Abstract
Feline immunodeficiency virus (FIV) infects domestic cats and at least 20 additional species of non-domestic felids throughout the world. Strains specific to domestic cat (FIVFca) produce AIDS-like disease progression, sequelae and pathology providing an informative model for HIV infection in humans. Less is known about the immunological and pathological influence of FIV in other felid species although multiple distinct strains of FIV circulate in natural populations. As in HIV-1 and HIV-2, multiple diverse cross-species infections may have occurred. In the Serengeti National Park, Tanzania, three divergent subtypes of lion FIV (FIVPle) are endemic, whereby 100% of adult lions are infected with one or more of these strains. Herein, the relative distribution of these subtypes in the population are surveyed and, combined with observed differences in lion mortality due to secondary infections based on FIVPle subtypes, the data suggest that FIVPle subtypes may have different patterns of pathogenicity and transmissibility among wild lion populations.
Keywords: FIVPle, lions, CDV, Babesia
Introduction
Feline immunodeficiency virus (FIV) is a lentivirus closely related to HIV and SIV. In domestic cats (Felis catus), FIV infection results in immune pathology, secondary infections, and death. The parallels between human and feline AIDS (FAIDS) have been explored for further understanding of HIV/AIDS transmission, infection, and pathology (Bendinelli et al., 1995; Burkhard and Dean, 2003; Elder et al., 1998; Elder et al., 2010; Henriksen et al., 1995; Stump and VandeWoude, 2007). As with HIV and SIV models, there is considerable variation in transmission, course of infection, and outcome of FIV infections in domestic cats. Some variation likely results from host genetic restriction factors that influence the viral life cycle, similar to those described in humans (Lochelt et al., 2005; Munk et al., 2008; Munk et al., 2007; Troyer et al., 2008; VandeWoude et al., 2010). However, differences in pathogenicity have also been demonstrated among genetically distinct subtypes of FIV that circulate in domestic cats (de Monte et al., 2002; Elder et al., 2010; Pedersen et al., 2001; Weaver, 2010).
Most experimental viruses representing FIVFca subtypes are cell-line adapted but nonetheless retain recognized differences in pathogenicity. For example, FIV-CPG derived strains generally result in high initial viral loads and a faster progression to disease, especially in young cats (de Rozieres et al., 2004a; de Rozieres et al., 2004b; de Rozieres et al., 2008). In contrast, cats infected with FIV-A strains often remain asymptomatic for longer periods of time, with lower initial viral loads, though viral growth kinetics are similar in adult cats once the acute stage of infection has passed (de Rozieres et al., 2008; Pedersen et al., 2001; Sparger et al., 1994). FIV subtype A strains are often neurotrophic and neurotoxic, producing CNS symptoms similar to those seen in HIV-1 infection (Gruol et al., 1998; Henriksen et al., 1995; Johnston et al., 2000; Meeker, 2007; Phillips et al., 1994; Phillips et al., 1996; Power et al., 1998).
Species-specific FIV viruses infect other felids and are distributed throughout the world, yet little is known about their immunological and pathological effects in wild populations (Brown et al., 2010; Carpenter et al., 1996; Franklin et al., 2008; Franklin et al., 2007; Olmsted et al., 1992; Troyer et al., 2005). Long term surveillance of non-domestic felids infected with FIV, as well as evidence from free-ranging populations of pumas (Puma concolor) and lions (Panthera leo), suggest that these viruses are ancient, host-adapted, and have little to no negative impact on life-history parameters such as longevity (Biek et al., 2006; Carpenter and O'Brien, 1995; Packer et al., 1999). However, a few clinical studies have revealed that individuals of these same species may demonstrate FIV-associated immune depletion and, in some cases, AIDS-like complications and death (Brennan et al., 2006; Brown et al., 2010; Bull et al., 2002; Bull et al., 2003; Roelke et al., 2009; Roelke et al., 2006). Data on life history and clinical parameters are rare, and seldom collected in the same population.
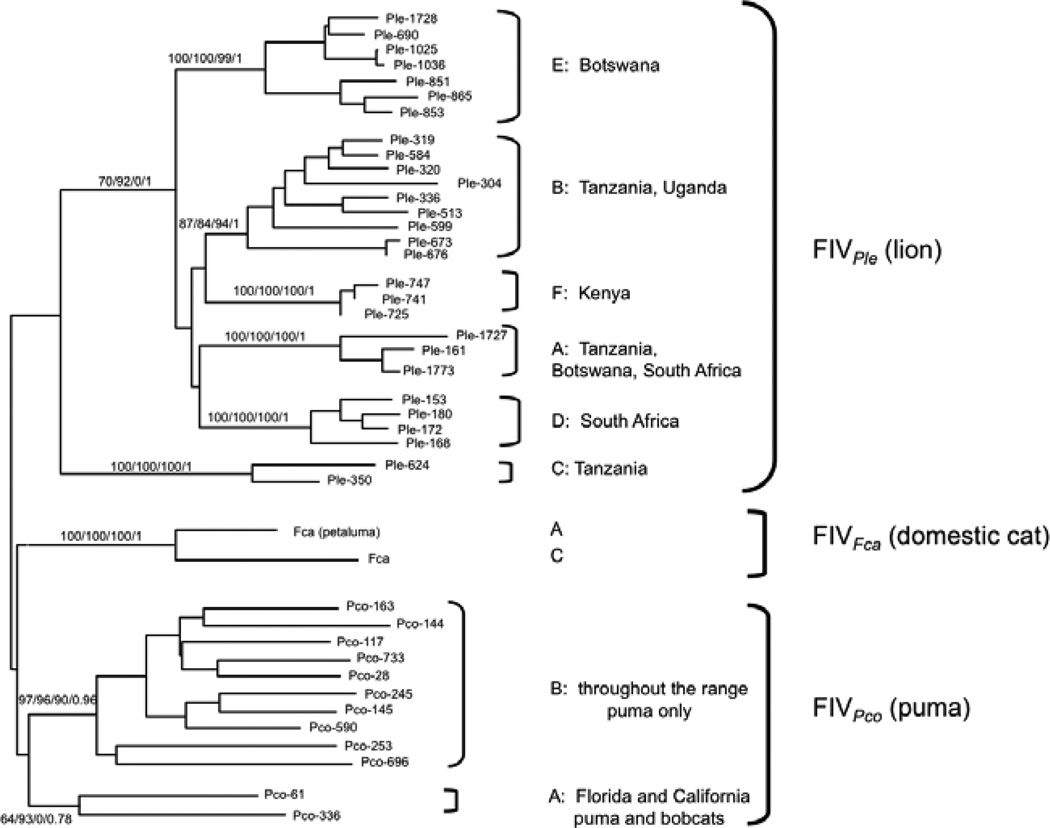
At least six genetically distinct strains of lion FIV (FIVPle) circulate in wild populations of African lions (Panthera leo) (figure 1; Antunes et al., 2008; O'Brien et al., 2006; Pecon-Slattery et al., 2008b; Troyer et al., 2005). FIVPle subtypes demonstrate distinct phylogeographic distributions, suggesting prolonged host association, perhaps predating the Late-Pleistocene expansions of lions (Antunes et al., 2008). Probably as a result of the highly social nature of lions, FIVPle-infected populations have high prevalence of seropositive individuals, often approaching 100% in adults, while other populations remain completely uninfected (Antunes et al., 2008; Brown et al., 1994; Troyer et al., 2005). This “all or nothing” distribution of FIV in lion population makes appropriate comparisons of infected vs. uninfected lions a challenge. Differences in FIVPle status may be confounded by important environmental parameters affecting lion health including other infectious agents, prey abundance, and water availability.
