Abstract
Nuclear protein import in eukaryotic cells is mediated by karyopherin proteins, which bind to specific nuclear localization signals on substrate proteins and transport them across the nuclear envelope and into the nucleus. Replication protein A (RPA) is a nuclear protein comprised of three subunits (termed Rfa1, Rfa2, and Rfa3 in Saccharomyces cerevisiae) that binds single-stranded DNA and is essential for DNA replication, recombination, and repair. RPA associates with two different karyopherins in yeast, Kap95, and Msn5/Kap142. However, it is unclear which of these karyopherins is responsible for RPA nuclear import. We have generated GFP fusion proteins with each of the RPA subunits and demonstrate that these Rfa-GFP chimeras are functional in yeast cells. The intracellular localization of the RPA proteins in live cells is similar in wild-type and msn5Δ deletion strains but becomes primarily cytoplasmic in cells lacking functional Kap95. Truncating the C-terminus of any of the RPA subunits results in mislocalization of the proteins to the cytoplasm and a loss of protein–protein interactions between the subunits. Our data indicate that Kap95 is likely the primary karyopherin responsible for RPA nuclear import in yeast and that the C-terminal regions of Rfa1, Rfa2, and Rfa3 are essential for efficient nucleocytoplasmic transport of each RPA subunit.
Introduction
Replication protein A (RPA) is a heterotrimeric protein complex that is essential for DNA replication, recombination, and repair in eukaryotic cells (reviewed in Wold, 1997; Iftode et al., 1999; Zou et al., 2006). RPA is a single-stranded DNA-binding protein that is comprised of three subunits: RPA70, RPA32, and RPA14. In the budding yeast Saccharomyces cerevisiae these subunits are referred to as Rfa1, Rfa2, and Rfa3, respectively. Each subunit is essential for viability and short truncations from the N- or C-termini of each are lethal (Heyer et al., 1990; Brill and Stillman, 1991; Philipova et al., 1996). The largest subunit, Rfa1, contains a protein–protein interaction domain at its N-terminus that associates with factors that transiently regulate its activity (see Fanning et al., 2006). The remainder of Rfa1 contains a series of DNA-binding domains, the most C-terminal of which also acts as a binding partner for the 34 kDa Rfa2 subunit (Philipova et al., 1996; Brill and Bastin-Shanower, 1998; Bochkareva et al., 2000, 2002). Rfa2 contains a single DNA-binding domain as well as a region toward its C-terminus that associates with other RPA subunits. However, Rfa2 can tolerate C-terminal truncations of more than 100 amino acids before completely losing function (Philipova et al., 1996). Rfa3 does not have strong intrinsic DNA-binding activity, but, like Rfa2, contains a C-terminal domain that is necessary for viability and for trimerization of the RPA complex (Philipova et al., 1996; Bochkareva et al., 2000). RPA subunits primarily localize to the nucleus, consistent with their role in DNA metabolism, although specific cytoplasmic localization is apparent upon overexpression and cell cycle progression (Murti et al., 1996; Sundin et al., 2004).
Eukaryotic cells regulate the transport of proteins between the nucleus and cytoplasm in an energy- and protein-dependent process (Tran and Wente, 2006). The double-membraned nuclear envelope (NE) that forms the barrier between the nuclear and cytoplasmic compartments is perforated by large, hetero-oligomeric structures termed nuclear pore complexes (NPCs), which serve as the only conduit for the movement of macromolecules between the nucleus and cytoplasm (reviewed in Fahrenkrog and Aebi, 2003; D'Angelo and Hetzer, 2008). Small molecules can diffuse freely through the aqueous channel at the center of the nuclear pores, whereas transport across the NE of proteins larger than ∼40 kDa is tightly regulated. Proteins transported into the nucleus through the NPCs contain nuclear localization signals (NLSs), short amino acid sequences or domains that are recognized by protein transport receptors (Lange et al., 2007). NLS sequences can be highly divergent, but function to mediate association with a specific soluble transport receptor that facilitates translocation through NPCs. Most transport receptors are members of a family of related proteins termed karyopherins that can either import NLS-containing substrates from the cytoplasm into the nucleus (i.e., importins) or export nuclear export signal containing cargoes from the nucleus to the cytoplasm (exportins) (Ström and Weis, 2001; Mosammaparast and Pemberton, 2004; Terry et al., 2007). The binding and release of substrates from karyopherins is regulated by the small GTPase Ran, which is in a predominantly GDP-bound form in the cytoplasm and GTP-bound in the nucleus (Izaurralde et al., 1997).
Fourteen karyopherins (Kaps) have been identified in the yeast S. cerevisiae, each of which recognizes a distinct but not mutually exclusive set of NLSs and thus transports a unique set of substrates across the NPC. The most well-characterized karyopherin forms a heterodimer of two importin subunits that cooperate to import substrates containing a related NLS sequence. These subunits have been termed importin-α (Imp-α or Kap60 in yeast) and importin-β (Imp-β/Kap95). Imp-α/Kap60 associates directly with the NLS of its cargoes, whereas Imp-β/Kap95 mediates movement of the cargo/importin complex through the NPC (Görlich et al., 1995; Enenkel et al., 1995; Conti et al., 1998). The NLS sequences recognized by Kap60 contain short clusters of basic amino acids and are referred to as classical NLS (cNLS) motifs. Over half of the yeast proteins that localize to the nucleus contain a predicted cNLS motif (Lange et al., 2007). However, Imp-β/Kap95 can also function as a monomeric importin, transporting some substrates into the nucleus without utilizing Imp-α as a cNLS-binding adaptor molecule (Palmeri and Malim, 1999; Nagoshi et al., 1999; Truant and Cullen, 1999; Lee et al., 2003; Strahl et al., 2005; Singhal et al., 2006; Fries et al., 2007).
The remaining karyopherins in yeast also function by associating as monomers directly with specific NLS or nuclear export signal sequences to mediate translocation of their substrates across the NPC (Ström and Weis, 2001; Madrid and Weis, 2006). With one exception, all of these karyopherins have thus far been shown to transport protein substrates in a single direction across the NPC, functioning either as importins or as exportins, but not as both (Harel and Forbes, 2004; Poon and Jans, 2005). The exception is Msn5/Kap142, which has been reported to have nuclear import and export activity for distinct protein substrates (Yoshida and Blobel, 2001). However, the role of Msn5 in nuclear protein import remains unclear (Belanger et al., 2004).
