Abstract
Phospholipase C-β (PLCβ) is a key regulator of intracellular calcium levels whose activity is controlled by heptahelical receptors that couple to Gq. We have determined atomic structures of two invertebrate homologs of PLCβ (PLC21) from cephalopod retina and identified a helix from the C-terminal regulatory region that interacts with a conserved surface of the catalytic core of the enzyme. Mutations designed to disrupt the analogous interaction in human PLCβ3 dramatically increase basal activity and diminish stimulation by Gαq. Gαq binding requires displacement of the autoinhibitory helix from the catalytic core, thus providing an allosteric mechanism for activation of PLCβ.
Phospholipase C-β (PLCβ) proteins form a highly conserved enzyme family that hydrolyze phosphatidylinositol 4,5-bisphosphate (PIP2) into inositol-1,4,5-triphosphate (IP3) and diacylglycerol, two key second messengers that mobilize intracellular calcium and stimulate the activity of protein kinase C1,2. PLCβ isoforms are potently activated via direct interactions with heterotrimeric G proteins of the Gαq family3,4, Gβγ heterodimers5–8, and small GTPases such as Rac19,10. The interaction between Gαq and PLCβ is of particular interest because regulation of PLCβ by Gq-coupled receptors is critical for normal cardiomyocyte function, and maladaptive changes in this pathway can result in the onset of cardiac arrhythmias, cardiac hypertrophy, and heart failure11–14.
PLCβ proteins and their invertebrate homologs NorpA15 and PLC2116,17 share a highly conserved catalytic core comprised of an N-terminal pleckstrin homology (PH) domain, followed by four EF hand domains, a triose-phosphate isomerase (TIM) barrel-like catalytic domain split into X and Y halves by a variable linker18,19, and a C2 domain1,20 (Fig. 1a)21,22,23. The X-Y linker is positioned adjacent to the active site, and its cleavage or truncation increases basal activity22,24,25. However, such activation is independent of both heterotrimeric G proteins and small GTPases22. The distinguishing feature of PLCβ enzymes is a ~400 amino acid C-terminal region (CTR) that is known to be important for membrane association as well as Gαq binding and activation1,26–29. Many of these functional properties have been mapped to residues within an extended coiled-coil domain found in the C-terminus, corresponding to residues 946 to 1200 in human PLCβ3 (Fig. 1a)1,26–29. Recently, a structure was reported for Gαq in complex with a human PLCβ3 truncation (Δ887) that includes a small portion of the CTR (residues 848 to 882). This region forms a helix-turn-helix motif (Hα1-Hα2) that docks with the effector-binding site of Gαq23. Although this structure revealed key interactions between PLCβ3 and Gαq, the activity of the Δ887 fragment was not shown to be regulated by Gαq and thus it remains unclear how Gαq enhances PLCβ3 activity and how other regions of the CTR contribute to its regulation27,30,31.
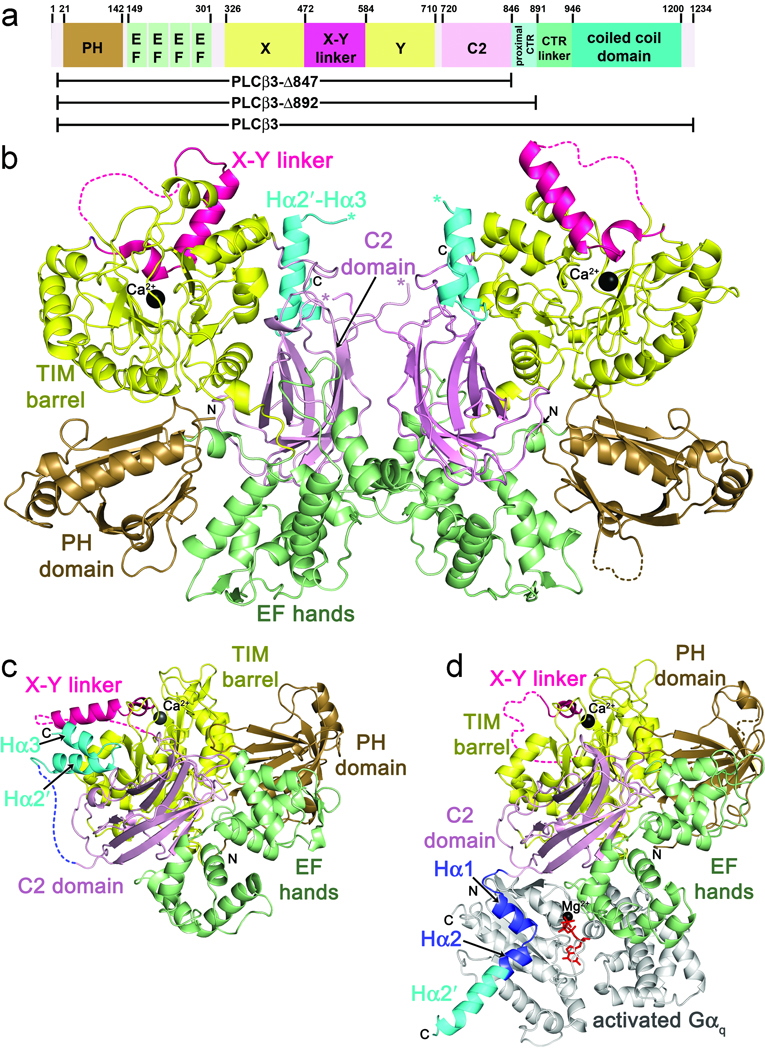
Figure 1.
Primary and tertiary structures of PLCβ family members, and comparison of cephalopod PLC21 with the Gαq–PLCβ3 complex. (a) Primary structure of human PLCβ3. PLCβ3 truncations used in this paper are indicated below the diagram. Numbers above the diagram correspond to amino acid positions at domain boundaries. (b) Crystal structure of LPLC21. LPLC21 crystallized as a dimer with pseudo two-fold symmetry. Domains are colored as in a. The Hα2′-Hα3 hairpin from the proximal CTR is shown in cyan, and the catalytic Ca2+ is shown as a black sphere. Disordered loops are drawn as dashed lines, with the exception of the connection between the C2 domain and the beginning of Hα2′, which is ambiguous in the dimer interface. The C-terminus of the C2 domain and start of Hα2′ are marked with pink and blue asterisks, respectively. N- and C-terminal ends of the protein fragment resolved in the crystal structure are labeled N and C, respectively. (c) Crystal structure of SPLC21. Domains are colored as in b. (d) Crystal structure of the Gαq–PLCβ3 complex (PDB entry 3OHM)23. Hα1 and Hα2, which form the primary Gαq binding site, are shown in dark blue. Residues corresponding to Hα2′ in the PLC21 structures are shown in cyan. Activated Gαq is shown in light gray, with GDP and AlF4 colored red, and Mg2+ black.
To better understand the activation mechanism of PLCβ, we solved crystal structures of two invertebrate, endogenously expressed PLCβ homologs from cephalopod retina. Within these structures, a helix located immediately C-terminal to the portion of the Hα1-Hα2 motif that directly interacts with Gαq is observed to dock with a conserved cleft on the PLCβ catalytic core. Perturbation of the analogous interaction in human PLCβ3 dramatically enhances basal activity, lowers the thermostability of the enzyme, increases Gαq affinity, and reduces the efficacy of Gαq activation. Our results are consistent with an allosteric mechanism in which Gαq binding displaces this inhibitory helix, leading to enhanced activity. Our studies also confirm that more distal regions of the CTR enhance the affinity of PLCβ3 for Gαq and facilitate PIP2 hydrolysis through a mechanism independent of Gαq.
