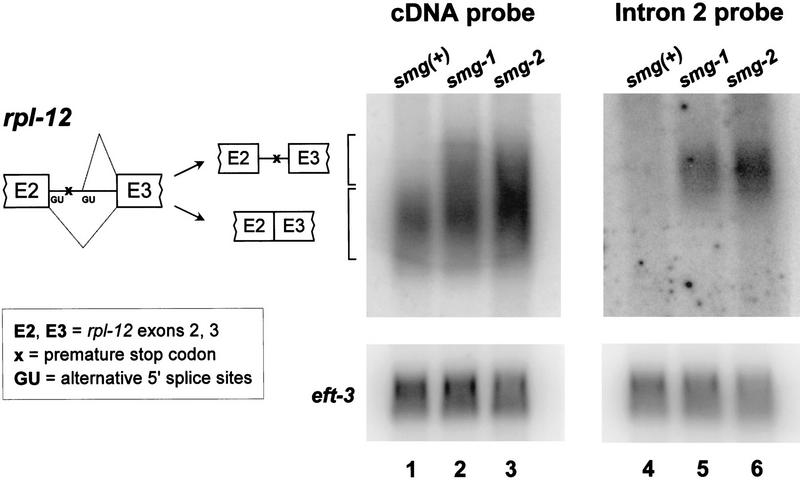
Figure 1.
rpl-12 mRNAs are alternatively spliced. Diagram depicts rpl-12 pre-mRNA and alternatively spliced products. Only exons two through three are shown. Northern blots were probed with an rpl-12 cDNA clone (lanes 1–3, upper panel), a 93-nucleotide cDNA fragment from the portion of rpl-12 intron 2 retained by alternative splicing (lanes 4–6, upper panel), or an eft-3 cDNA clone (loading control; lanes 1–6, lower panels). Strains used were N2 (wild type; lanes 1,4), smg-1(r861) (lanes 2,5) and unc-54(r315) smg-2(r863) (lanes 3,6).
Alternative splicing of rpl-12 is shown in Figure 1. Using rpl-12 cDNA sequences as a hybridization probe for Northern blots (lanes 1–3), a single rpl-12 mRNA is detected in wild type (lane 1), whereas two mRNA species are detected in smg-1(r861) (lane 2) and smg-2(r863) (lane 3). Using a hybridization probe specific for the portion of intron 2 retained by alternative splicing (lanes 4–6), only the larger of these mRNAs is detected, and only in smg(−) backgrounds. To ensure that the larger rpl-12 mRNA detected only in smg(−) mutants corresponds to the alternatively spliced mRNA identified by RDA, we analyzed by RT–PCR rpl-12 products that span intron 2. Using smg-1(r861) mRNA as template, we detected two distinct RT–PCR products. The sequences of these cDNAs confirmed that the two mRNA species detected on Northern blots (Fig. 1) represent the conventional and alternatively spliced mRNAs described above (data not shown). Analogous experiments with rpl-3, rpl-7a, and rpl-10a gave comparable results. In subsequent discussions, we refer to the conventional and alternative rpl mRNAs as resulting from productive and unproductive splicing, respectively.
To quantify the amounts of productively and unproductively spliced rpl mRNAs, we performed Northern blots of rpl mRNAs resolved on polyacrylamide gels after first removing their poly(A) tails and nearby 3′ sequences by oligo-directed RNase H cleavage. Removing the poly(A) tails reduces the size heterogeneity of rpl mRNAs evident in Figure 1. We hybridized equal amounts of total RNA from wild-type and smg-1(r861) nematodes with a DNA oligonucleotide complementary to an rpl exon downstream of each alternatively spliced intron and cleaved the hybridized mRNAs with RNase H. We resolved cleaved RNAs on polyacrylamide gels and probed Northern blots with rpl sequences upstream of the cleavage site. Parallel experiments were performed for rpl-3, rpl-7a, rpl-10a, and rpl-12, using a different cleavage-directing oligonucleotide for each. To control for the quantity of mRNAs analyzed in each experiment, we normalized rpl hybridization signals to those of translation elongation factor 1α (eft-3). Analysis of eft-3 mRNA levels relative to other loading controls (e.g., ribosomal RNA) demonstrates that eft-3 is not itself affected by smg mutations (data not shown). Representative Northern blots and a summary of measured mRNA quantities are shown in Figure 2. The data for each experiment represent five trials performed using three independent RNA samples from each strain.
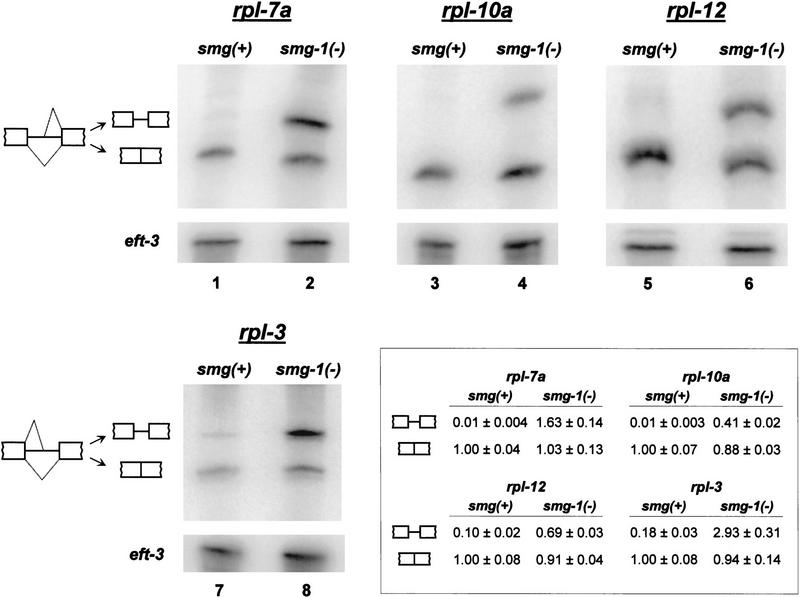
Figure 2.
The abundance of alternatively spliced forms of four ribosomal protein mRNAs is elevated in smg(−) mutants. Northern blots of oligonucleotide-cleaved total RNAs were probed with an eft-3 cDNA clone (lower panels) and the following ribosomal protein cDNAs (upper panels): rpl-7a (lanes 1–2); rpl-10a (lanes 3–4); rpl-12 (lanes 5–6); rpl-3 (lanes 7–8). Measured quantities of each mRNA species are summarized in the table inset. Each value is an average of five experiments ± standard error. For each gene, numbers in the upper rows quantify unproductively spliced mRNA, and numbers in the lower rows quantify productively spliced mRNA. Strains used were N2 (wild type; lanes 1,3,5,7) and smg-1(r861) (lanes 2,4,6,8).