MSN5 was initially identified in various genetic screens as a suppressor of Snf1 kinase mutants (Estruch and Carlson, 1990), modulator of pheromone response and calmodulin-dependent gene induction (Akada et al., 1996; Alepuz et al., 1999), and regulator of pseudohyphal differentiation (Lorenz and Heitman, 1998). Msn5 has subsequently been shown to function as an exportin for more than 15 different proteins that shuttle between the nucleus and cytoplasm, often performing an important role in regulating protein function by modulating intracellular localization through rapid nuclear export (Kaffman et al., 1998; Blondel et al., 1999; DeVit and Johnston, 1999; Mahanty et al., 1999; Komeili et al., 2000; Boustany and Cyert, 2002; Görner et al., 2002; Jaquenoud et al., 2002; Queralt and Igual, 2003; Strahl et al., 2005; Quan et al., 2006; Ueta et al., 2007; Bakhrat et al., 2008; Towpik et al., 2008). Interestingly, Msn5 was also reported to function as an importin for RPA subunits, which would make it the only yeast Kap to modulate transport of distinct protein substrates in different directions across the NPC (Yoshida and Blobel, 2001). Both Msn5 and Kap95 associate with the RPA complex in the presence of Ran-GDP, and immunofluorescence microscopy indicates that an Rfa1-Protein A chimera is mislocalized from the nucleus to the cytoplasm in cells lacking Msn5 (Yoshida and Blobel, 2001). However, kinetic analyses of Rfa2-GFP nuclear import do not reveal a decreased nuclear transport rate in the absence of Msn5 and suggest that Kap95 may be the primary karyopherin responsible for Rfa2 protein import (Belanger et al., 2004). An examination of nucleocytoplasmic diffusion rates suggests that Msn5 may function in Rfa2 nuclear retention rather than translocation (Belanger et al., 2004). While it is clear that both Kap95 and Msn5 associate with RPA complex members and modulate nucleocytoplasmic localization, the relative roles of these karyopherins in RPA transport remain unresolved.
Here we report the use of functional chimeric Rfa-GFP proteins expressed in S. cerevisiae to investigate the nuclear import of RPA. We observe that the karyopherin Kap95 is necessary for the nuclear accumulation of RPA subunits in the nucleus and that a deletion of Msn5/Kap142 does not detectably alter RPA localization. Using both full-length and truncated Rfa-GFP fusion proteins, we also show that the C-terminal trimerization domains of Rfa1, Rfa2, and Rfa3 are essential for intermolecular interactions among these fusions and their subsequent transport to the nucleus. Together, these data provide evidence for a Kap95-mediated nuclear import mechanism that requires Rfa domains important for RPA complex assembly for efficient nuclear targeting.
Materials and Methods
Yeast strains and plasmids used in this study are listed in Table 1. Manipulation and handling of yeast was performed as described (Guthrie and Fink, 1991). Yeast were grown at 30°C unless otherwise noted.
Table 1.
Yeast Strains and Plasmids Used in This Study
| Strain number | Genotype | Source |
|---|---|---|
| W303 | MATa ade2 trp1 leu2 his3 ura3 | R. Rothstein |
| BY4742 | MATα his3 leu2 lys2 ura3 | Open biosystems |
| HMY343 | MATa rfa2-210ts::HIS3 ura3 leu2 ade2 rfa2Δ::TRP1 | Maniar et al. (1997) |
| HMY353 | MATa rfa3-313ts::HIS3 ura3 leu2 ade2 rfa3Δ::TRP1 | Maniar et al. (1997) |
| PSY1102 | MATa rsl1-3/kap95ts ura3 leu2 trp1 | Koepp et al. (1996) |
| PSY1103 | MATa rsl1-4/kap95ts ura3 leu2 trp1 | Koepp et al. (1996) |
| KBY893 | MATa msn5Δ::KANr his3 leu2 ura3 met15 | Open biosystems |
| KBY1107 | MATα ybr137wΔ::KANr his3 leu2 lys2 ura3 | Open biosystems |
| KBY1076 | MATa RFA1-TAP::HIS3 his3 leu2 met15 ura3 | Open biosystems |
| KBY1077 | MATa RFA2-TAP::HIS3 his3 leu2 met15 ura3 | Open biosystems |
| KBY1078 | MATa RFA3-TAP::HIS3 his3 leu2 met15 ura3 | Open biosystems |
| KBY1249 | MATa/α rfa1Δ::KANr/RFA1 his3/his3 leu2/leu2 ura3/ura3 lys2/LYS2 met15/MET15 | Open biosystems |
| Plasmid number | Genotype | Source |
|---|---|---|
| pJM132 | CEN URA3 RFA1 RFA2 RFA3 Ampr | Maniar et al. (1997) |
| pKBB284 | RFA2-GFP URA3 CEN Ampr | Belanger et al. (2004) |
| pKBB285 | RFA3-GFP URA3 CEN Ampr | Belanger et al. (2004) |
| pKBB345 | rfa3Δ46-GFP URA3 CEN Ampr | This study |
| pKBB348 | rfa2Δ169-GFP URA3 CEN Ampr | This study |
| pKBB349 | rfa2Δ247-GFP URA3 CEN Ampr | This study |
| pKBB364 | GAL1::RFA1-GFP URA3 CEN Ampr | This study |
| pKBB372 | GAL1::rfa1[330–621]-GFP URA3 CEN Ampr | This study |
| pKBB438 | GAL1::rfa1[1–337,344–621]-GFP URA3 CEN Ampr | This study |
| pLDB350 | GFP URA3 CEN Ampr | L. Davis |
| pLDB351 | GAL1::GFP URA3 CEN Ampr | L. Davis |
Generation of RFA-GFP fusion proteins
Primers for generation of RFA2-GFP and RFA3-GFP fusions by homologous recombination in S. cerevisiae were generated by combining sequences flanking genomic RFA2 and RFA3 and sequences opposite translational start site of GFP in pLDB350 (CEN, URA3, AMPr, GFP). Primers for the amplification of RFA2 are KOL158 (5′-TACCGGGCCCCCCCTCGAGGTCGACGGTATCGATAAGCTTGATATCGAAATCCTGCAGGCCCATTTTGAAAACCTCAACG-3′) and KOL145 (5′-GAATTGGGACAACTCCAGTGAAGAGTTCTTCTCCTTTGCTAGCA GCAGCGGATCCAGCAGCTAGGGCAAAGTTATTGTC-3′). Primers for RFA3 are KOL159 (5′- CCGGGCCCCCCCTCGAGGTCGACGGTATCGATAAGCTTGATATC GAATTCCTGCAAGCAACGTCAAGATTGATCCTAT-3′) and KOL147 (5′-GAATTGGGACAACTCCAGTGAAGAGTTCTTCTCCTTTGCTAGCAGCAGCGGATCCAGCAGCAGCGTTATTTTCTGGGTATTTC-3′). Truncated rfa2Δ169 was generated with KOL 158 and KOL185 (5′-CGAACCGCCAATTTC-3′), and rfa2Δ247 with KOL158 and KOL186 (5′-GTTACGGACTGGAGCCGG-3′). A 600 bp fragment of RFA3 was amplified from plasmid pJM132 by PCR using KOL159 and KOL192 (5′-CTAATTCAACAAGAATTGGGACAACTCCAGTGAAGAGTTCTTCTCCTTTGCTTTTCGATGATATGGTTGGCG-3′). PCR products were inserted into pLDB350 by homologous recombination. pLDB350 (CEN URA3 GFP) was cut with SpeI (New England Biolabs, Beverly, MA) and cotransformed with RFA PCR product into W303 cells by LiAc transformation (Gietz and Woods, 2002). Transformants were selected for on SD-URA and grown at 30°C. RFA-GFP fusions were screened by whole cell PCR of potential fusions using T3 primer (5′-ATTAACCTCACTAAAG-3′) and GFP internal primer KOL152 (5′-CTGACAGAAAATTTGTGCCC-3′). Recombinant plasmids containing RFA-GFP fusions were isolated from yeast cells using EZ Yeast Plasmid Isolation Kit (Geno Technology, Inc., St. Louis, MO).