RESULTS
Structures of cephalopod PLC21
Crystal structures of endogenously expressed Loligo pealei (LPLC21) and Sepia officinales PLC21 (SPLC21) were solved by molecular replacment to 3.1 and 2.0 Å resolution, respectively (Fig. 1b,c and Table 1). For this work, it was necessary to sequence the coding region for SPLC21 from its endogenous source, which we found to be ~92% identical to that of LPLC21. Although full length proteins were subjected to crystallization trials, both SPLC21 and LPLC21 crystallized as ~95 kDa proteolytic fragments (Supplementary Methods online) that contain visible density for most of the catalytic core. In the higher resolution SPLC21 structure, residues 11–474 and 485–774 of the catalytic core as well as 26 residues from the CTR (residues 790–815) are visible. The catalytic cores of LPLC21 and SPLC21 are essentially identical, and superimpose with an r.m.s.d. value of 0.36 Å for 765 Cα atoms (out of 778 total in SPLC21). Within the catalytic core of SPLC21, only a short segment of the X-Y linker (amino acids 475–484) is missing electron density (Supplementary Fig. 1 online). There are only subtle differences in the domain arrangment of the catalytic core between cephalopod and human homologs. Most notably, the EF hand and C2 domains of SPLC21 are rotated by ~6° away from the TIM barrel and PH domains relative to their positions in the structures of PLCβ2 and PLCβ3.
Table 1.
Data collection and refinement statistics
| Sepia PLC21 | Loligo PLC21 | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 60.8, 83.4, 163.1 | 82.4, 148.9, 151.6 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 27.4-2.00 (2.04-2.00)* | 29.97-3.10 (3.15-3.10) |
| Rsym or Rmerge | 0.069 (0.679) | 0.146 (0.814) |
| I / σI | 17.4 (1.58) | 14.2 (2.3) |
| Completeness (%) | 97.7 (98.2) | 100 (100) |
| Redundancy | 3.7 (3.6) | 7.2 (7.2) |
| Refinement | ||
| Resolution (Å) | 27.4-2.00 | 30.0-3.20 |
| No. reflections | 53335 | 29832 |
| Rwork / Rfree | 0.176/0.208 | 0.232/0.256 |
| No. atoms | ||
| Protein | 12668 | 24953 |
| Ligand/ion | 126 | 2 |
| Water | 452 | 38 |
| B-factors | ||
| Protein | 20.2 | 61.5 |
| Ligand/ion | 45.9 | 60.2 |
| Water | 28.1 | 36.3 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 0.946 | 0.881 |
Each data set was collected from a single crystal.
Values in parentheses are for highest-resolution shell.
The TIM barrel-like domain of SPLC21 is ~55% identical in sequence with those of human PLCβ2 and PLCβ3, and can be superimposed with r.m.s.d. values of 0.38 and 0.39 Å for 232 and 234 Cα atoms, respectively, omitting the X–Y linker (residues 473–510 of SPLC21). In SPLC21, the C-terminal end of the linker (residues 500–510, corresponding to residues 576–586 in human PLCβ3) has essentially the same structure as in other PLCβ structures. However, residues 485–499 form a helix that extends from the catalytic site in a different direction than observed in previous PLCβ structures (Supplementary Fig. 1 online) 22,23. The signficance of this difference is not yet understood, but this unique conformation is also found in both unique chains of LPLC21 (Fig. 1b) suggesting that it is not dictated by the crystal packing environment.
In both PLC21 structures, strong electron density is observed for a segment of the CTR that can be modeled as a helical hairpin composed of Hα2′ and Hα3 helices (residues 790–816 of SPLC21) (Fig. 2a,b and Supplementary Fig. 2 online). The loop connecting the hairpin to the end of the C2 domain (residues 775–789) is disordered. The Hα2′ helix (residues 792–805) corresponds to the C-terminal end of the PLCβ3 Hα2 helix, just beyond the region of the Hα1-Hα2 motif in PLCβ3 that directly contacts Gαq (Fig. 1d and Supplementary Fig. 3a online). Hα2′ packs against a highly conserved, hydrophobic cleft formed between the TIM barrel and C2 domains of the catalytic core, in close proximity to the active site and X–Y linker, and buries 1200 Å2 of solvent accessible surface area (Fig. 2a,b). The side chains of Arg797, Leu801, and Phe804 form the principal interactions from Hα2′. The positively-charged guanidinium group of Arg797 caps the C-terminus of the Tα5 helix of the TIM barrel domain, and, along with Ala800 and Leu801, forms a hydrophobic pocket for the Phe639 side chain located in the linker between the TIM barrel and C2 domains of the catalytic core. The side chain of Phe804 packs in a hydrophobic cavity formed by the side chains of Val584 and Ile587 in the Tα6 helix of the TIM barrel domain and Pro679 and Thr682 of the C2 domain. The C-terminal end of Hα2′ is capped by the side chain of Arg684 from the C2 domain. The residues involved in these interactions are highly conserved among PLCβ enzymes, but poorly so in other PLC isozymes (Supplementary Fig. 3 online), consistent with the hypothesis that they contribute to a functionally important, PLCβ-specific intramolecular contact. Indeed, the same Hα2′-catalytic core interaction is observed in the SPLC21 structure (Fig. 1c and Fig. 2a,c) and the two independent chains of the LPLC21 structure (Fig. 1b), and a very similar interaction is formed in the Gαq–PLCβ3 crystal structure as an intermolecular crystal contact (Fig. 2c,d). The Hα2′-catalytic core interaction has thus persisted over 500 million years of evolution32.
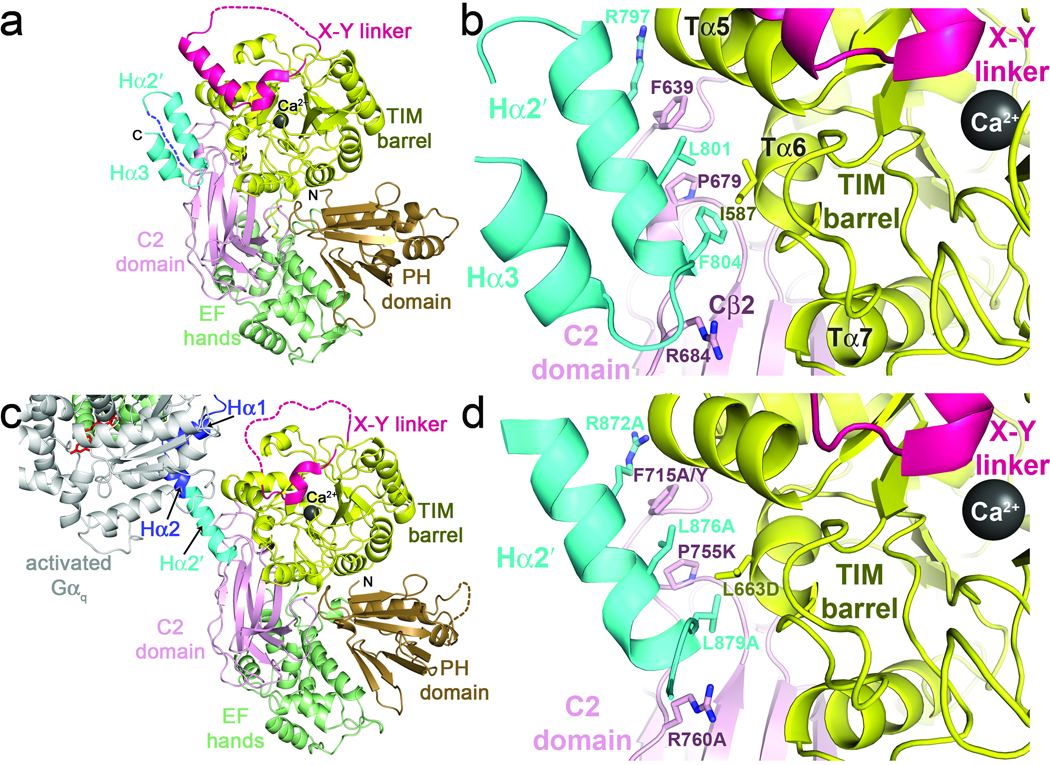
Figure 2.