All four rpl genes behave similarly in these analyses. In wild-type strain N2 (Fig. 2, lanes 1,3,5,7), the steady-state level of unproductively spliced rpl mRNA is low. Measured quantities range from an amount that is barely detectable (∼1% of the quantity of productively spliced mRNA; rpl-7a, lane 1 and rpl-10a, lane 3) to an amount that is ∼20% of the quantity of productively spliced mRNA (rpl-3, lane 7). In smg-1(r861) (lanes 2,4,6,8), the steady-state level of unproductively spliced rpl mRNAs is substantially elevated and often equals or exceeds that of the productively spliced counterpart (e.g., rpl-3, lane 8 and rpl-7a, lane 2). The steady-state quantities of productively spliced rpl mRNAs are approximately equal in N2 and smg-1(r861) (Fig. 2, table inset). We interpret these results to indicate that, in both smg(+) and smg(−) backgrounds, large amounts of both productively and unproductively spliced rpl mRNAs are produced. In smg(+) strains, unproductively spliced rpl mRNAs are unstable, such that they represent only a small fraction of steady-state rpl mRNA. In smg(−) strains, unproductively spliced rpl mRNAs are stabilized, such that they constitute, on average, over half of the steady-state rpl mRNA, ranging from 30% (rpl-10a) to 75% (rpl-3) of total rpl mRNA.
To investigate whether C. elegans ribosomal protein genes not identified by the above RDA protocol are alternatively spliced in a manner similar to that of rpl-3, rpl-7a, rpl-10a, and rpl-12, Northern blots of wild-type and smg-1(r861) mRNAs were probed for nine arbitrarily chosen ribosomal protein messages. No changes in either the quantity or sizes of mRNAs encoded by rps-7, rps-10, rps-13, rps-16, rpl-1, rpl-5, rpl-15, rpl-24, or rpl-32 were detected (A. Grimson and P. Anderson, unpubl.). We conclude that alternative splicing of ribosomal protein mRNAs is not a ubiquitous phenomenon in C. elegans, but is specific to a subset of these mRNAs.
In addition to the rpl mRNAs described above, we identified as potential natural targets mRNAs encoded by the novel genes Y41E3.16 and either F45D11.2, F45D11.3, or F45D11.4 (three genes of essentially identical sequence), and one of two alternative mRNAs encoded by the homolog of human HSR1, C53H9.2 (Vernet et al. 1994). The quantities of these mRNAs are substantially elevated in a smg(−) background, as determined by Northern blot analysis, but we have not characterized them further. The majority of mRNAs identified by our RDA clones were unaffected in a smg(−) background. These false positives represented highly expressed C. elegans RNAs not eliminated by the RDA procedures along with RNAs from Escherichia coli, the C. elegans food source.
Alternatively spliced rpl introns are highly conserved among nematodes
Alternative splicing of rpl-3, rpl-7a, rpl-10a, and rpl-12 could reflect an error-prone splicing system, or it could be functionally significant to expression of these genes. These introns may contain cryptic 5′ or 3′ splice sites that are functional but not significant with regard to rpl gene expression or ribosome biogenesis. Alternatively, productive versus unproductive splicing might be regulated to alter the quantity of full-length or, possibly, truncated RPL proteins. To address these and other possibilities, we cloned and sequenced rpl-3, rpl-7a, rpl-10a, and rpl-12 from related nematode species and investigated whether alternative splicing of the type described for C. elegans is evident in these species. We chose for analysis two species, Caenorhabditis briggsae and Caenorhabditis remanei, that are reasonably close relatives of C. elegans (Fitch et al. 1995; Fitch and Thomas 1997) and two species, Diploscapter coronata and Pratylenchus penetrans, that are more distant relatives (Blaxter et al. 1998).
C. elegans, C. briggsae, and C. remanei are morphologically similar, but 18S rDNA sequences indicate substantial molecular divergence, comparable to the divergence seen among different orders within tetrapod classes (Fitch et al. 1995). Coding sequences of these species are highly similar, but noncoding sequences generally share little sequence similarity (Fitch and Thomas 1997). We performed a survey of 249 arbitrarily chosen C. elegans introns (see Materials and Methods) and found that for 239 of these, an intron is also present at the same position within the equivalent C. briggsae gene. Neither the sizes nor the sequences of these introns, however, are well conserved. The correlation of intron size was low (correlation coefficient = 0.57). Using a global sequence alignment algorithm (Myers and Miller 1988), the average sequence identity for these 239 introns was 43.7%. A substantial portion of the observed identity may be because of general base sequence composition (e.g., AT richness) or conserved intron elements (e.g., splice site consensus sequences), as 110 randomly matched (i.e., nonequivalent) C. elegans and C. briggsae introns exhibited an average sequence identity of 36.0%. As expected, however, coding sequences are well conserved. We aligned 31 kb of coding sequence from 149 exons, and the average nucleotide identity was 74.7%. Although genomic sequence of C. remanei is limited, available data indicate that its divergence from C. elegans is similar to that of C. briggsae (Fitch et al. 1995; Fitch and Thomas 1997).
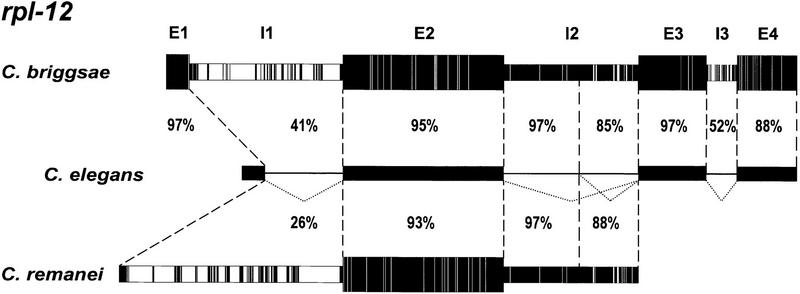
Apart from their alternatively spliced introns, rpl-3, rpl-7a, rpl-10a, and rpl-12 exhibit a typical conservation of gene sequence. Exons are highly similar, whereas introns are not. Introns that are alternatively spliced, however, are exceptional. These introns are highly conserved, ranging from 66% nucleotide identity (rpl-10a, C. elegans vs. C. briggsae) to 93% nucleotide identity (rpl-12, C. elegans vs. C. remanei). With the exception of rpl-10a, these introns are more highly conserved between C. elegans and C. briggsae than any of the 239 introns in the survey mentioned above. The portion of rpl-12 intron 2 retained by alternative splicing is, in fact, more highly conserved than the exons in which it is located (Fig. 3). The sequences of rpl-12 intron 2 are shown in Figure 4, and overall conservation of rpl-12 is diagrammed in Figure 3. Alternatively spliced introns of rpl-3, rpl-7a, and rpl-10a are similarly well conserved, although not quite as strikingly as that of rpl-12 (Fig. 4). The portions of these introns retained by alternative splicing are in all cases more highly conserved than the portions excised (Table 1).
Figure 3.
The alternatively spliced rpl-12 intron is well conserved within the Caenorhabditis genus. Diagram of rpl-12 pre-mRNA splicing and comparisons of nucleotide identity, with identical residues depicted in black and non-identical residues in white. Nucleotide identity between C. elegans and C. briggsae is shown for all exons (E1–E4) and introns (I1–I3); nucleotide identity between C. elegans and C. remanei is shown for exon 2 and introns 1 and 2.