Plasmids expressing Rfa1 under control of the GAL1 promoter were generated by cotransforming yeast strain BY4742 with plasmid LDB351 linearized with XbaI (New England Biolabs) and a PCR product containing a specific region of RFA1 and flanked by 45–50 nucleotide regions of homology with LDB351. Full-length RFA1 was generated by PCR using KOL222 (5′-CAACAAAAAATTGTTAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACTATAATGAGCAGTGTTCAACTTTCGAGG-3′) and KOL197 (5′-CTAATTCAACAAGAATTGGGACAACTCCAGTGAAGAGTTCTTCTCCTTTGCTAGCTAACAAAGCCTTGGATAACTC-3′). pKBB372 Rfa1[330–621]-GFP was made using KOL229 (5′-CAAAAAATTGTTAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACTATAATGTTTGAGCTAACTTCAAGGGCTGGG-3′) and KOL197. Sequence of the RFA region for each plasmid was confirmed by DNA sequencing with an ABI Prism 310 DNA sequencer.
Complementation of rfa1Δ mutants with GAL::RFA1-GFP
Yeast strain KBY1249 was transformed with pKBB364 and pKBB438, and diploid transformants were incubated on agarose sporulation media (Adams et al., 1997) for 5 days. The cells were then resuspended in 50 μL of 2 mg/mL Zymolyase and incubated at room temperature for 25 min. Tetrads were dissected by micromanipulation on media containing either 2% dextrose (YPD) or 2% galactose (YEPG), grown at 30°C for 4–6 days, and scored for growth. Yeast containing GAL::RFA1-GFP, GAL::rfa1Δ353-GFP, and GAL::rfa1[1–337,344–621]-GFP plasmids were tested for Rfa1-GFP expression by western blotting. Cells were cultured overnight in SD–Ura and split into two equivalent samples. Glucose was added to a final concentration of 2% to one; galactose was added to 2% to the other. After a 3–4-h incubation, cells were centrifuged, frozen, and lysed by agitation with acid-washed beads in a 50% trichloro-acetic acid (TCA) solution. Western blots were probed with mouse anti-GFP antibody (Roche, Basel, Switzerland), and detection was performed using Immun-Star AP chemiluminescence (BioRad, Hercules, CA).
Microscopy
All cells were grown in SD-Ura containing 0.6 mM adenine to early log phase (A600 0.1–0.2). Strains containing plasmids requiring galactose induction for GFP expression where incubated in selective media containing 2% galactose for 2–4 h before microscopy. Cells were centrifuged and resuspended in media at permissive (24°C) and nonpermissive (37°C) temperatures. Live cells were viewed under direct fluorescence using a Nikon E600 microscope. Images were captured using Hamamatsu 3CCD (Hamamatsu, Inc., Shizuoka, Japan) or SPOT RTKE (Diagnostic Instruments, Inc., Sterling Heights, MI) cameras. Final figures were generated using Adobe Photoshop (Adobe Systems Inc., Mountain View CA).
Immunoprecipitation and Western blot
The plasmids containing full-length and truncated Rfa1-, Rfa2-, and Rfa3-GFP fusions were transformed into KBY1076, KBY1077, and KBY1078 yeast strains containing TAP-tagged RFA1, RFA2, and RFA3, respectively. Protein extracts from each were isolated by acid-washed bead lysis. Extracts were incubated with IgG-agarose beads in ELB + NFDM (250 mM NaCl, 50 mM HEPES pH 7.0, 5 mM EDTA, 0.5 mM DTT, 1 mM PMSF, 0.1% Tween 20, and 4% nonfat dry milk) and bound proteins were washed six times with ELB + NFDM, eluted with Laemmli buffer, and subjected to Western blotting using anti-GFP antibody (Roche) and Immun-Star AP chemiluminescence (BioRad).
Results
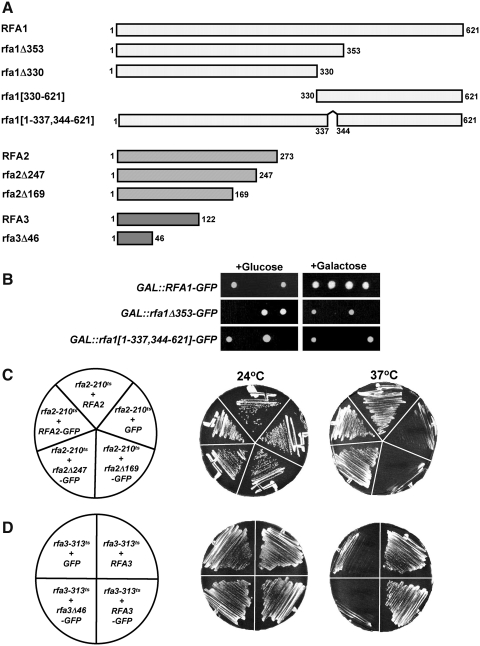
The RPA complex is a heterotrimeric protein complex involved in DNA replication, repair, and recombination within the nucleus of eukaryotic cells. To investigate RPA complex transport and intracomplex interactions, we synthesized a series of fusion proteins linking Rfa1, Rfa2, and Rfa3 with GFP at their C-termini. We then generated truncations and short deletions of regions of each protein to use for analysis of domain function in the assembly and nuclear import of RPA (Fig. 1A).
FIG. 1.
Complementation analysis of Rfa-GFP fusions. (A) Full-length and truncated Rfa1-, Rfa2-, and Rfa3-GFP chimeras were generated with GFP fused to the C-terminus of each. The numbers shown indicate the amino acid present at the N- and C-termini of each Rfa fusion encoded in each construct. (B) Diploid yeast heterozygous for an rfa1Δ deletion were transformed with full-length RFA1-GFP, rfa1Δ353-GFP, or rfa1[1–337,344–621]-GFP expressed under the control of the GAL1 promoter. Diploid transformants were sporulated and resulting tetrads dissected on selective media containing glucose (left) or galactose (right) as a carbon source. The growth of colonies from a single representative tetrad from each plate is depicted. (C) Haploid yeast containing the rfa2-210 temperature-sensitive mutation (Maniar et al., 1997) were transformed with plasmids expressing wild-type RFA2 (RFA2), empty vector (GFP), full-length RFA2 fused to GFP (RFA2-GFP), and two C-terminal rfa2 truncations fused with GFP (rfa2Δ169-GFP and rfa2Δ247-GFP). Transformants were streaked on selective media at permissive and restrictive temperatures assayed for growth. Images were taken 3 and 2 days after streaking to 24°C and 37°C, respectively. (D) Haploid cells containing rfa3-313ts were transformed with plasmids expressing wild-type RFA3 (RFA3), GFP alone (GFP), and full-length (RFA3-GFP) and truncated (rfa3Δ46-GFP) alleles of RFA3. Complementation was determined by assaying for growth at 24°C and 37°C after 48–72 h.