Interactions of Hα2′ with the catalytic core. (a) The SPLC21 Hα2′ helix docks in a conserved cleft formed between the TIM barrel and C2 domains, in close proximity to the active site and the X–Y linker. The shorter Hα3 helix forms a hairpin interaction with Hα2′ stabilized by hydrophobic interactions. Domains are colored as in 1a. (b) Specific interactions of SPLC21 Hα2′ with the catalytic core. Side chains that make large contributions to the binding interface are shown as sticks with carbons colored according to their respective domains and nitrogens blue. (c) The Hα2′-catalytic core interaction is recapitulated in a crystal contact of the Gαq–PLCβ3 structure (PDB entry 3OHM)23. Domains are colored as in 1d. The subunit of Gαq shown is in complex with a different catalytic core in the crystal lattice. (d) Specific interactions between Hα2′ and the catalytic core in human PLCβ3 (Fig. 1c). Residues analogous to those of SPLC21 shown in b are drawn as sticks, and site-directed mutations created in this study to perturb the interface are indicated. The SPLC21 Hα2′ helix is continuous, whereas Hα2′ in human PLCβ3 is kinked at Ala877, as if to optimize the interactions of the Leu879 side chain, which is smaller than that of the corresponding Phe804 residue in SPLC21.
Gαq binding to the Hα1-Hα2 motif is incompatible with the Hα2′ helix binding to the catalytic core in the same molecule. Indeed, the region is required to undergo a large conformational change upon Gαq binding, as the Cα atom of PLCβ3-Arg872 is translated ~50 Å away from its position when Hα2′ is docked with the catalytic core, as modeled by the equivalent atom of Arg797 in the SPLC21 structure (Fig. 1c,d). Given the high sequence conservation of Hα2′ among PLCβ isoforms, its close proximity to the active site, and its juxtaposition with the primary Gαq binding site, we hypothesized that the Hα2′ helix plays a role in the regulation of PLCβ by Gαq. For subsequent experiments, we focused on human PLCβ3 because it is readily expressed in baculovirus-infected insect cells and is potently and efficaciously regulated by Gαq7,33.
The Hα2′ helix modulates stability of the catalytic core
If the Hα2′ helix plays a regulatory role, then it should stably associate with the PLCβ catalytic core in solution. To detect this interaction, we used a ThermoFluor assay to measure the thermostability of three purified recombinant variants: PLCβ3 (which spans residues 10–1234 containing the complete CTR), PLCβ3-Δ892 (which terminates at residue 891 after the proximal CTR region observed in the PLC21 and PLCβ3 crystal structures), and PLCβ3-Δ847 (which terminates at residue 846 and lacks the entire CTR) (Fig. 1a). Whereas PLCβ3 and PLCβ3-Δ892 had a similar melting temperature (Tm) of 43 and 45 °C, respectively, PLCβ3-Δ847 had a markedly lower Tm of 38 °C, suggesting that the presence of the proximal CTR enhances the thermal stability of the catalytic core (Fig. 3a and Supplementary Table 1). We next tested if this thermostabilization could be accounted for by specific residues in the interface of Hα2′ with the catalytic core. Indeed, mutation of six individual residues in the context of the full-length protein (PLCβ3) reduced the Tm to 39–40 °C, similar to that of PLCβ3-Δ847 (Supplementary Table 1). Interface mutations in the background of PLCβ3-Δ892 similarly reduced the melting point by ~5 °C. In contrast to its effects in the context of PLCβ3 and PLCβ3-Δ892, the F715A mutation of PLCβ3-Δ847 did not significantly change thermostability, indicating that mutation of the catalytic core itself does not lead to lower stability. Thus, the higher thermostability exhibited by PLCβ3 and PLCβ3-Δ892 relative to PLCβ3-Δ847 is dependent on a specific interaction between the catalytic core and Hα2′.
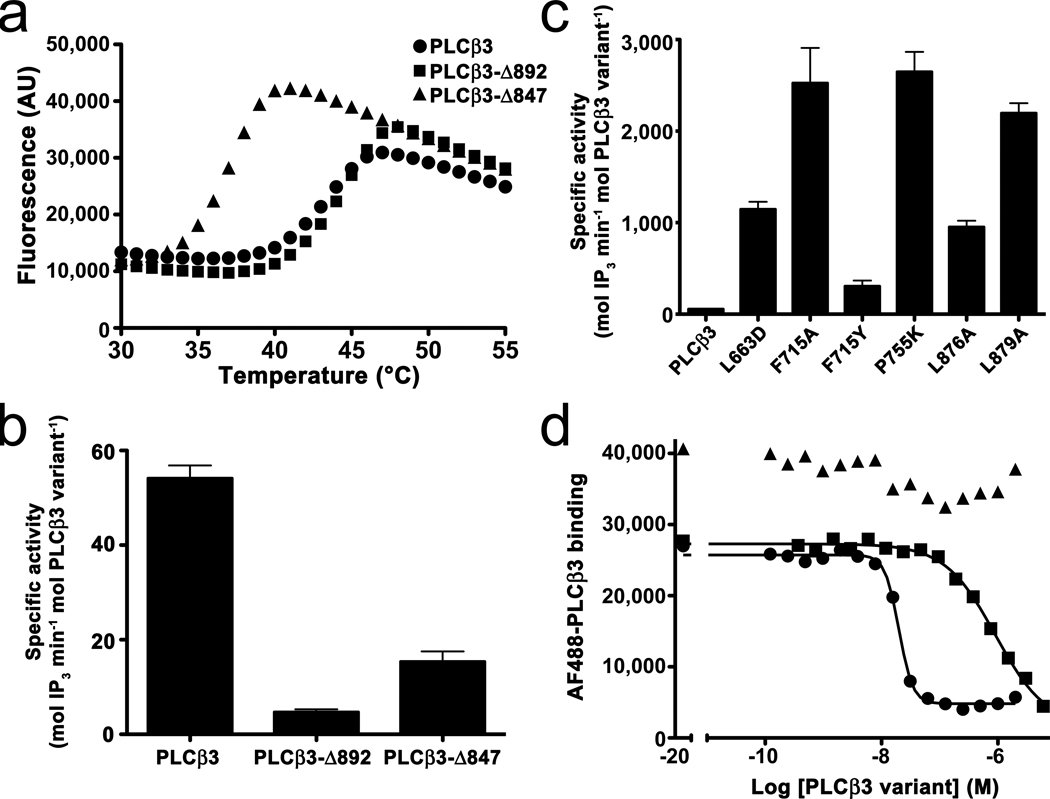
Figure 3.