Figure 1.
Phylogenetic relationship of geographically distributed FIV subtypes from lions, domestic cats, and puma adapted from O’Brien et al. (2006). Bootstraps and posterior probibilities of phylogentic reconstructions are shown (maximum likelihood/minimum evolution/maximum parsimony/Bayesian).
Epidemiological and life history vs. clinical and immunological studies on FIVPle - infected lions have been collected in different populations with only limited overlap. No evidence has been found of decreased lifespan in FIVPle-endemic populations in the Serengeti National Park and the Ngorongoro crater, where most lions are infected at an early age; no direct comparison with uninfected animals was possible (Packer et al., 1999). By contrast, infected individuals from Botswana and Tanzania demonstrated multiple clinical features of chronic immune depletion similar to human, simian, and domestic cat AIDS (Roelke et al., 2009; Roelke et al., 2006). The phylogeographic distribution of FIVPle raises the question as to whether FIVPle subtypes confer differential pathogenicity. However, to date, no study has correlated FIVPle subtypes with clinical or life history outcomes, in part because of the same geographic and environmental co-factors mentioned above.
Several lines of evidence suggest that FIVPle subtypes may be substantially different from each other. Two FIVPle strains, FIVPle subtype E and FIVPle subtype A, circulate in Botswana while three, FIVPle subtypes A, B, and C, occur in the Serengeti National Park (Antunes et al., 2007; Brown et al., 1994; O'Brien et al., 2006; Troyer et al., 2004; Troyer et al., 2005). Strains representing the predominant subtype in each of these populations, FIVPle-B from the Serengeti and FIVPle-E from Botswana, have been fully sequenced revealing remarkable differences between these subtypes (Pecon-Slattery et al., 2008a). While these two strains form a lion-specific clade when full length viruses are aligned, the envelope (env) sequence by itself displays a different phylogenetic relationship that suggests an historic recombination event between distantly related viruses, making FIVPle-E env seemingly more similar to domestic cat FIV than to FIVPle-B. In contrast, FIVPle-B env groups with other non-domestic cat env gene sequences (Carpenter and O'Brien, 1995; Pecon-Slattery et al., 2008a; Smirnova et al., 2005). The env gene is responsible for several aspects of lentiviral pathogenicity; changes in these sequences can affect receptor binding, antibody affinity, and target cell specificity. Therefore, these differences have been hypothesised to influence disease outcomes (Barlough et al., 1993; Burkhard and Dean, 2003; Elder et al., 2010; Patrick et al., 2002; VandeWoude and Apetrei, 2006).
The FIVPle subtypes circulating in Serengeti lions are more divergent then the FIVPle found in other African lion populations. Specifically, FIVPle-C pol is as different from the other two Serengeti subtypes as from FIV strains that infect other felid species (Troyer et al., 2005). Further, within-subtype diversity is much higher for FIVPle-B than for the other two subtypes. Phylogenetic reconstruction of the three Serengeti FIVPle subtypes suggest different ancestral evolutionary trajectories and/or selection pressures (Troyer et al., 2004). For example, FIVPle-B is representative of a widely distributed East African clade found across Tanzania, Uganda, and Kenya. FIVPle-A appears to have spread from Southern Africa as the most closely related lion viruses are found in South Africa and Botswana. Unique to the Serengeti, FIVPle-C is distantly related to other FIVPle viral subtypes (Antunes et al., 2008; O'Brien et al., 2006) and exhibits relatively low within strain diversity consistent with either recent introduction or stronger selective pressure from the host immune system. This pattern likely arose from three separate introductions of FIVPle to this population in the recent past, a hypothesis supported by population genetic analyses of lion microsatellite loci, autosomal sequences, and mitochondrial sequences (Antunes et al., 2008).
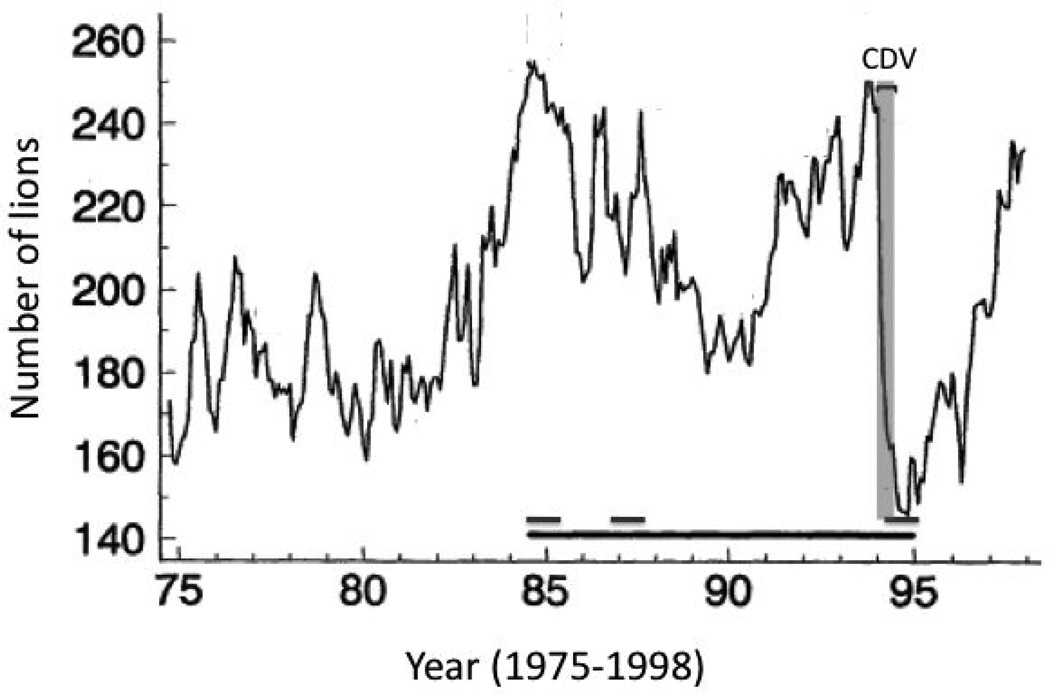
Because these three divergent strains exist in a single population, it is possible to compare both population dynamics and epidemiology of these strains while reducing the influence of confounding environmental factors. Here we present an analysis of FIVPle subtype composition throughout the lifespan of Serengeti lions and during a deadly disease outbreak. In late 1993, an unprecedented number of Serengeti lions died or disappeared; several were observed with neurological disorders including myoclonus ataxia and grand-mal seizures. From November 1993 to August 1994, the lion population in the long-term study area in the southeastern part of the Serengeti National Park plummeted by 39% with 78% of deaths occurring between the beginning of January and the end of March 1994 (Figure 2). The abruptness and extent of mortality indicated the emergence of a deadly pathogenic agent, initially suspected to be a neurotropic FIV strain. However, comprehensive clinical sampling and an assessment of over 100 lions during the outbreak revealed no relationship between disease and FIV infection, excluding FIVPle as the primary pathogen. The dead and sick lions showed reactivity to canine distemper virus (CDV) monoclonal antibodies, and post mortem immunohistopathology of brain tissue confirmed CDV as the causative agent. PCR detection and CDV viral sequences indicated that the strain had spread from domestic dogs living in villages adjacent to the National Park (Carpenter et al., 1998; Munson et al., 2008; Packer et al., 1999; Packer et al., 2005; Roelke-Parker et al., 1996).