Yeast cells lacking any of the three subunits of RPA are inviable (Heyer et al., 1990; Brill and Stillman, 1991). To determine if our full-length and truncated Rfa-GFP fusions retain function, we performed complementation analysis with each fusion. To investigate Rfa1-GFP function, we generated plasmids containing RFA1-GFP, rfa1Δ353-GFP, and rfa1[1–337,344–621]-GFP under control of the GAL1 promoter. Expression of the resulting Rfa1-GFP fusions was confirmed by fluorescence microscopy (see Fig. 2) and western blotting (data not shown). We transformed each plasmid into a/α diploid yeast heterozygous for an rfa1 deletion, induced the transformed diploid cells to undergo sporulation, and dissected the resulting tetrads on media containing either glucose or galactose as a carbon source (Fig. 1B). As expected, only two spores from each tetrad germinated on glucose, confirming that a deletion of RFA1 is lethal. However, all four spores from tetrads derived from the diploid strain containing GAL::RFA1-GFP and dissected on media containing galactose were able to germinate and form viable colonies. Their viability on galactose indicates that full-length RFA1-GFP complements rfa1Δ and thus encodes a functional Rfa1 protein. However, rfa1Δ353-GFP and rfa1[1–337,344–621]-GFP fail to complement rfa1Δ on either glucose or galactose, indicating that these rfa1 mutants no longer retain their essential Rfa1 activity. Thus, a full-length Rfa1-GFP fusion protein is functional, but the removal of amino acids 354–621 or 338–343 renders Rfa1 nonfunctional.
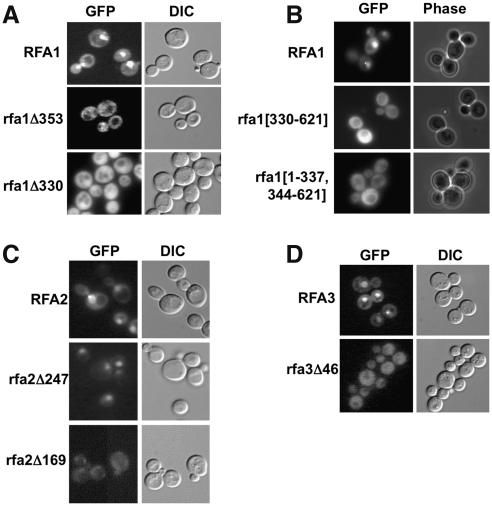
FIG. 2.
Functional alleles of RPA localize to the nucleus, whereas nonfunctional truncations do not. (A) RFA1-GFP fusions under control of their endogenous promoter were expressed in wild-type cells and localized by fluorescence microscopy (GFP) and DIC microscopy of live cells. Rfa1-GFP localized predominantly to the nucleus, whereas the truncations remained in the cytosol. (B) Full-length RFA1-GFP and rfa1 deletion mutants under control of the GAL1 promoter were induced for 2 h with 2% galactose and examined for intracellular localization. RFA1-GFP is primarily in the nucleus, whereas deletion mutants lacking residues 338–343 or 1–329 are retained within the cytoplasm. (C, D) Full-length and truncated RFA2-GFP and RFA3-GFP fusions were expressed in live cells and examined by live cell microscopy. For Rfa2, the full-length protein and the functional rfa2Δ247-GFP fusion localized to the nucleus, whereas the nonfunctional rfa2Δ169-GFP did not. Rfa3-GFP was efficiently imported into the nucleus, whereas the fusion encoded by the rfa3Δ46 truncation remained primarily cytosolic. DIC, differential interference contrast; RPA, replication protein A.
To investigate the functionality of Rfa2- and Rfa3-GFP fusions, we performed complementation analysis in rfa2 and rfa3 conditional mutant strains. Yeast containing rfa2-210 and rfa3-313 temperature-sensitive mutations (Maniar et al., 1997) were transformed with plasmids containing RFA2- and RFA3-GFP fusions under control of their endogenous promoter and assayed for growth at 24°C and 37°C (Fig. 1C, D). As reported previously (Belanger et al., 2004), RFA2-GFP complements rfa2-210 temperature sensitivity. Examination of rfa2Δ247-GFP, which encodes an rfa2 mutant lacking the C-terminal 25 amino acids of the protein, also complements rfa2-210 at 37°C without any apparent growth defect. rfa2Δ169, lacking the region encoding the C-terminal 103 amino acids, does not restore growth of an rfa2-210 strain at 37°C. These data suggest that some amino acids between 169 and 248 are necessary for Rfa2 function when fused to GFP and are similar to complementation results obtained for rfa2 truncations not linked to GFP (Wold, 1997). Full-length RFA3-GFP is also able to complement the conditional rfa3-313 mutant, whereas a truncation of the C-terminal 76 amino acids of Rfa3 eliminates viability at 37°C (Fig. 1D).
We next sought to determine whether the amino acids deleted from each RPA truncation were necessary for localization of the protein to the nucleus. In silico analysis of Rfa1, Rfa2, and Rfa3 using the PSORTII (Nakai and Horton, 1999) and PredictNLS (Cokol et al., 2000) programs failed to identify any sequences predicted to function as a cNLS in these proteins (data not shown). However, we identified a region from residues 338–343 within Rfa1 that contains the amino acid sequence KKFDRR, which includes a cluster of lysine and arginine residues similar to those found in a cNLS. To examine the subcellular localization of Rfa1 and the importance of this region for Rfa1 transport, we expressed full-length RFA1-GFP and rfa1-GFP truncations (rfa1Δ353-GFP and rfa1Δ330-GFP) under control of the endogenous RFA1 promoter (Fig. 2A). Only the fusion containing full-length Rfa1 accumulates primarily in the nucleus, exhibiting intensely fluorescent nuclei with much less intense cytosolic fluorescence. In contrast, both of the rfa1 truncations localize to the cytosol, with exclusion from the vacuole and with very few cells (<10%) exhibiting nuclear fluorescence of slightly greater intensity than the cytosol (see Fig. 2A, rfa1Δ330). Similarly, the C-terminal 291 amino acids of Rfa1, including the putative cNLS, are also retained in the cytoplasm and fail to accumulate in the nucleus (Fig. 2B, rfa1[330–621]).
To more carefully examine the potential cNLS at Rfa1 residues 338–343, we expressed full-length Rfa1-GFP and an Rfa1 mutant with the cNLS removed (rfa1[1–337,344–621]-GFP) in wild-type yeast under control of the GAL1 promoter and observed intracellular fluorescence (Fig. 2B). Induction of these fusions for <2 h reveals that full-length Rfa1-GFP accumulates within the nucleus, with only slightly more cytosolic fluorescence than we expressed from its endogenous promoter. In contrast, Rfa1[1–337,344–621] is almost exclusively cytosolic. Only when Rfa1-GFP is overexpressed for more than 2 h does significant cytoplasmic accumulation occur and this cytoplasmic fluorescence increases until it equals the nuclear fluorescence intensity by 4 h postinduction (data not shown). Rfa1[1–337,344–621] is cytoplasmic throughout its expression. Thus, both amino acid regions 338–343 and 354–621 are necessary for normal nuclear accumulation of Rfa1, although neither is sufficient for targeting a GFP fusion to the nucleus.