Functional studies of PLCβ3 variants. (a) The proximal CTR stabilizes the catalytic core. ThermoFluor assays were used to measure the melting point of three PLCβ3 variants by monitoring the change in fluorescence of ANS. Representative curves are shown for PLCβ3 (circles), PLCβ3-Δ892 (squares), and PLCβ3-Δ847 (triangles). PLCβ3-Δ847 is 5–7 °C less stable (left-shifted) than PLCβ3 or PLCβ3-Δ892. See Supplementary Table 1. AU, arbitrary units. (b) Comparison of the basal activity of PLCβ3 variants. Deletion of the proximal CTR in PLCβ3-Δ847 increases basal activity relative to PLCβ3-Δ892. The higher basal activity of PLCβ3 reflects the contribution of more distal regions of the CTR to maximal activity. Activity was measured by counting free [3H]-IP3 released from liposomes containing [3H]-PIP2 at 30 °C in the presence of ~200 nM free Ca2+ at 4–5 time points. The data shown represent at least four individual experiments performed in duplicate ± SEM. (c) Mutation of PLCβ3 at positions that contribute to the Hα2′-catalytic core interface dramatically increase basal activity, indicating that this interaction is involved in autoinhibition. (d) Distal regions of the PLCβ3 CTR enhance binding to Gαq. FCPIA was used to quantify the ability of PLCβ3 truncations to displace AlexaFluor488-labeled PLCβ3-Δ892 (R872A L876A L879A triple mutant) from biotinylated, AlF4-activated Gαi/q bound to avidin beads. Representative curves for PLCβ3 (circles), PLCβ3-Δ892 (squares) and PLCβ3-Δ847 (triangles) are shown. See Table 3 for measured inhibition constants.
The Hα2′ helix is an autoinhibitory element
Given the proximity of the Hα2′ helix to the active site and its contribution to protein stability, we hypothesized that its interactions with the core could influence catalytic activity. Because the CTR coiled-coil domain of PLCβ3 facilitates membrane recruitment of the enzyme to the substrate PIP2, the role of the Hα2′ helix in catalytic activity is most easily assessed by comparison of the truncations PLCβ3-Δ892 and PLCβ3-Δ847, or of site-directed mutations made in the full-length protein versus wild-type. PLCβ3 has 10-fold higher basal specific activity than PLCβ3-Δ892 (54 vs. 4.7 mol IP3 min−1 mol−1 PLCβ3, respectively), confirming that residues 892–1234 of the CTR strongly promote catalysis. The PLCβ3-Δ847 variant has 3-fold higher activity than PLCβ3-Δ892 (15 mol IP3 min−1 mol−1 PLCβ3), indicating that the presence of the proximal CTR inhibits the basal activity of PLCβ3 (Fig. 3b and Table 2). To confirm that specific interactions of Hα2′ with the catalytic core are responsible for the observed differences in basal activity, we measured the activities of point mutants within the Hα2′-catalytic core interface of PLCβ3. The L663D, F715A and P755K mutants in the catalytic core enhance basal activity by over an order of magnitude, as do point mutants of interacting residues in Hα2′ (Fig. 3c and Table 2). The conservative F715Y mutation leads to a more modest increase in activity (6-fold). The complimentary results obtained from mutation of residues on either Hα2′ or its binding site on the catalytic core strongly suggest that this interaction serves to repress the basal activity of PLCβ3. Interface mutations made in the background of PLCβ3-Δ892 yield smaller but consistent increases in activity, although these results should be considered along with the fact that the F715A mutation also causes a mild increase in activity in the context of PLCβ3-Δ847 (Table 2). Thus, mutations in the catalytic core itself can increase activity, but these effects are greatly amplified in the context of the full-length protein. The basal activity data of full-length PLCβ3 and the thermostability data are consistent with the interaction of Hα2′ with the catalytic core, thereby trapping the TIM barrel-like domain in a more quiescent state.
Table 2.
Basal Activity of PLCβ3 Variants
| Variant | Specific Activity ± SEMa (mol IP3 min−1 mol−1PLCβ3) |
Fold Increase Relative to Wild-Type |
|---|---|---|
| PLCβ3 (wt)b | 54 ± 2.7 | 1 |
| -L663D | 1150 ± 81 | 23 |
| -F715A | 2500 ± 380 | 51 |
| -F715Y | 310 ± 62 | 6 |
| -P755K | 2650 ± 216 | 53 |
| -L876A | 950 ± 69 | 19 |
| -L879A | 2000 ± 190 | 41 |
| PLCβ3-Δ892 (wt) | 4.7 ± 0.6 | 1 |
| -F715Ab | 9.5 ± 0.4 | 2 |
| -R760Ab | 7.2 ± 0.7 | 2 |
| -R872Ab | 22 ± 2.2 | 5 |
| -L876A | 9.9 ± 1.6 | 2 |
| -L879Ab | 13 ± 1.9 | 3 |
| -L876A L879Ab | 14 ± 1.1 | 3 |
| -AAAb,c | 14 ± 1.4 | 3 |
| PLCβ3-Δ847 (wt)b | 15 ± 2.1 | 1 |
| -F715A | 39 ± 4.4 | 2.3 |
At least five independent experiments performed in duplicate.
Includes samples from at least two independent protein purifications.
R872A L876A L879A
The Hα2′ helix and distal CTR modulate the affinity of Gαq for PLCβ3
The intermolecular interaction of the Hα1-Hα2 module with Gαq and the intramolecular interaction of Hα2′ with the catalytic core of PLCβ3 are expected to be competitive. If so, then disrupting the Hα2′-catalytic core interface should enhance the affinity of Gαq for PLCβ3. We used a flow cytometry protein interaction assay (FCPIA)34,35 to measure binding between Gαq and PLCβ3, in which the binding affinities of unlabeled PLCβ3 and its variants for Gαq are determined by their ability to displace a fluorescently-labeled variant of PLCβ3 from beads (Fig. 3d and Table 3). PLCβ3-Δ847 is unable to bind Gαq at any concentration tested, consistent with the absence of the proximal CTR. Although PLCβ3-Δ892 retains this region, it has nearly two orders of magnitude lower affinity for Gαq (270 nM) than full-length PLCβ3 (6 nM), suggesting that the more distal regions of the CTR contribute key interactions that enhance binding to Gαq1,27. The affinity of our truncation is consistent with that measured by surface plasmon resonance for the Δ887 truncation used in the crystallographic analysis of the Gαq–PLCβ3 complex23. However, these same studies indicated that full-length PLCβ3 has a similar binding affinity for Gαq. The 6 nM binding constant we measure for full-length PLCβ3 is consistent with previously reported EC50 values for Gαq stimulation36 and the EC50 values we report in Table 4.
Table 3.
Inhibition Constants of PLCβ3 Variants Measured by FCPIA
| Variant | KI ± SEM (nM) (n)a |
|---|---|
| PLCβ3 (wt)b | 6.0 ± 1. (5) |
| -F715A | 2.0 ± 0.3 (4) |
| -L876A | 3.1 ± 0.5 (4) |
| -L879A | 1.6 ± 0.1 (3) |
| PLCβ3-Δ892 (wt)b | 270 ± 60 (4) |
| -F715Ab | 31 ± 6 (3) |
| -AAAc | 26 ± 6 (3) |
| PLCβ3-Δ847 (wt)b | NBd (3) |
Number (n) of independent experiments performed in duplicate.
Includes samples from at least two independent protein purifications.
R872A L876A L879A
No binding detected.
Table 4.
Gαq Activation of PLCβ3 Variants in Vitroa
| Variant | EC50 (nM) | Increase in Activity (mol IP3 min−1 mol−1PLCβ3) |
Fold Max. Activity Over Basalb |
|---|---|---|---|
| PLCβ3 (wt) | 3.2 ± 0.6 | 1100 ± 100 | 21 |
| -F715A | 9.7 ± 1.4 | 5000 ± 690 | 3.0 |
| -P755K | 10. ± 1.8 | 3200 ± 230 | 2.2 |
| -L876A | 0.7 ± 0.1 | 1360 ± 120 | 2.4 |
| -L879A | 9.4 ± 1.7 | 4700 ± 310 | 3.3 |
| -AAAc | NDd | ND | ND |
| PLCβ3-Δ892 (wt) | 130 ± 19 | 30 ± 4 | 7.3 |
| -F715A | 54 ± 13 | 41 ± 3 | 3.9 |
| -AAAc | 67 ± 13 | 33 ± 2 | 3.3 |
| PLCβ3-Δ847 (wt) | --- | −9 ± 2 | 0.4 |
Three to 12 independent experiments, each performed in duplicate, ± SEM. See Supplementary Figure 4 online.