Figure 2.
Study population over time. In the time-period shown here, the population rose and fell cyclically until 1994, when it plummeted due to the high-mortality CDV epidemic shown in grey; in 1999 the population returned to previous levels (Packer et al., 2005). The black bar indicates the time period from 1984 through 1995 when samples were assessed for FIVPle subtype distribution. The three dark grey bars (above the back bar) represent periods of high intensity sampling.
A similar deadly outbreak of CDV occurred in the nearby Ngorongoro Crater in 2001, while 5 other CDV outbreaks in the area over a 20 year time span were not associated with any measureable mortality (Munson et al. 2008). The high-mortality of the 1994 Serengeti and 2001 Ngorongoro CDV outbreaks were apparently linked to co-infection with high levels of Babesia, a tick-borne hemoparasite that proliferated during severe droughts. The associated tick infestations of the primary prey species of lions is believed to have led to the high levels of Babesia observed during the CDV outbreak and to be an important co-factor in these mortality events (Munson et al., 2008).
Although FIVPle was not the primary cause of the fatal outbreaks, the clinical evidence that FIVPle can lead to immune suppression in lions raises the question of whether FIVPle could have played a supportive or interactive role. We therefore treated these die-offs as “naturalistic experiments”: challenges to lions’ health that allowed an opportunity to test whether different strains of FIVPle might have had differential influences on CDV alone or on CDV/Babesia combined pathogenesis.
Materials and Methods
FIVPle subtype designation
Blood samples from lions, had been used as a source of DNA, with subtype-specific PCR performed (reported in Troyer et al., 2004). The generated FIV sequences had been assigned to subtypes A, B, and C based on phylogenetic analysis of the resultant 300 bp sequences from the pol gene region produced by the strain-specific PCR primers (Troyer et al., 2004, Troyer et al., 2005, Antunes et al., 2008). For the current study, novel analyses used the previously generated FIVPle subtypes that had been determined for each lion at one collection time point and which were assumed to remain the same throughout the life of the lion. Whenever possible, a collection time point nearest to the CDV/Babesia high mortality outbreak was used. For three lions, subtype data was available at time points 5–10 years apart; in all three cases subtype composition was consistent over time. The novel analyses were as follows:
Age and longevity analysis
Birth dates, capture dates, and death dates were used to determine subtype distribution of the population as a function of age and longevity as a function of subtype. Because of the inherent difficulties of making meaningful comparisons of all possible subtype combinations given the large number of classes and relatively few individuals in each class, analysis was performed for simplified categories: presence of FIVPle subtype A, B, or C compared to each other or presence of FIVPle-B compared to presence of non-FIVPle-B subtypes.
Survival of CDV/Babesia outbreaks
Lions that were alive at the beginning of the outbreak were classified as surviving the outbreak if they lived through October of the following year. Any lion that died between January of the outbreak year and October of the following year were considered to have died during the outbreak. The majority of deaths were confined to a much narrower time window (figure 2).
Statistical analysis
Expected survival was determined empirically from the general and high-risk populations. Chi-squared analysis with 1 degree of freedom was performed for each possible FIVPle subtype category to determine deviations from expected values. Fisher’s exact test was used to compare 2X2 contingency tables of survival vs. death for subtype B vs. other subtypes. In all cases, two tailed p-values are reported. Survival analysis was performed using Gompertz regression (parametric hazard model) on survival curves produced by a LOWESS smoothing algorithm.
Results
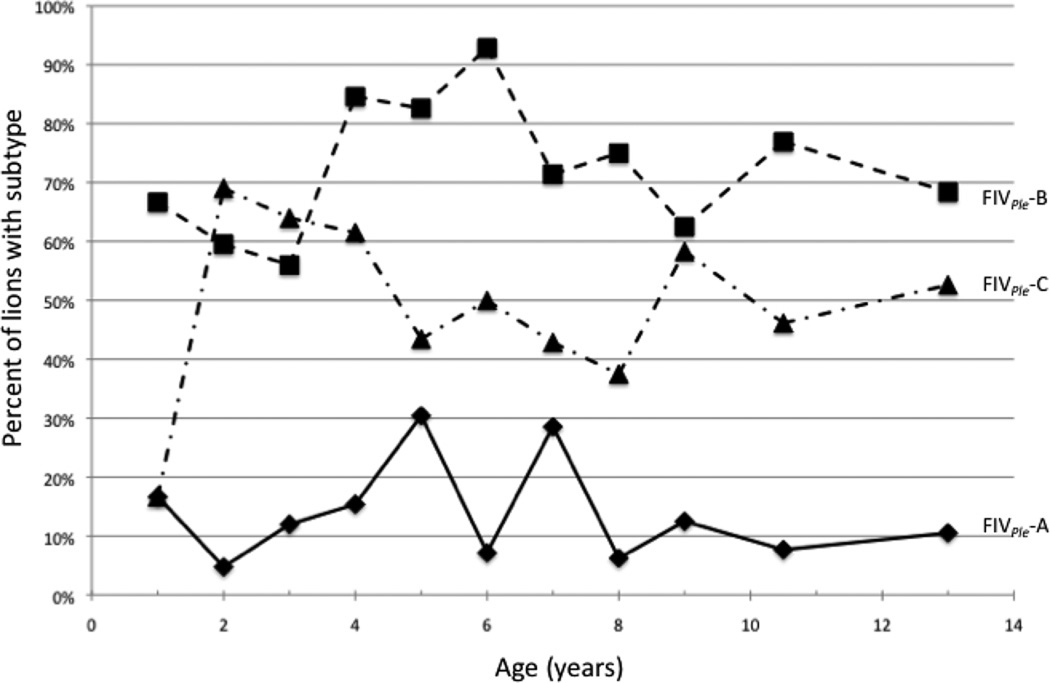
Subtype-specific primers (Troyer et al., 2004) were used to examine FIVPle circulating in blood samples from 216 lions collected over 15 years starting in 1984 and ending in 1999. As seen in a previous study of sixty-nine of these lions (Troyer, 2004), there was a high incidence of co-infections with multiple subtypes of FIV; 35% of the animals were infected with more than one subtype at the time of sampling. Overall there was a large difference in subtype frequency, with 12% of the population infected with FIVPle-A, 57% infected with FIVPle-C, and 69% infected with FIVPle-B. These frequencies varied substantially with age (figure 3): FIVPle-B was already common in cubs of one year of age (67%; N=6) and subsequently increased between the ages of 3 and 4 years old from close to 60% (N=25) to over 80% (N=13). In contrast the FIVPle-C strain was initially rare in yearlings (17%; N=6) and increased by the age of 2 years (69%; N=42). The incidence of FIVPle-A remained low throughout the lifespan of these lions.