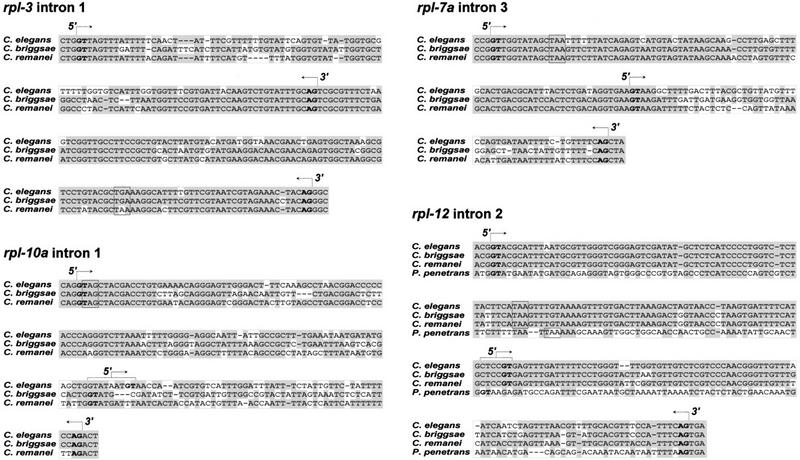
Figure 4.
Nucleotide alignments of nematode rpl-3, rpl-7a, rpl-10a, and rpl-12 alternatively spliced introns. Conserved residues are boxed in gray, non-conserved residues in white. Normal and alternative splice sites (GT/AG), as determined by cDNA sequence analysis, are shown in bold and indicated by 5′ or 3′ arrows. Nucleotides encoding the first in-frame termination codons in unproductively spliced mRNAs are boxed in black. Species are C. elegans, C. briggsae, C. remanei, and P. penetrans.
Table 1.
Conservation of alternatively spliced introns
|
|
Portion retained by alternative splicing (% identity)
|
Portion excised by alternative splicing (% identity)
|
|---|---|---|
| C. briggsae vs. C. elegans | ||
| rpl-3 | 88% | 70% |
| rpl-7a | 88% | 50% |
| rpl-10a | 71% | 46% |
| rpl-12 | 97% | 85% |
| C. remanei vs. C. elegans | ||
| rpl-3 | 88% | 71% |
| rpl-7a | 85% | 57% |
| rpl-10a | 75% | 57% |
| rpl-12 | 97% | 88% |
| P. penetrans vs. C. elegans | ||
| rpl-12 | 54% | 32% |
Alternative splicing of rpl introns occurs in widely diverged nematodes
To determine whether alternative splicing of the type described for C. elegans occurs in C. briggsae and C. remanei, we analyzed rpl-3, rpl-7a, rpl-10a, and rpl-12 mRNAs in these species by RT–PCR using primers that flank the conserved introns of each gene. For all four genes of both species, we identified two classes of cDNA comparable in size and abundance to those of C. elegans wild type (data not shown). Invariably, the smaller cDNAs were substantially more abundant than the larger cDNAs. We cloned and sequenced these cDNAs and established that they represent productively and unproductively spliced forms of rpl mRNAs equivalent to those of C. elegans. For rpl-3, rpl-7a, and rpl-12, the unproductively spliced mRNAs arise through the use of alternative splice sites equivalent in position to those of C. elegans (Fig. 4). The alternative 5′ splice site of rpl-10a in C. briggsae and C. remanei differs slightly from the site we identified in C. elegans (Fig. 4), but a C. elegans alternative 5′ splice site comparable to that of C. briggsae and C. remanei is, in fact, occasionally utilized, as demonstrated by C. elegans EST clones yk174a1 and yk527d7 (accession nos. C09644 and AV188689).
We examined the conservation of rpl-12 intron sequences and investigated possible alternative splicing in more distantly related nematode species. Using a degenerate PCR strategy, we cloned the relevant portion of rpl-12 from the soil nematode Diploscapter coronata and the more distantly related plant pathogenic nematode Pratylenchus penetrans (Blaxter et al. 1998). rpl-12 of P. penetrans contains an intron identical in size and position, and somewhat similar in sequence (44% identity), to intron 2 of C. elegans (Table 1). RT–PCR analysis similar to that described above demonstrates that the P. penetrans intron is alternatively spliced as in C. elegans (Fig. 4). rpl-12 of D. coronata contains an intron at the same position as C. elegans intron 2, but its size (367 nucleotides) and sequence are unlike the C. elegans intron. Analysis of D. coronata rpl-12 splicing via RT–PCR identified productively spliced rpl-12 mRNAs but failed to identify any unproductively or alternatively spliced mRNAs.
We conclude from these analyses that alternative splicing of rpl-12 likely arose in a species ancestral to the five species analyzed and that alternative splicing of the type described for C. elegans intron 2 was lost in D. coronata. We have not investigated, however, whether D. coronata contains additional rpl-12 genes in its genome or whether other introns of D. coronata rpl-12 are alternatively spliced.
The levels of RPL-12 expression autoregulate productive versus unproductive rpl-12 splicing
The conservation of alternatively spliced rpl introns suggests that the sequences of these introns are under strong selective pressure. We investigated two possible explanations for such conservation. First, we searched for small RNAs encoded within the alternatively spliced rpl-7a, rpl-10a, and rpl-12 introns. Small nucleolar RNAs, for example, are frequently encoded within introns of host ribosomal protein genes (Maxwell and Fournier 1995), and selection for such expressed sequences might, in principle, underlie rpl intron conservation. Using both hybridization and RT–PCR-based methods, which successfully detected control RNAs, we repeatedly failed to detect small RNAs emanating from the alternatively spliced rpl introns. C. elegans control RNAs included snoRNA U15 (accession no. Z75112) and the small, noncoding RNA encoded by lin-4 (Lee et al. 1993). We detected in the course of these experiments two previously uncharacterized small RNAs encoded within rpl-7a intron 2, which is not the alternatively spliced rpl-7a intron. rpl-7a intron 2 is 296 nt long and hybridizes to two small RNAs, a major species of approximately 250 nucleotides and a minor species of approximately 150 nucleotides. This intron is not similar to known small RNAs but does contain a 215-nt region that is 88% identical in C. briggsae. We have not investigated the function of these rpl-7a small RNAs, but our ability to identify them indicates that our methods of detection were robust. We therefore considered alternative explanations for sequence conservation of alternatively spliced rpl introns.
We investigated whether rpl alternative splicing is a mechanism to regulate the abundance of productively spliced rpl mRNAs. Specifically, we tested whether elevating the level of RPL-12 protein expression influences the balance of productive versus unproductive rpl-12 splicing. Feedback regulation by RPL-12 on rpl-12 splicing would provide an appealing mechanism to regulate productive rpl-12 mRNA and, hence, protein levels. For example, free nuclear RPL-12 (unassembled into ribosomes) might inhibit productive splicing or activate unproductive splicing and, thereby, posttranscriptionally regulate its own expression.