Fold activation is calculated as the (basal activity + increase in activity) × basal activity−1. Due to errors in determining very low basal activities in these dose response curves, basal activities are taken from Table 2.
R872A L876A L879A
Not determined. The full-length version of this protein was highly susceptible to proteolytic cleavage after the proximal CTR, and consequently the activity could not be unambiguously measured.
Mutation of residues in either Hα2′ or in its binding site on the catalytic core in PLCβ3-Δ892 enhances the apparent affinity for Gαq by 10-fold, consistent with competition between binding of the Hα1-Hα2 motif to Gαq and the binding of Hα2′ to the catalytic core. Analogous mutations in the longer PLCβ3 protein also seem to enhance affinity for Gαq, but only 2–3-fold, possibly because the more distal regions of the CTR play a compensatory role in binding to Gαq (Fig. 3d and Table 3). Our results are therefore consistent with a model in which Gαq binding to PLCβ3 displaces the proximal CTR from the catalytic core, leading to increases in both Gαq affinity and catalytic activity.
Mutations in the Hα2′-catalytic core interface reduce the efficacy of Gαq
The profound enhancement in basal activity observed for variants of PLCβ3 in which the Hα2′-catalytic core interface is disrupted is on the same scale as the activation of wild-type PLCβ3 by Gαq. We hypothesized that if Gαq activates PLCβ3 by displacing the autoinhibitory Hα2′ helix, then Gαq should be less efficacious at activating variants of PLCβ3 in which the Hα2′-catalytic core interface is disrupted. However, because Gαq should still be able to bind these variants (Table 3), residual Gαq activation effects might be observed at the same or lower EC50 values as measured for the wild-type proteins.
PLCβ3 is activated by Gαq 20-fold to 1100 mol IP3 min−1 mol−1 with an EC50 of 3 nM, consistent with the binding affinity we measure for these proteins by FCPIA (Table 3) and with previous studies7,36–38. PLCβ3-Δ847, which lacks the proximal CTR, is unresponsive to Gαq. However, PLCβ3-Δ892 is activated 7-fold by Gαq to 30 mol IP3 min−1 mol−1 (Table 4 and Supplementary Fig. 4 online). Considered along with the crystal structure of the Gαq–PLCβ3 complex23, our data firmly establishes that the proximal CTR region confers regulation by Gαq. However, the maximal Gαq-stimulated rate of hydrolysis catalyzed by PLCβ3 is 40-fold higher than that of PLCβ3-Δ892. Moreover, Gαq activates PLCβ3-Δ892 with a 30-fold higher EC50 (130 nM) than wild-type, consistent with the lower binding affinity of Gαq for PLCβ3-Δ892 we measure by FCPIA (Table 3). This data provides further evidence that the more distal regions of the CTR make important contributions to the activation mechanism by enhancing both the catalytic rate and the affinity for Gαq.
In the context of PLCβ3, mutations that disrupt the Hα2′-catalytic core interface are activated to a lesser extent by Gαq, with only 2–3 fold effects observed. Although we anticipated a small decrease in EC50 for these mutants because of their higher affinity for Gαq, this trend was only evident in the background of PLCβ3-Δ892. For PLCβ3 and its variants, the EC50 values remained in the low nM range and did not consistently decrease as we had measured in FCPIA (Table 3). Because our direct-binding assay is conducted in the presence of liposomes, these results may reflect the important role of the coiled-coil domain in mediating membrane association24,27,28,31. However, we did observe that Gαq has 2–3 higher potency when assayed against Hα2′ interface mutants in the background of PLCβ3-Δ892, which retains only the proximal CTR (Supplementary Fig. 4 online and Table 4).
DISCUSSION
Regulation of PLCβ isoforms is tightly controlled, and aberrations in this pathway or its components are associated with a number of pathophysiological processes, including heart failure11,13,39,40. One aspect of this control is their exceptionally low level of basal activity and their profound stimulation by Gq-coupled receptors. Previous studies have shown that deletion of the X–Y linker in a variety of PLC isoforms leads to increased basal activity, presumably because acidic charges in the linker electrostatically block access of PIP2 to the active site20,22,24. It has been proposed that membrane recruitment by activators of PLCβ, including Gαq, helps to displace this linker22,23. However, most PLCβ isoforms are already at least partially localized to the membrane in their basal state31,41, and deletion or perturbation of the X–Y linker, although profoundly activating, does not eliminate the ability to be activated by heterotrimeric G proteins in simple transfection assays22,24. This suggests that heterotrimeric G proteins can activate PLCβ isoforms through a mechanism that is not entirely reliant on the integrity of the X–Y linker.
Our structures of cephalopod PLC21 and functional studies of human PLCβ3 demonstrate that the Hα2′ helix in the proximal CTR interacts with a conserved cleft in the catalytic core. Disrupting the interactions of Hα2′ with the catalytic core dramatically increases basal activity up to ~50 fold (Table 2). The mechanism by which this occurs is as of yet unclear, but is possibly linked to the fact that Hα2′ is positioned on the same side of the TIM barrel as the X–Y linker, which may inhibit basal activity by sterically hindering the interaction of the active site with phospholipid bilayers. Alternatively, decreased thermal stability of the highly active mutants (Supplementary Table 1) implies greater dynamics in the catalytic core, which may lead to enhanced catalysis 42,43. A third possibility is that Hα2′ exerts a direct negative allosteric effect on the TIM barrel domain itself, although this seems less likely because the catalytic core has essentially the same structure with and without the bound Hα2′ element, as can be seen by comparing structures of PLCβ2 with that of PLCβ3 in the Gαq–PLCβ3 complex. Although these structural elements are in close proximity (Fig. 2), a direct functional relationship between displacement of Hα2′ and inhibition imposed by the X–Y linker remains to be explored, but one hypothesis is that disruption of either element leads to a less stable yet more dynamic enzyme with an active site that has easier access to its phospholipid substrates.
We also provide substantial evidence to support an allosteric mechanism by which Gαq modulates the interactions of the Hα2′ helix to activate PLCβ. In the absence of Gq-coupled receptor stimulation, the Hα2′ helix is bound to the catalytic core and suppresses basal activity (Fig. 2a,b and Fig. 3b,c). Our structures of PLC21 indicate that the residues in the Hα1-Hα2 motif that bind Gαq are disordered in this basal state and thus freely accessible to Gαq·GTP. Gαq·GTP binding to the Hα1-Hα2 motif leads to displacement of the Hα2′ helix from the catalytic core (cf. Fig. 1c and Fig. 1d) and dramatically enhanced PIP2 hydrolysis. Accordingly, disruption of the Hα2′-catalytic core interface by truncation or by site-directed mutagenesis also leads to a marked increase in basal activity (Fig. 3b,c and Table 2) at the expense of Gαq efficacy (Supplementary Fig. 4 and Table 4). However, even in these cases, Gαq binding still activates by ~3-fold, perhaps by increasing the affinity of the catalytic core for phospholipid membranes, or, more generally, by stabilizing a more catalytically competent state. Because our in vitro assays used Gαq isolated from the soluble fraction of cell lysates, this residual activation is unlikely to represent enhanced liposome association due to palmitoylation of the N-terminus of Gαq.