The Rfa2 and Rfa3 truncations exhibited a similar correlation between function and nuclear localization. All three complementing fusions—RFA2-GFP, rfa2Δ247-GFP, and RFA3-GFP—are present predominantly in the nucleus (Fig. 2C, D). Both inviable truncations—rfa2Δ169-GFP and rfa3Δ46-GFP—fail to accumulate in the nucleus at levels detectably greater than in the cytosol.
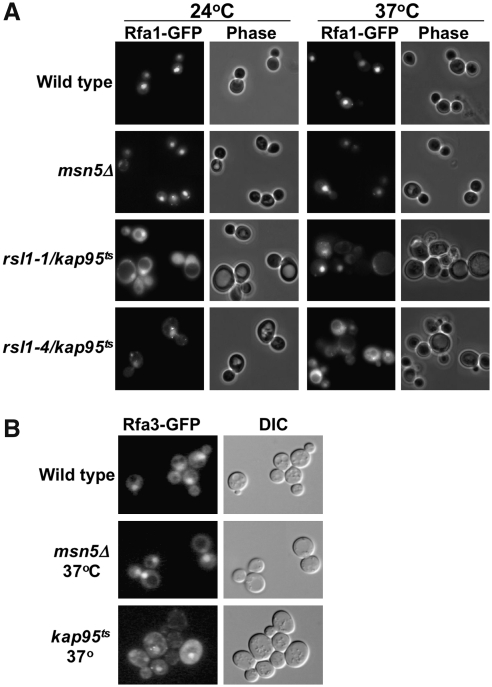
There is evidence that both Msn5/Kap142 and Kap95/Imp-β may participate in nucleocytoplasmic transport of RPA complex proteins, although the relative importance of each to RPA nuclear import is uncertain (Yoshida and Blobel, 2001; Belanger et al., 2004). To investigate the role of these karyopherins in the nuclear transport of the RPA complex, we examined the subcellular localization of functional, full-length Rfa1- and Rfa3-GFP in a number of karyopherin mutants. Rfa1, 2, and 3 all accumulate in the nucleus of kap114Δ, kap120Δ, kap121Δ, kap123Δ, nmd5Δ, and cse1-1 karyopherin mutants (data not shown). To specifically investigate Kap95 and Msn5 function in RPA complex transport, we expressed Rfa1-GFP and Rfa3-GFP in cells containing temperature-sensitive alleles of KAP95 or lacking MSN5 and observed the intracellular localization of both RPA subunits at 24°C and 37°C. To examine Rfa1 localization, GAL::RFA1-GFP expression was induced for 2 h in wild-type, msn5Δ, and two temperature-sensitive kap95 (rsl1-1 and rsl1-4) (Koepp et al., 1996) yeast strains. Cells were then cultured at either 24°C or 37°C and observed for Rfa1-GFP localization by live-cell fluorescence microscopy (Fig. 3). At 24°C, all strains exhibited a predominantly nuclear localization of Rfa1-GFP, although significant cytosolic fluorescence was observed in cells containing the rsl1-1 mutant of KAP95. At 37°C, the difference in localization between msn5Δ and kap95ts became much more pronounced. The msn5Δ mutant cells were indistinguishable from wild type, with nuclear accumulation of Rfa1-GFP in every cell and very little cytoplasmic fluorescence. The kap95ts strains had some cells with nuclear staining, but most cells exhibited a significant accumulation of fluorescence in the cytosol and many had no apparent nuclear fluorescence. These data suggest that Kap95 is important for the efficient import of Rfa1 into the yeast nucleus.
FIG. 3.
Kap95 is important for efficient RPA import, whereas Msn5 is not essential. (A) RFA1-GFP was expressed in wild-type, msn5 deletion (msn5Δ), and kap95 temperature-sensitive cells (rsl1-1 and rsl1-4 alleles of kap95ts) and grown at 24°C. Cells were retained at 24°C or shifted to 37°C for 2 h and observed for localization of Rfa1-GFP. Wild-type and msn5Δ cells contain Rfa1-GFP predominantly in the nucleus at both temperatures. kap95ts cells exhibit an increased level of cytoplasmic fluorescence at both temperatures and nearly equivalent nuclear and cytoplasmic fluorescence in the rsl1-4/kap95ts allele at 37°C. (B) The same experiment was performed with Rfa3-GFP expressed in wild-type, msn5Δ, and kap95ts cells. Rfa3-GFP remained apparent in the nucleus in all three strains, with increased cytoplasmic fluorescence in the kap95ts mutant.
We performed a similar localization experiment using Rfa3-GFP and observed that the pattern of Rfa3-GFP localization in wild-type, msn5Δ, and kap95ts cells is more similar than that observed for Rfa1. Rfa3-GFP localization is predominantly nuclear in all three genetic backgrounds at 24°C, with some cytosolic staining and complete exclusion from the vacuolar compartments (data not shown). At 37°C, wild-type, msn5Δ, and kap95ts cells retain this predominantly nuclear staining (Fig. 3B), although the nuclear accumulation is less distinctive and present in fewer cells in the kap95 strain. These observations suggest that neither Msn5 nor Kap95 is essential for Rfa3 import, possibly due to the small size of the Rfa3 protein.
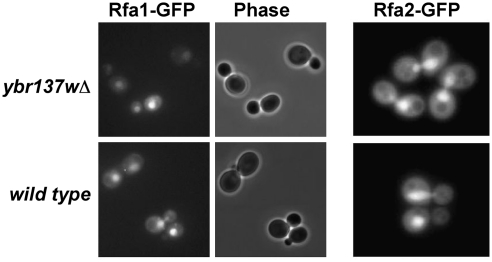
Yoshida and Blobel (2001) observed that a protein of unknown function encoded by the uncharacterized yeast ORF Ybr137w physically associates with the RPA complex in vitro. We sought to determine if a deletion of the Ybr137w open reading frame alters the localization of any of the Rfa-GFP chimeras we generated. We independently expressed Rfa1-GFP, Rfa2-GFP, and Rfa3-GFP in ybr137wΔ and wild-type yeast strains and observed GFP localization at 24°C and 37°C by live cell fluorescence microscopy. The localization of the Rfa-GFP fusions appeared the same between ybr137wΔ and wild-type cells at both temperatures for all three Rfa proteins (Fig. 4; data not shown). These observations confirm earlier data from Yoshida and Blobel (2001) obtained using an Rfa2-PrA fusion and fixed cells that Ybr137w is not essential for nuclear import of members of the RPA complex.
FIG. 4.
Ybr137w is not essential for import of Rfa1 or Rfa2. Full-length Rfa1-GFP and Rfa2-GFP fusions were expressed in cells lacking the RPA-binding protein Ybr137w (ybr137wΔ) and in wild-type cells. Both Rfa1-GFP and Rfa2-GFP are localized to the nucleus with no change in localization in the absence of Ybr137w.