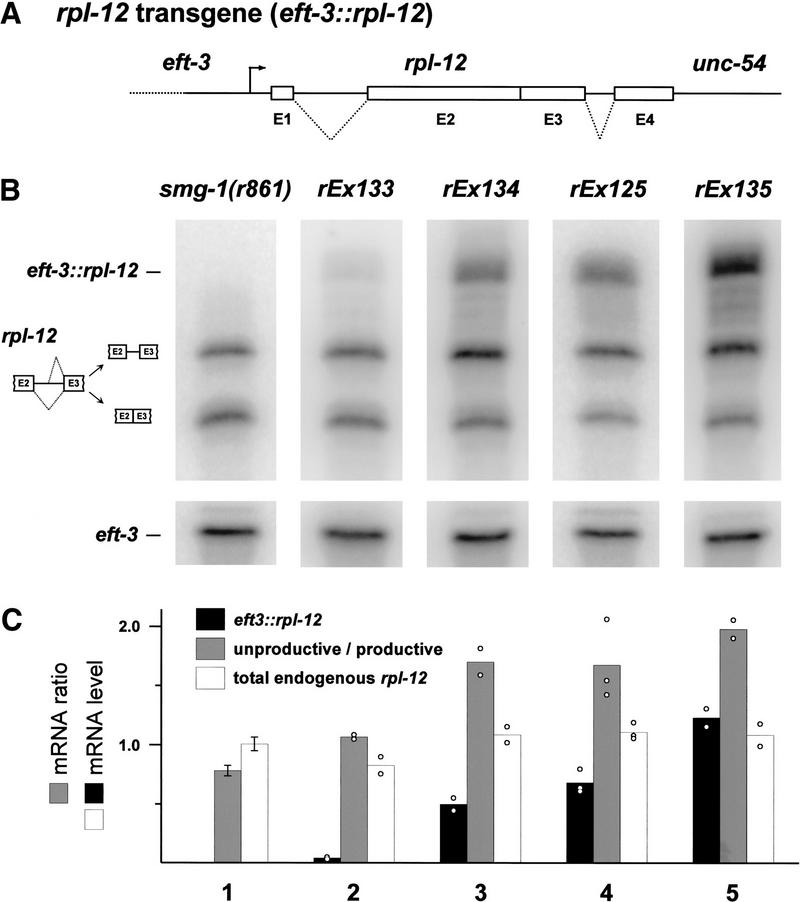
Plasmid TR#429, diagrammed in Figure 5A, is designed to express elevated quantities of RPL-12, whose amino acid sequence is identical to wild type. Features of this transgene (designated eft-3∷rpl-12) are: (1) RPL-12 is expressed under control of a promoter cloned from the C. elegans translation elongation factor 1-α (eft-3) gene. When fused to GFP, we found that an eft-3 promoter yielded high levels of expression throughout the soma. As with many C. elegans transgenes, eft-3∷GFP was expressed weakly or not at all in the germline (data not shown). (2) rpl-12 intron 2 has been removed from the transgene to avoid any potential complications caused by alternative splicing of the transgene message. (3) The rpl-12 3′ UTR has been replaced with that of unc-54, which causes transgene mRNA to be larger than endogenous rpl-12 mRNA and allows the mRNAs to be distinguished on Northern blots.
Figure 5.
Alternative splicing of rpl-12 message responds to RPL-12 protein levels. (A) Diagram of rpl-12 transgene (see text for description of elements). (B) Northern blots of oligonucleotide-cleaved total RNA probed with an rpl-12 cDNA clone (upper panels) and an eft-3 cDNA clone (lower panels). The oligonucleotide used for cleavage is complementary to rpl-12 3′UTR sequences, and thus does not affect the transgene mRNA. All transgenic strains (lanes 2–5) are derived from smg-1(r861) (lane 1). Four independent extrachromosomal array transformants are shown in lanes 2–5. (C) Quantitation of rpl-12 mRNAs exemplified in panel B. The quantity of transgene mRNA (black bars) and total endogenous mRNA (white bars) were normalized to eft-3 loading controls. Numerical values were scaled such that the total endogenous mRNA from the parental strain (lane 1) averages 1.0. Gray bars represent the ratio of unproductively to productively spliced endogenous rpl-12 mRNA. Open circles indicate individual data points (lanes 2–5), and error bars indicate ± standard error (lane 1; 6 independent trials).
We coinjected smg-1(r861) with eft-3∷rpl-12 and marker plasmids containing one or both of rol-6 (su1006) (Kramer et al. 1990) and eft-3∷GFP (a GFP gene driven by the eft-3 promoter), yielding extrachromosomal arrays rEx125, rEx133, rEx134, and rEx135. We also generated a control array, rEx132, marked with both rol-6 (su1006) and eft-3∷GFP but lacking an rpl-12 transgene.
Northern blots of transgenic and parental strains are shown in Figure 5B. The steady-state quantity of eft-3∷rpl-12 mRNA in strains carrying rEx125, rEx134, and rEx135 is comparable to, or somewhat higher than, that of endogenous, productively spliced rpl-12 mRNA (lanes 3–5). A strain carrying rEx133 (lane 2) expresses lower amounts of eft-3∷rpl-12 mRNA. We assume, but have not directly demonstrated, that strains expressing elevated levels of rpl-12 mRNA (derived from both eft-3∷rpl-12 and endogenous rpl-12) express elevated levels of RPL-12. We analyzed productive versus unproductive splicing of endogenous rpl-12 in such strains, using one (rEx135), two (rEx133 and rEx134), or three (rEx125) independent RNA samples for analysis (Fig. 5C). All strains in Figure 5 contain smg-1(r861). Thus, both productively and unproductively spliced rpl-12 mRNAs are expected to have normal stability.
In strains that overexpress RPL-12 (Fig. 5B, lanes 2–5), alternative splicing of endogenous rpl-12 mRNA is altered such that a greater proportion is spliced unproductively. In smg-1(r861) (lane 1) and in a control strain carrying rEx132, which does not contain an rpl-12 transgene (data not shown), ∼44% of endogenous rpl-12 mRNA is spliced unproductively (n = 7). In strains carrying rEx125, rEx134, or rEx135 (lanes 3–5), ∼64% of endogenous rpl-12 mRNA is spliced unproductively (n = 7). This represents a shift in splicing of ∼20% of rpl-12 pre-mRNA from the productive to the unproductive mode. The total quantity of endogenous rpl-12 mRNA is unchanged in array-containing strains (Fig. 5C, white bars). Only the ratio of productive and unproductive splicing is altered (Fig. 5C, gray bars). Strains that express the highest levels of eft-3∷rpl-12 mRNA (e.g., rEx135, lane 5) accumulate the highest proportion of unproductively spliced endogenous rpl-12 mRNA. As the eft-3 promoter does not yield germline expression of transgenes (see above), the data in Figure 5 likely underestimate the magnitude of RPL-12 effects on productive versus unproductive splicing. We conclude from these experiments that the level of RPL-12 expression regulates (either directly or indirectly) productive versus unproductive splicing of rpl-12 pre-mRNA. As discussed below, such feedback regulation might fine-tune the quantity of productive rpl-12 mRNA and, hence, RPL-12 itself.