Our data also strongly indicate that the more distal regions of the CTR contribute to high affinity Gαq binding and to basal and Gαq-stimulated activity (Fig. 3b,c,d and Table 4), as has been suggested by many other studies26,27,29,31,44. The coiled-coil domain in the CTR contains conserved basic regions important for function26,27, and plays a role in increasing the affinity of PLCβ for the cell membrane27,31. Residues in the coiled-coil domain have also been shown to be important for Gαq activation26,29,31, and our studies show that the distal CTR enhances the affinity of PLCβ3 for Gαq in protein binding assays conducted in the absence of phospholipid vesicles. The fact that the binding affinities of Gαq for PLCβ3 and PLCβ3-Δ892 (Table 3) are consistent with their respective EC50 values measured in our liposome-based hydrolysis activity assays (Table 4) supports the idea that there is a direct functional interaction between the distal regions of the CTR and Gαq which has yet to be resolved.
In summary, our PLC21 structures likely represent a snapshot of a fully inhibited PLCβ catalytic core, and when compared to the Gαq–PLCβ3 crystal structure (Fig. 1c,d) it provides a molecular mechanism for PLCβ activation in which Gαq effectively sequesters an autoinhibitory motif. The mechanism is reminiscent of the action of transducin on cGMP phosphodiesterase (PDE) in the visual signal transduction cascade, wherein activated Gαt sequesters the inhibitory PDEγ subunit45,46–47. As in other canonical heterotrimeric G protein effectors, such as adenylyl cyclase48, there are additional layers of regulation that are essential for full activity and add complexity. A better molecular understanding of how the distal regions of the CTR contribute to Gαq binding and enzyme activity is thus essential to composing a complete picture of how PLCβ activity is controlled and how synergy can be achieved in response to the binding of multiple activators38.
METHODS
Cloning, expression, and purification of human PLCβ3 proteins
DNAs encoding N-terminally His-tagged human PLCβ3 (amino acids 10–1234)49 and C-terminally truncated variants were cloned into pFastBac Dual (Invitrogen). Point mutations were introduced using QuikChange Site-Directed Mutagenesis (Stratagene) and confirmed by sequencing over the entire open reading frame. Baculovirus-infected High Five insect cells were resuspended in 20 mM HEPES pH 8, 200 mM NaCl, 10 mM β-mercaptoethanol (BME), 0.1 mM EDTA, 0.1 mM EGTA, and Roche EDTA-free protease inhibitor cocktail tablets. After sonication, the lysate was centrifuged and the supernatant was loaded on a Ni-NTA (Qiagen) column pre-equilibrated with buffer A (20 mM HEPES pH 8, 100 mM NaCl, 10 mM BME, 0.1 mM EGTA, and 0.1 mM EDTA). The column was washed with 10 column volumes of buffer A supplemented with 10 mM imidazole and 300 mM NaCl. PLCβ3 proteins were eluted with buffer A supplemented with 200 mM imidazole, and then concentrated and purified to homogeneity on two tandem Superdex S200 columns (GE Healthcare) equilibrated with 20 mM HEPES pH 8, 200 mM NaCl, 2 mM DTT, 0.1 mM EGTA, and 0.1 mM EDTA (Supplementary Fig. 5 online).
Expression and purification of Gαq
Activity assays were conducted with protein expressed from cDNA encoding murine Gαq amino acids 7–359 cloned into pFastBacHTA (Invitrogen) to produce an N-terminally His6-tagged protein. FCPIA was performed with a Gαi/q chimera in which the wild-type N-terminal helix of murine Gαq was replaced with Gαi, as described previously50. The expression and purification protocols for both Gαq variants are identical. Protein expression in High Five cells was increased upon co-infection of virus encoding Ric8A–GST51. The cell pellet was resuspended in 20 mM HEPES pH 8, 100 mM NaCl, 10 mM BME, 3 mM MgCl2, 10 µM GDP, 0.1 mM EDTA and protease inhibitors. After sonication, the lysate was centrifuged and the supernatant was loaded on a Ni-NTA (Qiagen) column. The column was washed with 20 column volumes of buffer B (20 mM HEPES pH 8, 100 mM NaCl, 10 mM BME, 1 mM MgCl2, 10 µM GDP), followed by 20 column volumes of buffer B supplemented with 10 mM imidazole and 300 mM NaCl. Gαq was eluted with buffer B supplemented with 150 mM imidazole. The sample was dialyzed to lower the NaCl concentration and applied to a MonoQ column. Gαq was eluted in 20 mM HEPES pH 8, 1 mM MgCl2, 10 µM GDP, and 1 mM DTT with a gradient ranging from 0–500 mM NaCl. Peak fractions were further purified on tandem S200 columns equilibrated in 20 mM HEPES pH 8, 100 mM NaCl, 1 mM MgCl2, 10 µM GDP, and 1 mM DTT.
Crystallization and structure determination
Endogenous SPLC21 and LPLC21 were purified from enucleated eye cups as described previously16. Full-length protein was further purified by gel filtration in 20 mM HEPES pH 8, 200 mM NaCl, and 2 mM DTT. Protein was supplemented with 5 mM CaCl2 and crystallized by the hanging drop method using a 1:1 mixture of 11 mg mL−1 protein and well solution containing 100 mM Bis-Tris pH 7, 100–300 mM NaCl, and 20–35% (w/v) PEG 3350. Diffraction data was collected at the Advanced Photon Source at LS-CAT beam line 21-ID-F from crystals maintained at 110 K using a wavelength of 0.979 Å and initial phases were derived by molecular replacement using the human PLCβ2 (PDB entry 2ZKM) structure22 as a search model.
Thermostability measurements
Melting temperatures (Tm) were determined by monitoring 1-anilinonaphthalene-8-sulfonic acid (ANS) binding to PLCβ3 variants during protein unfolding52. Wild type PLCβ3 and variants (0.3 mg mL−1) were incubated with 200 µm ANS in a total volume of 5 µL in triplicate in ABgene 384-well PCR microtiter plates (Thermo-Fisher). Fluorescence was measured as the temperature was increased from 20–80 °C in 1 °C intervals using a ThermoFluor 384-well plate reader (Johnson & Johnson).
PLCβ3 activity assays
PLCβ3 basal activity and Gαq-mediated activation was quantified by measuring the rate of hydrolysis of [3H]-labeled PIP2 in a liposome-based assay as previously described16,53. Briefly, lipid vesicles containing 200 µM phosphatidyl--ethanolamine, 50 µM PIP2, and ~4000–8000 cpm [3H]-labeled PIP2 per assay were mixed, dried under nitrogen, and resuspended by sonication in 50 mM HEPES pH 7, 80 mM KCl, 2 mM EGTA, and 1 mM DTT. PLCβ3 activity was assayed at 30 °C in 50 mM HEPES pH 7, 80 mM KCl, 15 mM NaCl, 0.83 mM MgCl2, 3 mM DTT, 1 mg mL−1 BSA, 2.5 mM EGTA, 0.2 mM EDTA with ~200 nM free Ca2+. Control reactions contained the same components, but lacked free Ca2+. Reactions were terminated by addition of BSA and 10% (w/v) ice-cold trichloroacetic acid. After centrifugation, free [3H]-IP3 in the supernatant was measured by scintillation counting16,53. To measure Gαq-stimulated activity, purified Gαq was activated with 50 mM HEPES pH 7, 80 mM KCl, 30 mM NaCl, 3 mM DTT, 10 mM NaF, 20 µM AlCl3, 50 µM GDP and 1.83 mM MgCl2 for 30 min on ice. Increasing amounts of activated Gαq were added to PLCβ3 proteins, and reactions initiated by addition of liposomes, and terminated after incubation at 30 °C for 5 min.