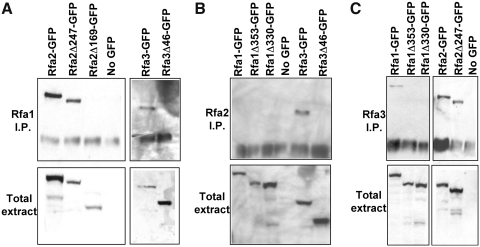
Because the truncations of Rfa1, Rfa2, and Rfa3 affect both protein localization and function, we utilized these truncation alleles and affinity chromatography to investigate intermolecular interactions among the three major proteins of the trimeric RPA complex. To this end, we expressed full-length and truncated fusions of Rfa1, Rfa2, and Rfa3 with GFP in yeast strains that also contained TAP-tagged, full-length alleles of each RPA complex member. We then immunoprecipitated the TAP-tagged full-length proteins from soluble protein extracts obtained from these strains and assayed for the coprecipitation of Rfa1, Rfa2, and Rfa3-GFP fusions (Fig. 5). As expected, immuno-precipitation of full-length Rfa1 results in coprecipitation of Rfa2-GFP and Rfa3-GFP (Fig. 5A). Rfa1 also associates with Rfa2Δ247-GFP with an apparent affinity similar to that of full-length Rfa2. However, Rfa2Δ169- and Rfa3Δ46-GFP cannot be precipitated in a complex with Rfa1, despite being present in detectable quantities in the total soluble protein extract. Full-length Rfa2 coprecipitated Rfa3-GFP, but not Rfa3Δ46-GFP or any of the Rfa1-GFP constructs, despite their presence in abundance in total extract (Fig. 5B). Further, Rfa3 coprecipitated Rfa1-GFP, Rfa2-GFP, and Rfa2Δ247-GFP, but not the nonfunctional Rfa1Δ353- or Rfa1Δ330-GFP truncations (Fig. 5C). Thus, each full-length RPA subunit is able to associate either directly or indirectly with each of the functional Rfa-GFP fusion proteins (excepting Rfa2 not precipitating any Rfa1-GFP chimeras), but fails to coprecipitate those truncations that are not functional and do not localize to the nucleus.
FIG. 5.
Truncated Rfa-GFP fusions that fail to enter the nucleus also fail to bind other members of the RPA complex. Whole cell protein extracts containing one Rfa protein fused to protein A (PrA) and another linked to GFP were immunoprecipitated using IgG-sepharose beads and western blotted using anti-GFP antibodies. For each panel, the top blot contains proteins isolated by immunoprecipitation (I.P.) and the bottom blot contains total soluble protein (total extract). (A) Full-length Rfa1 precipitates functional Rfa2- and Rfa3-GFP fusions (Rfa2-GFP, Rfa2Δ247-GFP, and Rfa3-GFP) but does not associate with nonfunctional C-terminal truncations (Rfa2Δ169-GFP and Rfa3Δ46-GFP). (B) Only full-length Rfa3-GFP was precipitated by Rfa2-PrA. (C) Rfa3-PrA binds functional Rfa1- and Rfa2-GFP fusions (Rfa1-GFP, Rfa2-GFP, and Rfa2Δ247-GFP) but not inviable truncations (Rfa1Δ353-GFP, Rfa1Δ330-GFP, and GFP).
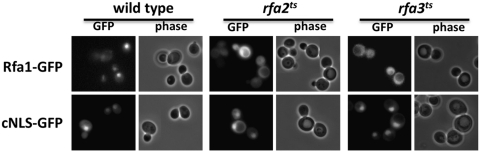
To determine if the absence of a functional subunit of the RPA complex affects the nuclear targeting of other components, we expressed Rfa1-GFP in rfa2-215ts and rfa3-313ts strains under control of the GAL promoter (Fig. 6). Induction of Rfa1-GFP expression results in fluorescence in the cytoplasm of rfa2ts and rfa3ts mutant cells, whereas nuclear localization of Rfa1-GFP is observed in wild-type cells. A cNLS-GFP reporter containing a cNLS is localized to the nucleus in all three strains. These data indicate that the presence of all three RPA complex subunits is important for the nuclear localization of Rfa1.
FIG. 6.
Rfa1-GFP is mislocalized to the cytoplasm in rfa2ts and rfa3ts mutant cells. Plasmids expressing Rfa1-GFP or cNLS-GFP were transformed into wild-type, rfa2-215ts, and rfa3-313ts cells. Cells were grown overnight in media lacking galactose, and then induced for Rfa1-GFP expression by addition of galactose 3 h before microscopy. Rfa1-GFP is detectable in the nucleus of wild-type cells, but remains in the cytoplasm in rfa2ts and rfa3ts cells. The cNLS-GFP control is nuclear in all three strains. cNLS, classical nuclear localization signal.
Discussion
While it is essential for RPA to enter the nucleus to perform its roles in DNA recombination, repair, and replication, the mechanism by which the constituent proteins of the RPA complex traverse the NE is not clear. In this study, we report the use of GFP fusions with the Rfa1, Rfa2, and Rfa3 proteins to investigate the nucleocytoplasmic trafficking of the RPA complex in the yeast S. cerevisiae. This study provides the first real-time examination of Rfa1 and Rfa3 protein localization in yeast using Rfa-GFP constructs that have been shown to be functionally competent. Previous studies have provided evidence that RPA complex proteins interact physically and functionally with Kap95 and Msn5/Kap142 (Yoshida and Blobel, 2001; Belanger et al., 2004), but the role for these karyopherins in RPA transport remains unclear. In this report, we provide additional evidence that Kap95 is the karyopherin primarily responsible for the nuclear accumulation of RPA complex proteins in yeast. We also observe that Rfa1, Rfa2, and Rfa3 truncations lacking function in vivo also fail to accumulate in the nucleus, whereas full-length Rfa-GFP fusions and truncations that complement rfa mutations exhibit predominantly nuclear localization. Finally, we observe that Rfa proteins that are not efficiently imported into the nucleus do not associate with other members of the RPA complex with high affinity in vitro.
All three subunits of RPA are essential for viability (Heyer et al., 1990; Brill and Stillman, 1991; Wold, 1997) and deletions of specific regions of each result in loss of function. We have shown here that the attachment of the 27 kDa GFP protein to the C-terminus of each of the RPA subunits does not compromise their functionality to an extent that results in a decreased growth phenotype at any temperature tested (Fig. 1). In this study, we also confirm that the C-terminal region of both Rfa1 and Rfa3 is essential for function even when the truncated protein terminates with GFP. The C-terminal 103 amino acids of Rfa2 are not necessary for function in the endogenous protein (Philipova et al., 1996) or in our Rfa2-GFP fusion (Fig. 1). These complementation analyses confirm that our C-terminal GFP fusions have retained the function of endogenous Rfa1, Rfa2, and Rfa3. Investigation of RPA intracellular localization has revealed that each subunit localizes to the nucleus, as would be expected of a protein complex that functions in DNA metabolism. Interestingly, others have shown that RPA70 and RPA14 undergo cell cycle-dependent changes in nuclear concentration in metazoans, suggesting that under some conditions the entire complex may not undergo transport or intranuclear anchoring as a trimeric complex (Murti et al., 1996; Sundin et al., 2004). Some independent regulation of RPA subunit assembly and localization is likely in response to intra- and extracellular signals.