Figure 3.
FIVPle subtype distribution by age of lion at time of sampling. An average of 19 lions were included per age class with a range of 6 (for yearlings) to 42 (for 2-year-olds). Because of co-infections, some lions were included in multiple subtype categories.
Long-term survival analysis was performed for lions that were not involved in the high mortality CDV outbreaks (N=97). Measuring longer-term mortality must control for the sex of the sampled animal as males have shorter life expectancies than females and males often disperse from the study area where their fates cannot be monitored. Females infected with subtype B demonstrated significantly longer post-sampling lifespan than those infected with A or C (Gompertz parametric hazard model; z=2.36, P=0.018; data not shown), though this effect was not seen in males (data not shown).
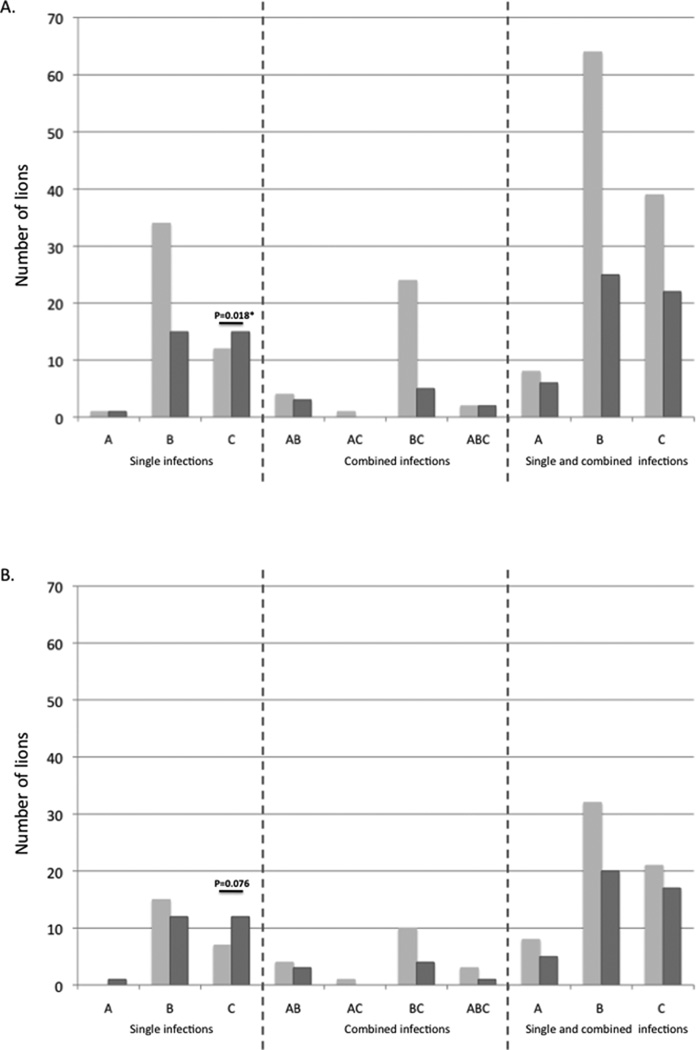
Short-term survival of lions during the 1994 Serengeti and 2001 Ngorongoro Crater CDV outbreaks was compared with infection status of FIVPle-A, FIVPle-B and FIVPle-C (Figure 4). Two different sets of animals were examined; all lions with FIVPle subtype data that were alive in the Serengeti (N-115) and Ngorongoro (N=4) study areas at the beginning of their respective outbreaks and a subset of these lions that were shown to be in areas with elevated levels of Babesia and had known CDV exposure (N=73). Overall, 66% of lions survived the outbreak. However, only 57% of the higher-risk subset survived. In all cases, lions infected with FIVPle-B singly or in co-infections with other subtypes exhibited mortality rates similar to the population at large. In contrast, lions infected with C singly had higher than expected mortality (Chi squared=5.591, two tailed p=0.0181; figure 4). The survival disadvantage exhibited by lions infected with subtype C was most pronounced when all lions in the population were considered; in the high-risk subset, the trend remained, but was not significant (Chi squared=3.1, two tailed p=0.076; figure 4b).
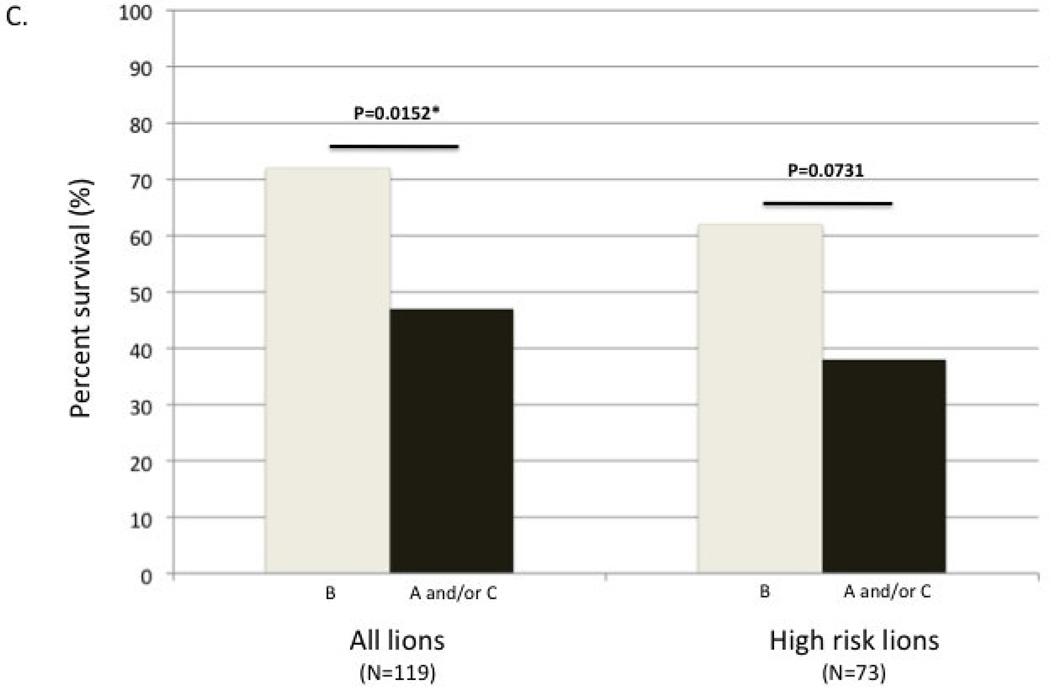
Figure 4.