FCPIA
The (R872A L876A L879A) triple mutant of PLCβ3-Δ892, which was considered to be the least likely to be autoinhibited by Hα2′, was fluorescently labeled with AlexaFluor-488 (AF488) C5-maleimide. Gαi/q was biotinylated and linked to streptavidin coated beads as previously described34. Unlabeled PLCβ3 variants were added at increasing concentrations to bead-bound Gαi/q, followed by addition of the AF488-labeled PLCβ3-Δ892 variant at its measured KD (20.5 nM). The resulting mixtures were incubated for 1 hr, and then analyzed in duplicate with an Accuri C6 Flow Cytometer. Competition data was fit by nonlinear regression using a variable slope fit (PLCβ3 and mutants) or standard slope (other variants) using GraphPad Prism (version 5.0a). KI values were estimated from fitted IC50 values using the Cheng-Prussof equation.
Statistical methods
Statistical analyses used ANOVA with a Dunnett’s post-test as implemented in GraphPad Prism (version 5.0a).
Additional and more detailed methods are provided in the Supplementary Methods online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Elliott Ross (University of Texas Southwestern Medical Center at Dallas) for the vector encoding human PLCβ3, and Dr. Greg Tall (University of Rochester) for the baculovirus vector expressing GST-Ric8A and insight into how to increase yields of Gαq before publication of his work. We also thank Dr. Peter Backlund, Section on Mass Spectrometry and Metabolism (NICHD) for mass spectrometry of PLC21 samples. This work was supported by National Institutes of Health grants HL071818 and HL086865 (J.J.G.T.), and by the Intramural Research program of the National Institute on Deafness and Other Communication Disorders, National Institutes of Health (J.K.N.). Our research used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center supported by DK20572. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Footnotes
Accession codes. The SPLC21 and LPLC21 structures and their structure factor amplitudes are deposited in the Protein Data Bank with accession codes 3RQ0 and 3RQ1, respectively.
AUTHOR CONTRIBUTIONS
A.M.L, V.M.T., J.K.N, and J.J.G.T. designed the overall experimental approach. J.G., S.C., and J.K.N. purified LPLC21 and SPLC21, and cloned and sequenced cDNA encoding SPLC21. K.C.S. crystallized LPLC21. A.M.L. crystallized SPLC21 and determined the crystal structures of LPLC21 and SPLC21. A.M.L. and V.M.T. cloned, expressed, and purified human PLCβ3 variants. V.M.T. cloned, expressed, and purified Gαq. A.M.L. conducted all activity-based assays. D.M.T helped design and together with V. D. D. conducted ThermoFluor and FCPIA assays. A.M.L, V.M.T., and J.J.G.T co-wrote the manuscript.
REFERENCES
- 1.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SJ, Exton JH. Two a subunits of the Gq class of G proteins stimulate phosphoinositide phospholipase C-β1 activity. FEBS Lett. 1991;286:214–216. doi: 10.1016/0014-5793(91)80976-a. [DOI] [PubMed] [Google Scholar]
- 4.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 5.Park D, Jhon DY, Lee CW, Lee KH, Rhee SG. Activation of phospholipase C isozymes by G protein βγ subunits. J Biol Chem. 1993;268:4573–4576. [PubMed] [Google Scholar]
- 6.Boyer JL, Waldo GL, Harden TK. βγ-subunit activation of G-protein-regulated phospholipase C. J Biol Chem. 1992;267:25451–25456. [PubMed] [Google Scholar]
- 7.Smrcka AV, Sternweis PC. Regulation of purified subtypes of phosphatidylinositol-specific phospholipase C β by G protein α and βγ subunits. J Biol Chem. 1993;268:9667–9674. [PubMed] [Google Scholar]
- 8.Sternweis PC, Smrcka AV. G proteins in signal transduction: the regulation of phospholipase C. Ciba Found Symp. 1993;176:96–106. doi: 10.1002/9780470514450.ch7. discussion 106–11. [DOI] [PubMed] [Google Scholar]
- 9.Illenberger D, et al. Stimulation of phospholipase C-β2 by the Rho GTPases Cdc42Hs and Rac1. EMBO J. 1998;17:6241–6249. doi: 10.1093/emboj/17.21.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harden TK, Hicks SN, Sondek J. Phospholipase C isozymes as effectors of Ras superfamily GTPases. J Lipid Res. 2009;50(Suppl):S243–S248. doi: 10.1194/jlr.R800045-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugden PH, Clerk A. Cellular mechanisms of cardiac hypertrophy. J Mol Med. 1998;76:725–746. doi: 10.1007/s001090050275. [DOI] [PubMed] [Google Scholar]
- 12.Berridge MJ. Cardiac calcium signalling. Biochem Soc Trans. 2003;31:930–933. doi: 10.1042/bst0310930. [DOI] [PubMed] [Google Scholar]
- 13.Ju H, Zhao S, Tappia PS, Panagia V, Dixon IM. Expression of Gqα and PLC-β in scar and border tissue in heart failure due to myocardial infarction. Circulation. 1998;97:892–899. doi: 10.1161/01.cir.97.9.892. [DOI] [PubMed] [Google Scholar]
- 14.Woodcock EA, Kistler PM, Ju Y-K. Phosphoinositide signalling and cardiac arrhythmias. Cardiovasc. Res. 2009;82:286–295. doi: 10.1093/cvr/cvn283. [DOI] [PubMed] [Google Scholar]
- 15.Schneuwly S, Burg MG, Lending C, Perdew MH, Pak WL. Properties of photoreceptor-specific phospholipase C encoded by the norpA gene of Drosophila melanogaster. J Biol Chem. 1991;266:24314–24319. [PubMed] [Google Scholar]
- 16.Mitchell J, Gutierrez J, Northup JK. Purification, characterization, and partial amino acid sequence of a G protein-activated phospholipase C from squid photoreceptors. J Biol Chem. 1995;270:854–859. doi: 10.1074/jbc.270.2.854. [DOI] [PubMed] [Google Scholar]
- 17.Shortridge RD, et al. A Drosophila phospholipase C gene that is expressed in the central nervous system. J Biol Chem. 1991;266:12474–12480. [PubMed] [Google Scholar]
- 18.Essen LO, et al. Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry. 1997;36:1704–1718. doi: 10.1021/bi962512p. [DOI] [PubMed] [Google Scholar]
- 19.Ellis MV, et al. Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of PLCδ1. J Biol Chem. 1998;273:11650–11659. doi: 10.1074/jbc.273.19.11650. [DOI] [PubMed] [Google Scholar]
- 20.Suh PG, et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008;41:415–434. doi: 10.5483/bmbrep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- 21.Jezyk MR, et al. Crystal structure of Rac1 bound to its effector phospholipase C-β2. Nat Struct Mol Biol. 2006;13:1135–1140. doi: 10.1038/nsmb1175. [DOI] [PubMed] [Google Scholar]
- 22.Hicks SN, et al. General and versatile autoinhibition of PLC isozymes. Mol Cell. 2008;31:383–394. doi: 10.1016/j.molcel.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldo GL, et al. Kinetic Scaffolding mediated by a phospholipase C-β and Gq signaling complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Neer EJ. Reassembly of phospholipase C-β2 from separated domains: analysis of basal and G protein-stimulated activities. J Biol Chem. 2001;276:2503–2508. doi: 10.1074/jbc.M003562200. [DOI] [PubMed] [Google Scholar]