The components of the RPA complex associate with the karyopherins Kap95 and Msn5/Kap142 (Yoshida and Blobel, 2001). Kap95 in yeast and its importin-β1 homologs in higher eukaryotes have been shown to function as the primary importers of nuclear proteins with hundreds of transport substrates identified as being dependent upon Kap95 for transport from the cytoplasm into the nucleus (reviewed in Lange et al., 2007). Kap95 has two distinct mechanisms by which it associates with transport substrates: indirect and direct. The indirect association is dependent upon the formation of a Kap95/Kap60 (importin-β1/importin-α) heterodimer. While Kap60 binds to a cNLS in its cargo, Kap95 associates with Kap60 and is primarily responsible for translocation of the complex across the NE. Conversely, direct association involves Kap95 binding directly to its cargo and is not dependent upon Kap60. The RPA complex has been shown to bind directly to Kap95 without requiring Kap60 for association (Yoshida and Blobel, 2001) and therefore probably does not use the cNLS-mediated transport mechanism. That Rfa1, Rfa2, and Rfa3 all undergo changes in nucleocytoplasmic localization in kap95 mutants (Fig. 3) (Belanger et al., 2004) provides further evidence that Kap95 is important for RPA nuclear import.
Msn5/Kap142 has been shown to bind directly to the RPA complex and a deletion of MSN5 results in Rfa2 tagged with Protein A being localized throughout the cell when observed by immunofluorescence microscopy (Yoshida and Blobel, 2001). However, a functional Rfa2-GFP fusion localizes predominantly to the nucleus and undergoes nuclear import with the same kinetics in both wild-type and msn5Δ cells, while failing to be imported efficiently in a kap95 mutant (Belanger et al., 2004). In addition, a deletion of MSN5 results in an altered rate of diffusion of nuclear Rfa2 through the NPC (Belanger et al., 2004), suggesting that Msn5 may be involved in protein tethering within the nucleus or regulation of RPA nuclear export rather than facilitating nuclear import. While Msn5 functions as the karyopherin mediating the nuclear export of >15 proteins that shuttle between the nucleus and cytoplasm (Kaffman et al., 1998; Blondel et al., 1999; DeVit and Johnston, 1999; Mahanty et al., 1999; Komeili et al., 2000; Boustany and Cyert, 2002; Görner et al., 2002; Jaquenoud et al., 2002; Queralt and Igual, 2003; Strahl et al., 2005; Quan et al., 2006; Ueta et al., 2007; Bakhrat et al., 2008; Towpik et al., 2008), it is unlikely that Msn5 functions to export the RPA complex. Msn5 associates with RPA proteins at a high affinity in the presence of Ran-GDP, which is predominant in the cytoplasm, whereas dissociation of Msn5 from the RPA complex takes place upon addition of Ran-GTP, which is most prevalent in the nucleus (Yoshida and Blobel, 2001). If Msn5 were to function as an exportin for RPA complex proteins, it would be required to bind in the nucleus and release in the cytoplasm. However, the changes in intracellular localization observed for Rfa1 in yeast and the Rfa3 homolog in mammalian cells (Murti et al., 1996; Sundin et al., 2004) during different stages of the cell cycle implicate subcellular localization in the regulation of the activity of RPA subunits. It remains to be determined whether Msn5 or some other factor is involved in maintaining these patterns of localization.
Here and in earlier studies (Belanger et al., 2004) we report that a loss of Kap95 activity leads to a change in localization of Rfa1- and Rfa2-GFP from nuclear to cytosolic, whereas Rfa1 and Rfa2 remains accumulated in the nucleus of cells lacking Msn5. These data suggest that Kap95 is the karyopherin primarily responsible for nuclear import of the Rfa1 and Rfa2 subunits of RPA, either independently or as part of a larger complex. It is beyond the resolution of our assay to show that RPA proteins are excluded from the nucleus in a kap95 mutant, thus making the import of some Rfa1 and Rfa2 by Msn5 a possibility. Rfa3-GFP is also localized similarly in wild-type and msn5Δ cells, but undergoes a more subtle change in localization in kap95 mutants, remaining at a higher level in the nucleus after shift to the nonpermissive temperature. This nuclear accumulation could be due to import by another transporter, such as Msn5, in the absence of Kap95. Alternatively, Rfa3 may continue to selectively accumulate in the nucleus due to diffusion across the NE and anchoring within the nucleus. At 17 kDa, monomeric Rfa3 is below the diffusion limit for NPC and can travel freely through nuclear pores independent of karyopherins. Even fused to GFP, the Rfa3-GFP chimera remains small enough to undergo limited nucleocytoplasmic diffusion. Any free Rfa3 monomers could diffuse into the nucleus and be retained there by association with other proteins, DNA, or any other nuclear molecule for which it may have affinity. RPA subunits associate and dissociate in a regulated manner throughout the cell cycle (Cardoso, 1993) and evidence that the small subunit of RPA can move independently in mammalian cells is provided by its relocation to the cytoplasm in M-phase, whereas the larger subunits remain nuclear (Murti et al., 1996). Experimental identification of the sequences necessary and sufficient for mediating RPA import and association with karyopherins will be necessary to completely resolve the Kap(s) responsible for RPA targeting.
To investigate regions of RPA complex proteins important for nuclear import, we examined the intracellular localization of Rfa-GFP truncations. We observed that Rfa-GFP fusions that were functional in vivo were localized to the nucleus, with lesser concentrations in the cytosol and exclusion from the vacuole. These functional nuclear fusions included full-length Rfa1, Rfa2, and Rfa3 chimeras with GFP, plus the Rfa2 truncation lacking 26 amino acids from the C-terminal end. Deletion of regions of Rfa1-, Rfa2-, and Rfa3-GFP fusions that resulted in nonfunctional protein also resulted in mislocalization of the chimeric polypeptide to the cytoplasm. These deleted C-terminal regions of Rfa1 and Rfa2 are essential for complex formation with each other and with Rfa3 (Lin et al., 1996; Bochkareva et al., 2000) (Fig. 5 this paper) for generation of the RPA heterotrimer. In addition, Rfa1-GFP does not localize to the nucleus in rfa2ts or rfa3ts mutant cells (Fig. 6). Taken together, these localization and association data suggest that either an intact heterotrimer must form for each of the subunits to be targeted to the nucleus or that each subunit has its own NLS located within its C-terminal protein–protein interaction domain. Given that Rfa2 and Rfa3 are essential for the proper folding of Rfa1 in vitro (Stigger et al., 1994), it is likely that the fully assembled heterotrimer contains clusters of residues within its tertiary structure that serve as an NLS (or NLSs) for karyopherin recognition and subsequent nuclear import.
Here we have provided data that support a model for RPA nucleocytoplasmic transport that includes Kap95 as the predominant karyopherin and the C-terminus of each RPA subunit as essential for efficient nuclear import. However, it remains possible that multiple karyopherins are involved in the nuclear import of RPA, and even that karyopherin-mediated shuttling of the entire complex or individual subunits is important for RPA function. The generation of additional mutants will be necessary to identify the regions of each subunit that function as NLSs and, eventually, to carefully investigate which karyopherins associate with those domains to mediate RPA transport.
Acknowledgments
The authors would like to acknowledge Shouhei Yamagami, Vanessa Obourn, and Colgate students from Biology 483 for preliminary experiments and insightful discussions. Plasmids and strains were kindly shared by Pamela Silver, Steve Brill, and Laura Davis. Invaluable technical assistance by Susan Geier and Karyn Belanger is greatly appreciated. Funding was provided by Colgate University and NIH-AREA grant GM065107 to K.D.B.