Number of lions by subtype composition that survived (light grey bars) or died/disappeared (dark grey bars) during the high mortality CDV/Babesia outbreaks. Lions were infected with one, two, or three subtypes. (A) Total number of lions in each possible subtype category at the onset of the outbreak, divided by survival class. (B) Number of high-risk lions in each possible subtype category at the onset of the outbreak, divided by survival class. The scale is set the same as part A for comparison. Two-tailed Chi-squared p-values are shown in (A) and (B); expected values were based on the survival rates of the population in question. (C) Survival rates comparing lions infected with B alone or in any combination to those infected with A and/or C but not B. Two-tailed Fisher’s exact test p-values are shown.
The primary disparity in survival observed was between subtypes B and C; subtype A was rare and usually in combination with other subtypes, complicating analysis. However, since lions with subtype A were also more likely to die than those with B, subsequent analysis compared lions infected with subtype B alone or in combination to those infected with subtype A and/or C, but not with B. Overall survival of the CDV/Babesia outbreak was significantly greater for lions infected with subtype B compared to those infected with either or both of the other subtypes (figure 4c; N=119; Fisher’s exact test, two tailed p=0.0152). The trend remained the same for the high-risk subset (figure 4c; N=73; Fisher’s exact test, two tailed p=0.0731).
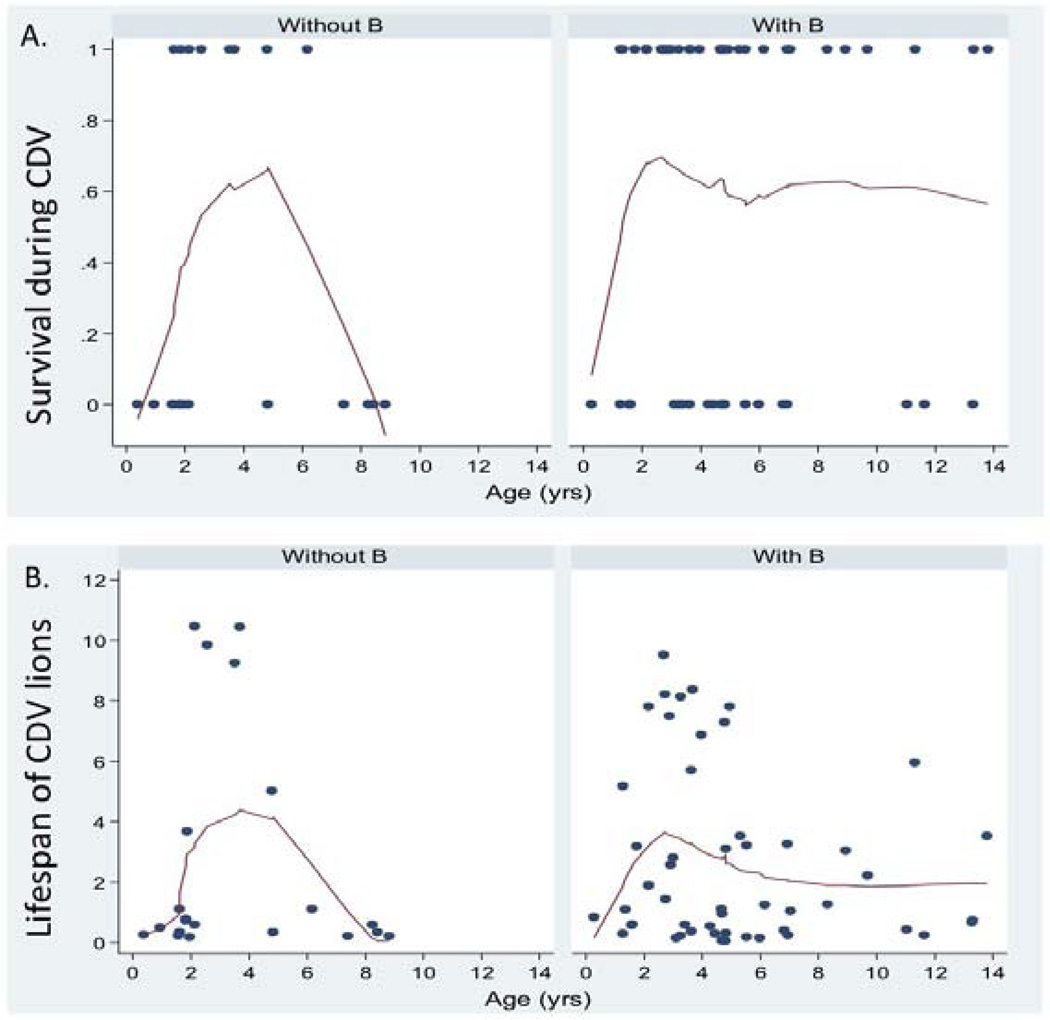
The major difference between the whole data set and the high-risk lions consisted of lions from 4 prides that had negative CDV titers or low Babesia levels (N=46); a majority of the lions from these 4 prides (70%) survived and were infected with either subtype B or subtypes B and C together. The overall effect of subtype on surviving these outbreaks was apparently due to the lower survival of adults between 5–10 yrs of age that lacked the B subtype. FIVPle subtype may also have had an effect on the survival of younger animals: survival of non-B individuals peaked at 3 yrs of age (each arm of the non-B spline was significant at P<0.05; Figure 5a). In addition (similar to lions that were not involved in the outbreak) there was a trend for the subsequent lifespan of FIVPle-B infected lions that survived the outbreak to be greater than that of lions with the other strains (figure 5b).
Figure 5.
Lowess curves of (A) short-term age-specific survival of lions that lived (1) or died (0) between the onset of CDV and the following October and (B) post-CDV onset lifespan (in years) of lions sampled during the CDV/Babesia outbreaks for FIV+ animals that were not infected with the B-subtype (left) and those that were infected with B (right).