- 25.Schnabel P, Camps M. Activation of a phospholipase Cβ2 deletion mutant by limited proteolysis. Biochem J. 1998;330(Pt 1):461–468. doi: 10.1042/bj3300461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilkaeva O, Kinch LN, Paulssen RH, Ross EM. Mutations in the carboxyl-terminal domain of phospholipase C-β1 delineate the dimer interface and a potential Gαq interaction site. J Biol Chem. 2002;277:4294–4300. doi: 10.1074/jbc.M109612200. [DOI] [PubMed] [Google Scholar]
- 27.Kim CG, Park D, Rhee SG. The role of carboxyl-terminal basic amino acids in Gqα-dependent activation, particulate association, and nuclear localization of phospholipase C-β1. J Biol Chem. 1996;271:21187–21192. doi: 10.1074/jbc.271.35.21187. [DOI] [PubMed] [Google Scholar]
- 28.Park D, Jhon DY, Lee CW, Ryu SH, Rhee SG. Removal of the carboxyl-terminal region of phospholipase C-β1 by calpain abolishes activation by Gαq. J Biol Chem. 1993;268:3710–3714. [PubMed] [Google Scholar]
- 29.Singer AU, Waldo GL, Harden TK, Sondek J. A unique fold of phospholipase C-β mediates dimerization and interaction with Gαq. Nat Struct Biol. 2002;9:32–36. doi: 10.1038/nsb731. [DOI] [PubMed] [Google Scholar]
- 30.Paulssen RH, Woodson J, Liu Z, Ross EM. Carboxyl-terminal fragments of phospholipase C-β1 with intrinsic Gq GTPase-activating protein (GAP) activity. J Biol Chem. 1996;271:26622–26629. doi: 10.1074/jbc.271.43.26622. [DOI] [PubMed] [Google Scholar]
- 31.Wu D, Jiang H, Katz A, Simon MI. Identification of critical regions on phospholipase C-β1 required for activation by G-proteins. J Biol Chem. 1993;268:3704–3709. [PubMed] [Google Scholar]
- 32.Koyanagi M, Ono K, Suga H, Iwabe N, Miyata T. Phospholipase C cDNAs from sponge and hydra: antiquity of genes involved in the inositol phospholipid signaling pathway. FEBS Lett. 1998;439:66–70. doi: 10.1016/s0014-5793(98)01339-8. [DOI] [PubMed] [Google Scholar]
- 33.Jhon DY, et al. Cloning, sequencing, purification, and Gq-dependent activation of phospholipase C-β3. J Biol Chem. 1993;268:6654–6661. [PubMed] [Google Scholar]
- 34.Shankaranarayanan A, et al. Assembly of high order Gαq-effector complexes with RGS proteins. J Biol Chem. 2008;283:34923–34934. doi: 10.1074/jbc.M805860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu S, et al. Unique hydrophobic extension of the RGS2 amphipathic helix domain imparts increased plasma membrane binding and function relative to other RGS R4/B subfamily members. J Biol Chem. 2007;282:33064–33075. doi: 10.1074/jbc.M702685200. [DOI] [PubMed] [Google Scholar]
- 36.Hepler JR, et al. Purification from Sf9 cells and characterization of recombinant Gqα, G11α.Activation of purified phospholipase C isozymes by Gα subunits. J Biol Chem. 1993;268:14367–14375. [PubMed] [Google Scholar]
- 37.Lee SB, Shin SH, Hepler JR, Gilman AG, Rhee SG. Activation of phospholipase C-β2 Mutants by G Protein αq, βγ Subunits. J. Biol. Chem. 1993;268:25952–25957. [PubMed] [Google Scholar]
- 38.Philip F, Kadamur G, Silos RG, Woodson J, Ross EM. Synergistic activation of phospholipase C-β3 by Gαq and Gβγ describes a simple two-state coincidence detector. Curr Biol. 2010;20:1327–1335. doi: 10.1016/j.cub.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodcock EA, et al. Selective activation of the "b" splice variant of phospholipase Cβ1 in chronically dilated human and mouse atria. J Mol Cell Cardiol. 2009;47:676–683. doi: 10.1016/j.yjmcc.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 40.Achour L, Labbe-Jullie C, Scott MG, Marullo S. An escort for GPCRs: implications for regulation of receptor density at the cell surface. Trends Pharmacol Sci. 2008;29:528–535. doi: 10.1016/j.tips.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Jenco JM, Becker KP, Morris AJ. Membrane-binding properties of phospholipase C-β1 and phospholipase C-β2: role of the C-terminus and effects of polyphosphoinositides, G-proteins and Ca2+ Biochem J. 1997;327(Pt 2):431–437. doi: 10.1042/bj3270431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson CM, Zucker FH, Steitz TA. Space-filling models of kinase clefts and conformation changes. Science. 1979;204:375–380. doi: 10.1126/science.220706. [DOI] [PubMed] [Google Scholar]
- 43.Daniel RM. The upper limits of enzyme thermal stability. Enzyme and Microbial Technology. 1996;19:74–79. [Google Scholar]
- 44.Wu D, Katz A, Simon MI. Activation of phospholipase Cβ2 by the α and βγ subunits of trimeric GTP-binding protein. Proc Natl Acad Sci U S A. 1993;90:5297–5301. doi: 10.1073/pnas.90.11.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurley JB. Molecular properties of the cGMP cascade of vertebrate photoreceptors. Annu Rev Physiol. 1987;49:793–812. doi: 10.1146/annurev.ph.49.030187.004045. [DOI] [PubMed] [Google Scholar]
- 46.Slep KC, et al. Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 Å. Nature. 2001;409:1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 47.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 48.Sunahara RK, Taussig R. Isoforms of mammalian adenylyl cyclase: multiplicities of signaling. Mol Interv. 2002;2:168–184. doi: 10.1124/mi.2.3.168. [DOI] [PubMed] [Google Scholar]
METHODS ONLY REFERENCES
- 49.Biddlecome GH, Berstein G, Ross EM. Regulation of phospholipase C-β1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J Biol Chem. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- 50.Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Snapshot of activated G proteins at the membrane: the Gαq-GRK2-Gβγ complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- 51.Chan P, et al. Purification of heterotrimeric G protein α subunits by GST-Ric-8 association: primary characterization of purified Gαolf. J Biol Chem. 2011;286:2625–2635. doi: 10.1074/jbc.M110.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mezzasalma TM, et al. Enhancing recombinant protein quality and yield by protein stability profiling. J Biomol Screen. 2007;12:418–428. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- 53.Ghosh M, Smrcka AV. Assay for G protein-dependent activation of phospholipase Cβ using purified protein components. Methods Mol Biol. 2004;237:67–75. doi: 10.1385/1-59259-430-1:67. [DOI] [PubMed] [Google Scholar]
METHODS ONLY REFERENCES
- 49.Biddlecome GH, Berstein G, Ross EM. Regulation of phospholipase C-β1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J Biol Chem. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- 50.Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Snapshot of activated G proteins at the membrane: the Gαq-GRK2-Gβγ complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- 51.Chan P, et al. Purification of heterotrimeric G protein α subunits by GST-Ric-8 association: primary characterization of purified Gαolf. J Biol Chem. 2011;286:2625–2635. doi: 10.1074/jbc.M110.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mezzasalma TM, et al. Enhancing recombinant protein quality and yield by protein stability profiling. J Biomol Screen. 2007;12:418–428. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- 53.Ghosh M, Smrcka AV. Assay for G protein-dependent activation of phospholipase Cβ using purified protein components. Methods Mol Biol. 2004;237:67–75. doi: 10.1385/1-59259-430-1:67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.