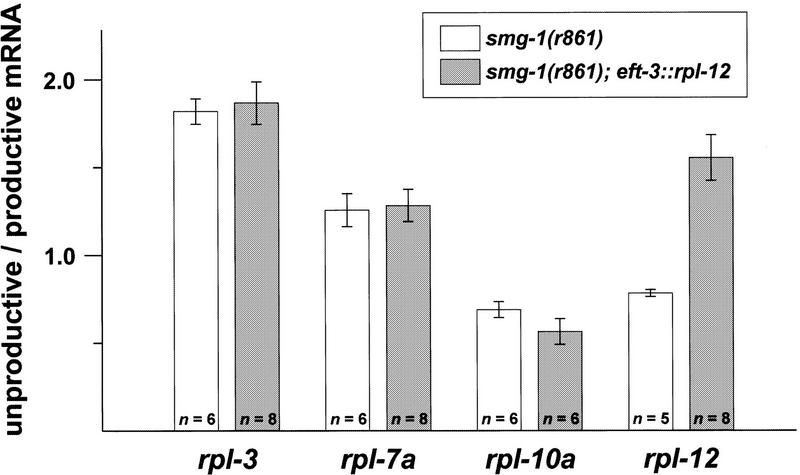
Levels of RPL-12 expression do not influence splicing of rpl-3, rpl-7a, and rpl-10a
The strong conservation of alternatively spliced rpl-3, rpl-7a, and rpl-10a introns suggests that expression of these genes will be regulated posttranscriptionally in a manner similar to that of rpl-12. Such regulation might occur by each RPL protein autoregulating its own splicing or, alternatively, all four genes might respond to a common regulatory signal. We therefore tested whether alternative splicing of rpl-3, rpl-7a, and rpl-10a is influenced by overexpression of RPL-12. In experiments analogous to those described above, we analyzed productive versus unproductive splicing of rpl-3, rpl-7a, and rpl-10a in strains that overexpress RPL-12. In contrast to our observations with rpl-12 mRNAs, we detected no significant change in the ratios of productively and unproductively spliced rpl-3, rpl-7a, and rpl-10a mRNAs (Fig. 6). We conclude that the level of RPL-12 expression does not regulate productive versus unproductive splicing of rpl-3, rpl-7a, or rpl-10a.
Figure 6.
Overexpression of RPL-12 does not influence alternative splicing of rpl-3, rpl-7a, and rpl-10a. Bars indicate the ratios of unproductively to productively spliced mRNA for each rpl gene; error bars indicate ± standard error. The ratios of alternatively spliced products were determined for the parental strain [smg-1(r861); white bars] and for four independent transgenic arrays that over-express RPL-12 (rEx125, rEx133, rEx134, and rEx135 considered as a group; gray bars). The number of independent trials is indicated within each bar.
Discussion
To better understand the role of mRNA surveillance in wild-type organisms, we have undertaken a screen for naturally occurring targets of this mRNA decay system. We used a cDNA-based representational difference analysis or RDA (Hubank and Schatz 1994) to identify C. elegans mRNAs whose abundance increases in smg(−) mutants, reasoning that such mRNAs should include natural targets of mRNA surveillance. We identified in such experiments alternatively spliced forms of four ribosomal protein mRNAs (rpl-3, rpl-7a, rpl-10a, and rpl-12). For each gene, alternative splicing of a key intron yields two distinct mRNAs. One message, resulting from productive splicing, encodes the expected full-length RPL protein. A second message, resulting from unproductive splicing, is exceptional in that a portion of the key intron is retained in the mature mRNA. Such messages contain in-frame stop codons and are consequently degraded by mRNA surveillance. Unproductively spliced rpl mRNAs are barely detectable in smg(+) animals, but their abundance often equals or exceeds that of their productively spliced counterparts in smg(−) mutants (Fig. 2). We interpret these results to indicate that wild-type strains express large amounts of productively and unproductively spliced rpl mRNAs but that unproductively spliced mRNAs are rapidly degraded by mRNA surveillance.
We believe for two reasons that smg(−) mutations affect the stability of unproductively spliced rpl mRNAs, rather than directly affecting rpl splicing. First, the structure of unproductively spliced rpl mRNAs predicts strongly that they should be substrates of mRNA surveillance (Hilleren and Parker 1999). Second, the quantity of productively spliced rpl mRNA is unchanged in smg(−) mutants, whereas the quantity of unproductively spliced rpl mRNA increases dramatically. Thus, the total quantity of rpl mRNA increases as much as threefold (rpl-3) in smg(−) mutants. If smg mutations directly affected splicing (e.g., by influencing splice site choice), the ratio of spliced products might be altered in smg(−) mutants, but the total quantity of rpl mRNA should be unchanged. Our results, therefore, are most consistent with interpretations in which smg(−) mutations affect the stability of, not the formation of, unproductively spliced rpl mRNAs.
Messenger RNA surveillance was initially suggested to be a proofreading system whereby the effects of erroneous gene expression are alleviated through the preemptive elimination of aberrant messages (Pulak and Anderson 1993). Unproductively spliced rpl mRNAs are, indeed, aberrant, but the conservation of both intron sequences and alternative splicing in other nematodes indicates that such mRNAs are unlikely to be mistakes of an error-prone splicing apparatus. What then is the function of alternative rpl splicing? One possibility is that alternatively spliced rpl mRNAs encode truncated RPL proteins that have an undiscovered role in translation. This model, however, seems insufficient to explain the striking conservation of alternatively spliced introns. The entirety of these introns (not just translated portions) is well conserved. Translation of unproductively spliced rpl-10a and rpl-7a mRNA terminates one and five codons, respectively, downstream of the exon/intron junction. Most of the retained intron sequences are therefore noncoding, yet they are highly conserved. Even the excised portions of rpl-3 and rpl-12 introns are well conserved, although such sequences are not present in mature mRNA. Taken together, these observations suggest that, even if truncated RPL proteins are functionally significant, there are likely to be additional evolutionary constraints on these introns.
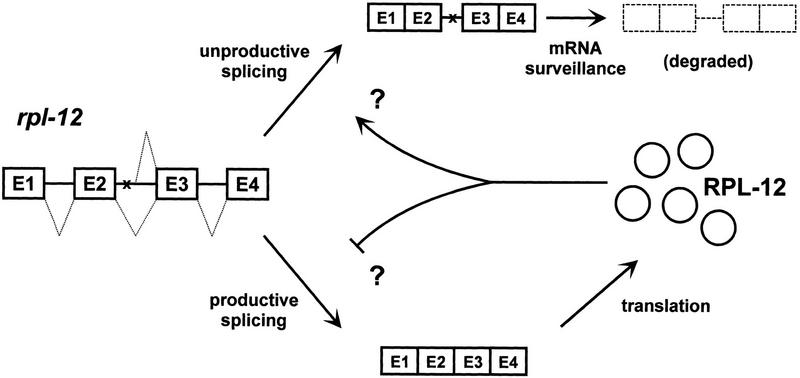
We believe a more likely function for alternative splicing of rpl mRNAs is regulation of rpl gene expression. Our studies indicate that a sensor of RPL-12 expression, perhaps the level of RPL-12 itself, affects rpl-12 splicing by either negatively regulating productive splicing or positively regulating unproductive splicing. Such negative feedback would provide a mechanism to control the levels of free RPL-12 (Fig. 7).