Discussion
Lentiviruses evolve rapidly and demonstrate strong species fidelity. However, divergent strains infect individual species, sometimes as a result of multiple introductions and sometimes through viral evolution post-introduction. These strains may differ in virulence, host-cell tropism, pathogenicity and clinical outcome. In free-ranging African lions, FIVPle-A, FIVPle-B, and FIVPle-C demonstrated differential pathology and transmission. Older lions (>5 years old) infected with FIVPle-C or A alone were more likely to die than those infected with FIVPle-B. The differences in outcome between subtypes were less apparent in lions with high Babesia levels and CDV then in the population at large. This suggests the possibility of increased immune suppression with these subtype compared with FIVPle-B. It also indicates that chronic effects of FIVPle are important to long-term survival and differ according to subtype. Phylogenetic evidence supports the notion that subtype B has been in residence longer and therefore it is possible that it is more host-adapted than the other two subtypes. While speculative, this could explain the lack of FIV-related pathology in the lions of the Serengeti, where FIVPle-B is the predominant circulating strain.
The change in subtype frequency relative to age observed here raises the intriguing possibility that FIVPle subtypes may also have different modes of transmission. 67% of lions were infected with FIVPle –B by their first year, suggesting possible maternal transmission of this subtype. In contrast, the prevalence of FIVPle –C rose substantially between the ages of one and two years, when non-sexual social contact and aggression begins, suggesting biting as the primary mode of transmission. Finally FIVPle –B prevalence rose between the ages of two and three years. Since this corresponds to the age of sexual maturity, it suggests sexual transmission of FIVPle –B. The dearth of FIVPle subtypes A and C in older lions combined with the disproportionate mortality of very young and very old lions may account for the subtype differences observed for disease outcome.
The slight decline of FIVPle –C as lions age may be stochastic, but could also reflect increased mortality or be due to competition by FIVPle –B. Anecdotal evidence from cell culture suggests that B is better able than C to propagate in PBMCs and a variety of cat cell lines (Roelke, unpublished data). Perhaps a more host-adapted, less pathogenic FIVPle –B is even “protective” against immunosuppressive effects of more pathogenic strains. In this study, animals co-infected with FIVPle –B and another strain had similar mortality to those infected with B alone and were more likely to survive then lions infected with either or both of the other strains. This model is not without precedent; in domestic cats, non-pathogenic FIV infections (FIVPco or FIVPle) have been shown to impart resistance to infection by pathogenic FIVFca in vitro, and prior FIVPco infection ameliorates immunosuppressive effects of pathogenic FIVFca in vivo, though the mechanisms behind this protection are, as yet, poorly understood (Terwee et al., 2008; VandeWoude et al., 2002; VandeWoude et al., 2003).
Although these conclusions are largely speculative, careful examination of the evidence suggests that subtype differences are important and interactions between subtypes may be complex. Because of limitations of sampling wild species, our data set is necessarily small and imperfect; in many cases subtype compositions at one time point were extrapolated to a later date. In addition, low-abundance viral sequences may be missed during PCR amplification. Finally, cause of death was not always known for animals that died; often bodies were not recovered or were putrescent. We have made the assumption that animals that died during the outbreak died because of it. However, despite these caveats, the data clearly support further investigation and independent verification including clinical assessment of infected lions, analysis of lymphocyte profiles of lions infected with each subtype, and full-length genetic sequencing of each subtype to elucidate viral correlates of pathogenicity.
Acknowledgements
We thank Randy Johnson and Rachel Simmons for statistical assistance and consultation. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and has also been funded in part with federal funds from the National Cancer Institute and National Institutes of Health, under contract HHSN26120080001E. Samples were collected in full compliance with specific federal permits (CITES; Endangered and Threatened Species) issued to the National Cancer Institute, principal investigator S. J. O'Brien, by the U.S. Fish and Wildlife Service of the Department of the Interior. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: All authors declare no conflict of interest.
Bibliography
- Antunes A, Pontius J, Ramos MJ, O'Brien SJ, Johnson WE. Mitochondrial introgressions into the nuclear genome of the domestic cat. J Hered. 2007;98:414–420. doi: 10.1093/jhered/esm062. [DOI] [PubMed] [Google Scholar]
- Antunes A, Troyer JL, Roelke ME, Pecon-Slattery J, Packer C, Winterbach C, Winterbach H, Hemson G, Frank L, Stander P, Siefert L, Driciru M, Funston PJ, Alexander KA, Prager KC, Mills G, Wildt D, Bush M, O'Brien SJ, Johnson WE. The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS Genet. 2008;4:e1000251. doi: 10.1371/journal.pgen.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlough JE, North TW, Oxford CL, Remington KM, Dandekar S, Ellis MN, Pedersen NC. Feline immunodeficiency virus infection of cats as a model to test the effect of certain in vitro selection pressures on the infectivity and virulence of resultant lentivirus variants. Antiviral Res. 1993;22:259–272. doi: 10.1016/0166-3542(93)90036-i. [DOI] [PubMed] [Google Scholar]
- Bendinelli M, Pistello M, Lombardi S, Poli A, Garzelli C, Matteucci D, Ceccherini-Nelli L, Malvaldi G, Tozzini F. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev. 1995;8:87–112. doi: 10.1128/cmr.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R, Ruth TK, Murphy KM, Anderson CR, Jr, Johnson M, DeSimone R, Gray R, Hornocker MG, Gillin CM, Poss M. Factors associated with pathogen seroprevalence and infection in Rocky Mountain cougars. J Wildl Dis. 2006;42:606–615. doi: 10.7589/0090-3558-42.3.606. [DOI] [PubMed] [Google Scholar]