Disclosure Statement
No competing financial interests exist.
References
- Adams A. Gottschling D.E. Kaiser C.A. Stearns T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. Methods in Yeast Genetics. [Google Scholar]
- Akada R. Kallal L. Johnson D.I. Kurjan J. Genetic relationships between the G protein beta gamma complex, Ste5p, Ste20p, and Cdc42p: investigation of effector roles in the yeast pheromone response pathway. Genetics. 1996;143:103–117. doi: 10.1093/genetics/143.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz P.M. Matheos D. Cunningham K.W. Estruch F. The Saccharomyces cerevisiae RanGTP-binding protein Msn5p is involved in different signal transduction pathways. Genetics. 1999;153:1219–1231. doi: 10.1093/genetics/153.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhrat A. Baranes-Bachar K. Reshef D. Voloshin O. Krichevsky O. Raveh D. Nuclear export of Ho endonuclease of yeast via Msn5. Curr Genet. 2008;54:271–281. doi: 10.1007/s00294-008-0216-8. [DOI] [PubMed] [Google Scholar]
- Belanger K.D. Simmons L.A. Roth J.K. VanderPloeg K.A. Lichten L.B. Fahrenkrog B. The karyopherin Msn5/Kap142 requires Nup82 for nuclear export and performs a function distinct from translocation in RPA protein import. J Biol Chem. 2004;279:43530–43539. doi: 10.1074/jbc.M407641200. [DOI] [PubMed] [Google Scholar]
- Blondel M. Alepuz P.M. Huang L.S. Shaham S. Ammerer G. Peter M. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 1999;13:2284–2300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E. Korolev S. Bochkarev A. The role for zinc in replication protein A. J Biol Chem. 2000;275:27332–27338. doi: 10.1074/jbc.M000620200. [DOI] [PubMed] [Google Scholar]
- Bochkareva E. Korolev S. Lees-Miller S.P. Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boustany L.M. Cyert M.S. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 2002;16:608–619. doi: 10.1101/gad.967602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S.J. Bastin-Shanower S. Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol Cell Biol. 1998;18:7225–7234. doi: 10.1128/mcb.18.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S.J. Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Cardoso M.C. Leonhardt H. Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- Cokol M. Nair R. Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E. Uy M. Leighton L. Blobel G. Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- D'Angelo M.A. Hetzer M.W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVit M.J. Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- Enenkel C. Blobel G. Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Estruch F. Carlson M. Increased dosage of the MSN1 gene restores invertase expression in yeast mutants defective in the SNF1 protein kinase. Nucleic Acids Res. 1990;18:6959–6964. doi: 10.1093/nar/18.23.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B. Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- Fanning E. Klimovich V. Nager A.R. A dynamic mnodel for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries T. Betz C. Sohn K. Caesar S. Schlenstedt G. Bailer S.M. A novel conserved nuclear localization signal is recognized by a group of yeast importins. J Biol Chem. 2007;282:19292–19301. doi: 10.1074/jbc.M700217200. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. Woods R.A. Transformation of yeast by the Liac/ss carrier DNA/PEG method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Görlich D. Kostka S. Kraft R. Dingwall C. Laskey R.A. Hartmann E. Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Görner W. Durchschlag E. Wolf J. Brown E.L. Ammerer G. Ruis H. Schüller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. Fink G.R. Guide to Yeast Genetics and Molecular Biology. Academic Press; San Diego: 1991. [Google Scholar]
- Harel A. Forbes D.J. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Heyer W.D., et al. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode C. Daniely Y. Borowiec J.A. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. Kutay U. von Kobbe C. Mattaj I.W. Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M. van Drogen F. Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C (Cdh1) EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A. Rank N.M. O'Neill E.M. Huang L.S. O'Shea E.K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Koepp D.M. Wong D.H. Corbett A.H. Silver P.A. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A. Wedaman K.P. O'Shea E.K. Powers T. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A. Mills R.E. Lange C.J. Stewart M. Devine S.E. Corbett A.H. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J. Sekimoto T. Yamashita E. Nagoshi E. Nakagawa A. Imamoto N. Yoshimura M. Sakai H. Chong K.T. Tsukihara T. Yoneda Y. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- Lin Y.L. Chen C. Keshav K.F. Winchester E. Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C. Heitman J. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics. 1998;150:1443–1457. doi: 10.1093/genetics/150.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A.S. Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- Mahanty S.K. Wang Y. Farley F.W. Elion E.A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- Maniar H.S. Wilson R. Brill S.J. Roles of replication protein-A subunits 2 and 3 in DNA replication fork movement in Saccharomyces cerevisiae. Genetics. 1997;145:891–902. doi: 10.1093/genetics/145.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N. Pemberton L.F. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Murti K.G. He D.C. Brinkley B.R. Scott R. Lee S.H. Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp Cell Res. 1996;223:279–289. doi: 10.1006/excr.1996.0083. [DOI] [PubMed] [Google Scholar]
- Nagoshi E. Imamoto N. Sato R. Yoneda Y. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol Biol Cell. 1999;10:2221–2233. doi: 10.1091/mbc.10.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K. Horton P. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Palmeri D. Malim M.H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipova D. Mullen J.R. Maniar H.S. Lu J. Gu C. Brill S.J. A hierarchy of SSB protomers in replication protein A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- Poon I.K. Jans D.A. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Quan X. Tsoulos P. Kuritzky A. Zhang R. Stochaj U. The carrier Msn5p/Kap142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol Microbiol. 2006;62:592–609. doi: 10.1111/j.1365-2958.2006.05395.x. [DOI] [PubMed] [Google Scholar]
- Queralt E. Igual J.C. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol Cell Biol. 2003;23:3126–3140. doi: 10.1128/MCB.23.9.3126-3140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal P.K. Kumar P.R. Rao M.R. Kyasani M. Mahalingam S. Simian immunodeficiency virus Vpx is imported into the nucleus via importin alpha-dependent and -independent pathways. J Virol. 2006;80:526–536. doi: 10.1128/JVI.80.1.526-536.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigger E. Dean F.B. Hurwitz J. Lee S.H. Reconstitution of functional human single-stranded DNA-binding protein from individual subunits expressed by recombinant baculoviruses. Proc Natl Acad Sci USA. 1994;91:579–583. doi: 10.1073/pnas.91.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T. Hama H. DeWald D.B. Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–979. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström A.C. Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin B.A. Chiu C.H. Riffle M. Davis T.N. Muller E.G. Localization of proteins that are coordinately expressed with Cln2 during the cell cycle. Yeast. 2004;21:793–800. doi: 10.1002/yea.1133. [DOI] [PubMed] [Google Scholar]
- Terry L.J. Shows E.B. Wente S.R. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Towpik J. Graczyk D. Gajda A. Lefebvre O. Boguta M. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J Biol Chem. 2008;283:17168–17174. doi: 10.1074/jbc.M709157200. [DOI] [PubMed] [Google Scholar]
- Tran E.J. Wente S.R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Truant R. Cullen B.R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta R. Fujiwara N. Iwai K. Yamaguchi-Iwai Y. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2980–2990. doi: 10.1091/mbc.E06-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Yoshida K. Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y. Liu Y. Wu X. Shell S.M. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Phys. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]