Figure 7.
Model for RPL-12 feedback regulation. Productively spliced mRNA encodes for RPL-12. When present in excess, RPL-12 either positively or negatively regulates splicing of its own pre-mRNA, as shown. This reduces the level of productively spliced mRNA and increases the level of unproductively spliced mRNA. The unproductively spliced mRNA is subsequently eliminated by mRNA surveillance.
How might such autoregulation be accomplished? RPL-12 binds rRNA as part of the 60S ribosomal subunit (Uchiumi and Kominami 1997). Perhaps RPL-12 also binds its own pre-mRNA in or near intron 2 and, in doing so, modulates rpl-12 splicing. Ribosomal subunits are assembled within nuclei, and RPL proteins transported to the nucleus are, in principle, available to modulate splicing. Perhaps the quantity of free RPL-12 (unassembled into ribosomal subunits) regulates directly its own pre-mRNA splicing. Under conditions of RPL-12 excess or deficit, splicing would be directed toward the unproductive or productive modes, respectively (Fig. 7). The striking sequence conservation of alternatively spliced rpl introns may indicate that the alternatively spliced introns fold into specific structures to be bound by RPL-12 or other relevant regulatory proteins. Secondary structures for the alternatively spliced Caenorhabditis rpl-12 introns, predicted by m-fold (Mathews et al. 1999), share similarities with the rRNA binding site of RPL-12. The P. penetrans intron, however, is not predicted to form a similar structure, and it remains to be seen whether the predicted Caenorhabditis structures are of functional relevance.
The quantity of individual ribosomal proteins is tightly regulated, ensuring an appropriate stoichiometry for ribosome assembly. In prokaryotes, such regulation occurs through both transcriptional and translational mechanisms (for review, see Nomura et al. 1984). In eukaryotes, additional control mechanisms have been described. For example, two yeast ribosomal proteins, rpL30 and rpS14B (encoded by RPL30 [RPL32] and RPS14B [CRY2]), autoregulate their expression by inhibiting their own splicing. Such inhibition decreases the quantity of mature, productive mRNA and increases the quantity of pre-mRNA (Vilardell and Warner 1994; Li et al. 1995). Unspliced RPS14B pre-mRNAs are cytoplasmic and subjected to mRNA surveillance, as their quantity is elevated in upf1− mutants (Li et al. 1995). Interestingly, two additional natural targets of yeast mRNA surveillance are pre-mRNAs of the ribosomal protein genes RPL28 [CYH2] and RPS17B [RP51B] (He et al. 1993). Perhaps RPL28 and RPS17B are regulated in a manner similar to RPL30 and RPS14B.
Expression of ribosomal protein genes is regulated broadly by transcription, but the posttranscriptional mechanisms described in yeast and nematodes may fine-tune ribosomal protein gene expression. In yeast, the decision to splice or not to splice certain ribosomal protein pre-mRNAs is regulated. In C. elegans, the decision to splice productively or unproductively is regulated. In both cases, however, unproductive and unneeded rpl mRNAs are degraded by mRNA surveillance. Surveillance contributes to the process only indirectly and after the important regulatory decisions have been made. However, if unproductively spliced mRNAs are merely by-products of gene regulation, why are they degraded? Many mRNAs that contain nonsense mutations encode disruptive polypeptide fragments (Cali and Anderson 1998; Pulak and Anderson 1993). Messenger RNA surveillance lessens the deleterious consequences of such mRNAs by ensuring their rapid degradation. Were it not for mRNA surveillance, unproductively spliced rpl mRNAs would be among the most abundant mRNAs in a cell. Perhaps some or all of the morphogenetic phenotypes of smg(−) mutants are caused by the accumulated effects of translating these and other unproductively spliced mRNAs.
Messenger RNAs encoding SRp20 and SRp30b, two C. elegans serine/arginine-rich (SR) proteins, are alternatively spliced (Morrison et al. 1997) in a manner reminiscent of rpl mRNAs. For both SRp20 and SRp30b, an alternatively spliced exon contains an upstream stop codon, and these mRNAs are selectively destabilized in a smg-dependent manner (Morrison et al. 1997). As SR proteins are splicing regulatory factors (Tacke and Manley 1999), perhaps this is another example of C. elegans autoregulation mediated via unproductive splicing coupled with mRNA surveillance. The murine homolog of SRp20 is alternatively spliced in a similar manner, and alternative splicing is, in fact, responsive to SRp20 protein levels (Jumaa and Nielsen 1997).
How common are unproductively spliced mRNAs like those described here? The method of representational difference analysis is naturally biased toward identifying abundant transcripts. Such bias may have contributed to our identifying ribosomal protein mRNAs in preference to less abundant transcripts, like the SR protein mRNAs mentioned above. Regulatory mechanisms that yield unproductively spliced mRNAs may be more common than has been appreciated. Identifying such mRNAs may require the use of organisms deficient for mRNA surveillance, as the unproductively spliced mRNAs will be unstable in wild-type backgrounds.
Previous discussions of the in vivo role of mRNA surveillance focused on the elimination of aberrant mRNAs arising through somatic mutation or errors in gene expression. The natural targets that we have found, however, are alternative transcripts that arise not through errors but through feedback regulation of splicing. Other natural targets, discussed above (He et al. 1993; Li et al. 1995; Morrison et al. 1997), may reflect similar regulatory processes. An important unanswered question is how significant are the errors of gene expression, as originally proposed for surveillance, compared to the unneeded byproducts of regulated mRNA processing? Unproductively spliced rpl mRNAs appear to be the remnants of posttranscriptional regulation, and we suggest that an important function of mRNA surveillance is to rid the cell of such remnant RNAs.
Materials and methods
Nematode strains
C. elegans nematodes were raised at 20°C as described by Lewis and Fleming (1995). Strains used were N2 (wild type), TR374 [unc-54(r315)], TR1331 [smg-1(r861)], and TR1533 [unc-54(r315) smg-2(r863)]. Transgenic strains generated were TR2401 [smg-1(r861); rEx125], TR2402 (smg-1(r861); rEx132], TR2403 [smg-1(r861); rEx133], TR2404 [smg-1(r861); rEx134], and TR2405 [smg-1(r861); rEx135].
C. briggsae (strain AF16) and C. remanei (strain SB146) DNAs and mRNAs were gifts from E. Haag, A. Puoti, and D. Rudel (HHMI/UW, Madison, Wisconsin). Wild isolates of D. coronata and P. penetrans were gifts from A. MacGuidwin (UW, Madison, Wisconsin).