- Brennan G, Podell MD, Wack R, Kraft S, Troyer JL, Bielefeldt-Ohmann H, VandeWoude S. Neurologic disease in captive lions (Panthera leo) with low-titer lion lentivirus infection. J Clin Microbiol. 2006;44:4345–4352. doi: 10.1128/JCM.00577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EW, Yuhki N, Packer C, O'Brien SJ. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J Virol. 1994;68:5953–5968. doi: 10.1128/jvi.68.9.5953-5968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Munkhtsog B, Troyer JL, Ross S, Sellers R, Fine AE, Swanson WF, Roelke ME, O'Brien SJ. Feline immunodeficiency virus (FIV) in wild Pallas' cats. Vet Immunol Immunopathol. 2010;134:90–95. doi: 10.1016/j.vetimm.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull ME, Gebhard DG, Tompkins WA, Kennedy-Stoskopf S. Polymorphic expression in the CD8alpha chain surface receptor of African lions (Panthera leo) Vet Immunol Immunopathol. 2002;84:181–189. doi: 10.1016/s0165-2427(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Bull ME, Kennedy-Stoskopf S, Levine JF, Loomis M, Gebhard DG, Tompkins WA. Evaluation of T lymphocytes in captive african lions (Panthera leo) infected with feline immunodeficiency virus. Am J Vet Res. 2003;64:1293–1300. doi: 10.2460/ajvr.2003.64.1293. [DOI] [PubMed] [Google Scholar]
- Burkhard MJ, Dean GA. Transmission and immunopathogenesis of FIV in cats as a model for HIV. Curr HIV Res. 2003;1:15–29. doi: 10.2174/1570162033352101. [DOI] [PubMed] [Google Scholar]
- Carpenter MA, Appel MJ, Roelke-Parker ME, Munson L, Hofer H, East M, O'Brien SJ. Genetic characterization of canine distemper virus in Serengeti carnivores. Vet Immunol Immunopathol. 1998;65:259–266. doi: 10.1016/s0165-2427(98)00159-7. [DOI] [PubMed] [Google Scholar]
- Carpenter MA, Brown EW, Culver M, Johnson WE, Pecon-Slattery J, Brousset D, O'Brien SJ. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor) J Virol. 1996;70:6682–6693. doi: 10.1128/jvi.70.10.6682-6693.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, O'Brien SJ. Coadaptation and immunodeficiency virus: lessons from the Felidae. Curr Opin Genet Dev. 1995;5:739–745. doi: 10.1016/0959-437x(95)80006-q. [DOI] [PubMed] [Google Scholar]
- de Monte M, Nonnenmacher H, Brignon N, Ullmann M, Martin JP. A multivariate statistical analysis to follow the course of disease after infection of cats with different strains of the feline immunodeficiency virus (FIV) J Virol Methods. 2002;103:157–170. doi: 10.1016/s0166-0934(02)00024-1. [DOI] [PubMed] [Google Scholar]
- de Rozieres S, Mathiason CK, Rolston MR, Chatterji U, Hoover EA, Elder JH. Characterization of a highly pathogenic molecular clone of feline immunodeficiency virus clade C. J Virol. 2004a;78:8971–8982. doi: 10.1128/JVI.78.17.8971-8982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rozieres S, Swan CH, Sheeter DA, Clingerman KJ, Lin YC, Huitron-Resendiz S, Henriksen S, Torbett BE, Elder JH. Assessment of FIV-C infection of cats as a function of treatment with the protease inhibitor, TL-3. Retrovirology. 2004b;1:38. doi: 10.1186/1742-4690-1-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rozieres S, Thompson J, Sundstrom M, Gruber J, Stump DS, de Parseval AP, VandeWoude S, Elder JH. Replication properties of clade A/C chimeric feline immunodeficiency viruses and evaluation of infection kinetics in the domestic cat. J Virol. 2008;82:7953–7963. doi: 10.1128/JVI.00337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JH, Dean GA, Hoover EA, Hoxie JA, Malim MH, Mathes L, Neil JC, North TW, Sparger E, Tompkins MB, Tompkins WA, Yamamoto J, Yuhki N, Pedersen NC, Miller RH. Lessons from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1998;14:797–801. doi: 10.1089/aid.1998.14.797. [DOI] [PubMed] [Google Scholar]
- Elder JH, Lin YC, Fink E, Grant CK. Feline immunodeficiency virus (FIV) as a model for study of lentivirus infections: parallels with HIV. Curr HIV Res. 2010;8:73–80. doi: 10.2174/157016210790416389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin SP, Kays RW, Moreno R, TerWee JA, Troyer JL, VandeWoude S. Ocelots on Barro Colorado Island are infected with feline immunodeficiency virus but not other common feline and canine viruses. J Wildl Dis. 2008;44:760–765. doi: 10.7589/0090-3558-44.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin SP, Troyer JL, Terwee JA, Lyren LM, Boyce WM, Riley SP, Roelke ME, Crooks KR, Vandewoude S. Frequent transmission of immunodeficiency viruses among bobcats and pumas. J Virol. 2007;81:10961–10969. doi: 10.1128/JVI.00997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruol DL, Yu N, Parsons KL, Billaud JN, Elder JH, Phillips TR. Neurotoxic effects of feline immunodeficiency virus, FIV-PPR. J Neurovirol. 1998;4:415–425. doi: 10.3109/13550289809114540. [DOI] [PubMed] [Google Scholar]
- Henriksen SJ, Prospero-Garcia O, Phillips TR, Fox HS, Bloom FE, Elder JH. Feline immunodeficiency virus as a model for study of lentivirus infection of the central nervous system. Curr Top Microbiol Immunol. 1995;202:167–186. doi: 10.1007/978-3-642-79657-9_12. [DOI] [PubMed] [Google Scholar]
- Johnston JB, Jiang Y, van Marle G, Mayne MB, Ni W, Holden J, McArthur JC, Power C. Lentivirus infection in the brain induces matrix metalloproteinase expression: role of envelope diversity. J Virol. 2000;74:7211–7220. doi: 10.1128/jvi.74.16.7211-7220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochelt M, Romen F, Bastone P, Muckenfuss H, Kirchner N, Kim YB, Truyen U, Rosler U, Battenberg M, Saib A, Flory E, Cichutek K, Munk C. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc Natl Acad Sci U S A. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker RB. Feline immunodeficiency virus neuropathogenesis: from cats to calcium. J Neuroimmune Pharmacol. 2007;2:154–170. doi: 10.1007/s11481-006-9045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O'Brien SJ, Lochelt M, Yuhki N. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C, Zielonka J, Constabel H, Kloke BP, Rengstl B, Battenberg M, Bonci F, Pistello M, Lochelt M, Cichutek K. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J Virol. 2007;81:7048–7060. doi: 10.1128/JVI.02714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson L, Terio KA, Kock R, Mlengeya T, Roelke ME, Dubovi E, Summers B, Sinclair AR, Packer C. Climate extremes promote fatal co-infections during canine distemper epidemics in African lions. PLoS One. 2008;3:e2545. doi: 10.1371/journal.pone.0002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Troyer JL, Roelke M, Marker L, Pecon-Slattery J. Plagues and adaptation: Lessons from the Felidae models for SARS and AIDS. Biological Conservation. 2006;131:255–267. doi: 10.1016/j.biocon.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted RA, Langley R, Roelke ME, Goeken RM, Adgerjohnson D, Goff JP, Albert JP, Packer C, Laurenson MK, Caro TM, Scheepers L, Wildt DE, Bush M, Martenson JS, Obrien SJ. Worldwide Prevalence of Lentivirus Infection in Wild Feline Species - Epidemiologic and Phylogenetic Aspects. Journal of Virology. 1992;66:6008–6018. doi: 10.1128/jvi.66.10.6008-6018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer C, Altizer S, Appel M, Brown E, Martenson J, O'Brien SJ, Roelke-Parker M, Hofmann-Lehmann R, Lutz H. Viruses of the Serengeti: patterns of infection and mortality in African lions. Journal of Animal Ecology. 1999;68:1161–1178. [Google Scholar]