RNA preparation and cDNA synthesis
C. elegans RNA for RDA and Northern analysis was prepared from worms grown in liquid culture (Lewis and Fleming 1995). For RDA, 2.5 volumes sonication buffer (2% SDS, 50 mm Tris, 10 mm EDTA at pH 8.0) and 2.5 volumes phenol/chloroform (1:1) were added to frozen worm pellets. Immediate sonication of pellets, RNA extraction, and poly(A) selection were carried out as described by Klein and Meyer (1993). For Northern analysis, frozen worm pellets were ground by mortar and pestle in liquid Nitrogen, then RNA was extracted using Trizol, following manufacturer's recommendations, including an optional centrifugation step to remove insoluble material prior to phase separation (GIBCO BRL). Poly(A) selection was performed using the PolyATtract mRNA Isolation System (Promega).
Nematodes for rpl-12 overexpression analysis were grown on plates as described by Lewis and Fleming (1995). The proportion of transgenic worms was increased by manually removing worms that had lost marker expression during population growth. Worms were washed from plates in M9 buffer (3 g/l KH2PO4, 6 g/l Na2HPO4, 0.5 g/l NaCl, 1 mm MgSO4) and concentrated by centrifugation (5 min at 1000g). Worm pellets and remaining buffer were diluted in four volumes Trizol, frozen at −80°C, heated to 65°C for 90 sec, vortexed briefly, then incubated at room temperature for 15 min. Insoluble material was removed by centrifugation (5 min at 15,800g, 4°C), and RNA was extracted from aqueous phase following manufacturer's recommendations (GIBCO BRL).
RNAs from D. coronata and P. penetrans were also isolated from plate-grown worms using Trizol reagent. P. penetrans nematodes required sonication in Trizol before RNA isolation.
First-strand cDNA was prepared from total RNA (D. coronata and P. penetrans) or poly(A)+ RNA (C. briggsae, C. elegans, and C. remanei) using (dN)6 or (dT)14 oligonucleotides and the Ready-To-Go You-Prime First-Strand cDNA synthesis kit according to manufacturer's recommendations (Pharmacia). Second-strand cDNA was synthesized as described by Gubler (1987).
Representational difference analysis
Representational difference analysis (RDA) was performed by subtracting unc-54(r315) driver cDNAs against unc-54(r315) smg-2(r863) tester cDNAs, essentially as described for DNA analysis by Lisitsyn and Wigler (1995), and modified for cDNA analysis, essentially as described by Hubank and Schatz (1994). Tester:driver ratios used were 1:40, 1:400, and 1:8000 for first, second, and third round subtractions, respectively.
The rpl-12 alternative transcript was identified from second-round products using DpnII-digested representations and the Bgl primers (at stock concentrations of 62 pmole/μl) described by Lisitsyn and Wigler (1995). The rpl-3 and rpl-7a alternative transcripts were identified from second-round products and the rpl-10a alternative transcript from third round products, using TaqαI-digested representations. For these latter representations, primer pair set one was replaced with 5′-GCACTCTCCAGCCTCTCACCGCAT-3′ and 5′-CGATGCGGTGA-3′, set two with 5′-CCGACGTGCACTATCCATGAACAT-3′ and 5′-CGATGTTCATG-3′, and set three with 5′-GGCAACTGTGCTATCCGAGGGAAT-3′ and 5′-CGATTCCCTCG-3′.
Northern analysis
RNAs were separated for Northern analysis on 1.4% agarose/6.6% formaldehyde gels and transferred to Zeta-Probe nylon membranes (Bio-Rad) as described by Rosen and Villa-Komaroff (1990). Oligonucleotide-cleaved RNAs were separated on 4.5% polyacrylamide/8m urea gels and transferred to GeneScreen Plus nylon membranes (NEN) as described by Rasmussen and Culbertson (1996).
Radiolabeled DNA probes were generated by amplifying plasmid sequences for 25 cycles under standard PCR conditions, but using 5 μm each dCTP, dGTP, and dTTP, and 0.2 μm [α-32P]dATP (6,000 Ci/mmole). Probes were hybridized to Northern blots as described by Church and Gilbert (1984). Signals were detected using a STORM 860 PhosphorImager and quantified using ImageQuant software v5.0 (Molecular Dynamics).
RNA cleavage
RNAs were cleaved using a modification of the procedure described by Shibahara et al. (1987). The amount of total RNA to be analyzed by Northern (2.5–5 μg) was added to a 9-μl reaction containing 1.1× RNase H buffer (USB) and 10 pmole oligonucleotide specific to the RNA to be cleaved (see below). The reaction was incubated at 65°C for 2 min, then slow-cooled over 1 hr to 30°C. 0.1 units Ribonuclease H (USB) in a 1μl volume was added, and the reaction was incubated at 30°C for 30 min. The reaction was stopped by adding an equal volume of formamide loading buffer (Sambrook et al. 1989).
Oligonucleotides used for cleavage were: 5′-GCCACTTCTTGGCGTACTTG-3′ for rpl-3, 5′-CGGCACGAGCACGGAGAC-3′ for rpl-7a, 5′-CTTCAAGTCATCGGCGCTC-3′ for rpl-10a, and 5′-GGAACAACAAAAACAAGTGACAG-3′ for rpl-12.
DNA cloning
Sequence identity within rpl coding sequences was high enough among the Caenorhabditis species to allow PCR cloning of C. briggsae and C. remanei rpl DNA sequences using oligonucleotides designed for C. elegans. Alternative cDNAs from the C. briggsae and C. remanei rpl genes were PCR amplified using coding sequence oligonucleotides flanking the alternatively spliced introns.
rpl-12 DNA from D. coronata and P. penetrans was amplified using nested degenerate oligonucleotides. The initial primer pair was 5′-CATGCCNCCNAARTTYGAYCC-3′ and 5′-GCCNGGDATYTCDATYTCNCC-3′; the nested primer pair was 5′-CGGNGANGAYATHGCNAARGC-3′ and 5′-CTCDATDATRTCRTGNGGRTG-3′. To amplify putative alternative cDNA products, forward primers were designed with sequences early in the relevant introns and reverse primers were designed with downstream coding sequences.
Plasmids TR#428 and TR#429 were constructed for expression of GFP and rpl-12, respectively, in transformed nematodes. Both plasmids contain a pBluescript II KS(+) backbone (Stratagene) and approximately 600 nucleotides of eft-3 upstream sequence to drive transgene expression. TR#428 contains the GFP coding sequence, including artificial introns, and the unc-54 3′UTR and CPS cloned from plasmid pPD93.48 (A. Fire, S. Xu, J. Ahnn, and G. Seydoux, pers. comm.). TR#429 contains an rpl-12 DNA/cDNA fusion generated by replacing an internal DNA fragment spanning the second intron with the comparable cDNA fragment from productively spliced mRNA. TR#429 also contains the unc-54 3′UTR and CPS.