- Packer C, Hilborn R, Mosser A, Kissui B, Borner M, Hopcraft G, Wilmshurst J, Mduma S, Sinclair ARE. Ecological change, group territoriality, and population dynamics in Serengeti lions. Science. 2005;307:390–393. doi: 10.1126/science.1105122. [DOI] [PubMed] [Google Scholar]
- Patrick MK, Johnston JB, Power C. Lentiviral neuropathogenesis: comparative neuroinvasion, neurotropism, neurovirulence, and host neurosusceptibility. J Virol. 2002;76:7923–7931. doi: 10.1128/JVI.76.16.7923-7931.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery J, McCracken CL, Troyer JL, VandeWoude S, Roelke M, Sondgeroth K, Winterbach C, Winterbach H, O'Brien SJ. Genomic organization, sequence divergence, and recombination of feline immunodeficiency virus from lions in the wild. BMC Genomics. 2008a;9:66. doi: 10.1186/1471-2164-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery J, Troyer JL, Johnson WE, O'Brien SJ. Evolution of feline immunodeficiency virus in Felidae: Implications for human health and wildlife ecology. Vet Immunol Immunopathol. 2008b;123:32–44. doi: 10.1016/j.vetimm.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NC, Leutenegger CM, Woo J, Higgins J. Virulence differences between two field isolates of feline immunodeficiency virus (FIV-APetaluma and FIV-CPGammar) in young adult specific pathogen free cats. Vet Immunol Immunopathol. 2001;79:53–67. doi: 10.1016/s0165-2427(01)00252-5. [DOI] [PubMed] [Google Scholar]
- Phillips TR, Prospero-Garcia O, Puaoi DL, Lerner DL, Fox HS, Olmsted RA, Bloom FE, Henriksen SJ, Elder JH. Neurological abnormalities associated with feline immunodeficiency virus infection. J Gen Virol. 1994;75 ( Pt 5):979–987. doi: 10.1099/0022-1317-75-5-979. [DOI] [PubMed] [Google Scholar]
- Phillips TR, Prospero-Garcia O, Wheeler DW, Wagaman PC, Lerner DL, Fox HS, Whalen LR, Bloom FE, Elder JH, Henriksen SJ. Neurologic dysfunctions caused by a molecular clone of feline immunodeficiency virus, FIV-PPR. J Neurovirol. 1996;2:388–396. doi: 10.3109/13550289609146904. [DOI] [PubMed] [Google Scholar]
- Power C, Buist R, Johnston JB, Del Bigio MR, Ni W, Dawood MR, Peeling J. Neurovirulence in feline immunodeficiency virus-infected neonatal cats is viral strain specific and dependent on systemic immune suppression. J Virol. 1998;72:9109–9115. doi: 10.1128/jvi.72.11.9109-9115.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke ME, Brown MA, Troyer JL, Winterbach H, Winterbach C, Hemson G, Smith D, Johnson RC, Pecon-Slattery J, Roca AL, Alexander KA, Klein L, Martelli P, Krishnasamy K, O'Brien SJ. Pathological manifestations of feline immunodeficiency virus (FIV) infection in wild African lions. Virology. 2009;390:1–12. doi: 10.1016/j.virol.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke ME, Pecon-Slattery J, Taylor S, Citino S, Brown E, Packer C, VandeWoude S, O'Brien SJ. T-lymphocyte profiles in FIV-infected wild lions and pumas reveal CD4 depletion. Journal of Wildlife Diseases. 2006;42:234–248. doi: 10.7589/0090-3558-42.2.234. [DOI] [PubMed] [Google Scholar]
- Roelke-Parker ME, Munson L, Packer C, Kock R, Cleaveland S, Carpenter M, O'Brien SJ, Pospischil A, Hofmann-Lehmann R, Lutz H, Mwamengele GL, Mgasa MN, Machange GA, Summers BA, Appel MJ. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996;379:441–445. doi: 10.1038/379441a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova N, Troyer JL, Schissler J, Terwee J, Poss M, VandeWoude S. Feline lentiviruses demonstrate differences in receptor repertoire and envelope structural elements. Virology. 2005;342:60–76. doi: 10.1016/j.virol.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Sparger EE, Beebe AM, Dua N, Himathongkam S, Elder JH, Torten M, Higgins J. Infection of cats with molecularly cloned and biological isolates of the feline immunodeficiency virus. Virology. 1994;205:546–553. doi: 10.1006/viro.1994.1677. [DOI] [PubMed] [Google Scholar]
- Stump DS, VandeWoude S. Animal models for HIV AIDS: a comparative review. Comp Med. 2007;57:33–43. [PubMed] [Google Scholar]
- Terwee JA, Carlson JK, Sprague WS, Sondgeroth KS, Shropshire SB, Troyer JL, Vandewoude S. Prevention of immunodeficiency virus induced CD4+ T-cell depletion by prior infection with a non-pathogenic virus. Virology. 2008 doi: 10.1016/j.virol.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JL, Pecon-Slattery J, Roelke ME, Black L, Packer C, O'Brien SJ. Patterns of feline immunodeficiency virus multiple infection and genome divergence in a free-ranging population of African lions. J Virol. 2004;78:3777–3791. doi: 10.1128/JVI.78.7.3777-3791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JL, Pecon-Slattery J, Roelke ME, Johnson W, VandeWoude S, Vazquez-Salat N, Brown M, Frank L, Woodroffe R, Winterbach C, Winterbach H, Hemson G, Bush M, Alexander KA, Revilla E, O'Brien SJ. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JL, Vandewoude S, Pecon-Slattery J, McIntosh C, Franklin S, Antunes A, Johnson W, O'Brien SJ. FIV cross-species transmission: An evolutionary prospective. Vet Immunol Immunopathol. 2008;123:159–166. doi: 10.1016/j.vetimm.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWoude S, Hageman CA, O'Brien SJ, Hoover EA. Nonpathogenic lion and puma lentiviruses impart resistance to superinfection by virulent feline immunodeficiency virus. J Acquir Immune Defic Syndr. 2002;29:1–10. doi: 10.1097/00126334-200201010-00001. [DOI] [PubMed] [Google Scholar]
- VandeWoude S, Hageman CL, Hoover EA. Domestic cats infected with lion or puma lentivirus develop anti-feline immunodeficiency virus immune responses. J Acquir Immune Defic Syndr. 2003;34:20–31. doi: 10.1097/00126334-200309010-00003. [DOI] [PubMed] [Google Scholar]
- VandeWoude S, Troyer J, Poss M. Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV. Vet Immunol Immunopathol. 2010;134:25–32. doi: 10.1016/j.vetimm.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver EA. A detailed phylogenetic analysis of FIV in the United States. PLoS One. 2010;5:e12004. doi: 10.1371/journal.pone.0012004. [DOI] [PMC free article] [PubMed] [Google Scholar]