Nematode transformation
Nematodes were transformed by microinjection using standard procedures (Mello and Fire 1995). PvuII-digested N2 DNA (100 ng/μl) was used as carrier to increase complexity of resulting extra-chromosomal arrays (S. Xu, W. Kelly, and A. Fire, unpubl.). All injection mixes also contained SmaI-digested plasmid pRF4 (Kramer et al. 1990) at 4 ng/μl (rEx132) or 2 ng/μl (all other arrays: not expressed from rEx134). To generate arrays rEx132, rEx133, rEx134, and rEx135, ScaI-digested plasmid TR#428 (GFP) was included at 4 ng/μl, and for arrays rEx125, rEx133, rEx134, and rEx135, SmaI-digested plasmid TR#429 (rpl-12) was included at 1 ng/μl.
Sequence analysis
Caenorhabditis and Pratylenchus rpl intron sequences were aligned using the CLUSTAL V algorithm (Higgins and Sharp 1989), then adjusted manually.
For the general survey of C. elegans and C. briggsae sequence conservation, we analyzed C. briggsae fosmids for which synteny with C. elegans had been established (see http://genome.wustl.edu). Corresponding coding and intron sequences from the two species were identified using the web-based Intronerator (Kent and Zahler 2000). Sequence conservation for individual introns and exons was established by performing simple optimal global alignments (Myers and Miller 1988) using the Web-based ALIGN program from the Institut de Génétique Humaine (see http://www2.igh.cnrs.fr).
Accession numbers for C. elegans sequences: rpl-3 (Z66495 and Z49936, gene F13B10.2), rpl-7a (AC006729, coordinates 12440–13828), rpl-10a (AC024851, coordinates 15471–16342), and rpl-12 (Z82274, gene JC8.3). C. briggsae sequences: rpl-3 (AF247847), rpl-7a (AF247849), rpl-10a (AF247848), and rpl-12 (AF246691). C. remanei sequences: rpl-3 (AF247852), rpl-7a (AF247853), rpl-10a (AF247854), and rpl-12 (AF247851). P. penetrans sequence: rpl-12 (AF247850). The nomenclature for rpl-7a and rpl-10a follows the convention for ribosomal proteins in higher eukaryotes and refers to genes that are not homologous with rpl-7 and rpl-10 (encoded in C. elegans by F53G12.10 and F10B5.1, respectively).
Acknowledgments
We thank E. Haag, A. Puoti, and D. Rudel for providing nucleic acids, A. MacGuidwin for providing nematodes, A. Grimson for sharing unpublished data and RNA samples, and members of the Anderson lab for their support. This work was supported by a National Institutes of Health individual research grant (GM50933), the University of Wisconsin Training Grant in Genetics, a University of Wisconsin WARF Graduate Fellowship, and a Predoctoral Graduate Fellowship from the National Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL andersn@facstaff.wisc.edu; FAX (608) 262-2976.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.819900.
References
- Ainscough R, Bardill S, Barlow K, Basham V, Baynes C, Beard L, Beasley A, Berks M, Bonfield J, Brown J, et al. Genome sequence of the nematode Caenorhabditis elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Altamura N, Groudinsky O, Dujardin G, Slonimski PP. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. doi: 10.1016/0022-2836(92)90545-u. [DOI] [PubMed] [Google Scholar]
- Baumann B, Potash MJ, Kohler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985;4:351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejsovec A, Anderson P. Functions of the myosin ATP and actin binding sites are required for C. elegans thick filament assembly. Cell. 1990;60:133–140. doi: 10.1016/0092-8674(90)90723-r. [DOI] [PubMed] [Google Scholar]
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Steward K. RNA processing and gene structure. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- Cali BM, Anderson P. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol Gen Genet. 1998;260:176–184. doi: 10.1007/s004380050883. [DOI] [PubMed] [Google Scholar]
- Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in C. elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, González CI, Peltz SW. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays. 1999;21:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fitch DHA, Bugaj-Gaweda B, Emmons SW. 18S ribosomal RNA gene phylogeny for some Rhabditidae related to Caenorhabditis. Mol Biol Evol. 1995;12:346–358. doi: 10.1093/oxfordjournals.molbev.a040207. [DOI] [PubMed] [Google Scholar]
- Fitch DHA, Thomas WK. Evolution. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1997. pp. 815–850. [PubMed] [Google Scholar]
- Gubler U. Second-strand cDNA synthesis: mRNA fragments as primers. Methods Enzymol. 1987;152:330–335. doi: 10.1016/0076-6879(87)52038-9. [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Hilleren P, Parker R. Mechanisms of mRNA surveillance in eukaryotes. Annu Rev Genet. 1999;33:229–260. doi: 10.1146/annurev.genet.33.1.229. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM. The Intronerator: exploring introns and alternative splicing in C. elegans. Nucleic Acids Res. 2000;28:91–93. doi: 10.1093/nar/28.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RD, Meyer BJ. Independent domains of the sdc-3 protein control sex determination and dosage compensation in C. elegans. Cell. 1993;72:349–364. doi: 10.1016/0092-8674(93)90113-5. [DOI] [PubMed] [Google Scholar]
- Kramer J, French RP, Park E, Johnson JJ. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–2090. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The heterochronic gene lin-4 of C. elegans encodes two small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee B-S, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- Li Z, Paulovich AG, Woolford JL., Jr Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol Cell Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsyn N, Wigler M. Representational difference analysis in detection of genetic lesions in cancer. Methods Enzymol. 1995;254:291–304. doi: 10.1016/0076-6879(95)54021-0. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M, Harris KS, Roth MB. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci. 1997;94:9782–9785. doi: 10.1073/pnas.94.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E, Miller W. Optimal alignments in linear space. CABIOS. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Rasmussen TP, Culbertson MR. Analysis of yeast trimethylguanosine-capped RNAs by Midwestern blotting. Gene. 1996;182:89–96. doi: 10.1016/s0378-1119(96)00519-7. [DOI] [PubMed] [Google Scholar]
- Rosen KM, Villa-Komaroff L. An alternative method for the visualization of RNA in formaldehyde agarose gels. Focus. 1990;12:23–24. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shibahara S, Mukai S, Nishihara T, Inoue H, Ohtsuka E, Morisawa H. Site-directed cleavage of RNA. Nucleic Acids Res. 1987;15:4403–4415. doi: 10.1093/nar/15.11.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Manley JL. Determinants of SR protein specificity. Curr Opin Cell Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Kominami R. Binding of mammalian ribosomal protein complex P0·P1·P2 and protein L12 to the GTPase-associated domain of 28S ribosomal RNA and effect on the accessibility to anti-28S RNA autoantibody. J Biol Chem. 1997;272:3302–3308. doi: 10.1074/jbc.272.6.3302. [DOI] [PubMed] [Google Scholar]
- Vernet C, Ribouchon M-T, Chimini G, Pontarotti P. Structure and evolution of a member of a new subfamily of GTP-binding proteins mapping to the human MHC class I region. Mamm Genome. 1994;5:100–105. doi: 10.1007/BF00292335. [DOI] [PubMed] [Google Scholar]
- Vilardell J, Warner JR. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes & Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- Welch EM, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]