Abstract
The first DNA joint formed in homologous recombination processes is a D-loop. Saccharomyces cerevisiae RDH54/TID1-encoded product, a Swi2/Snf2-like factor involved in recombination, is shown here to promote D-loop formation with Rad51 recombinase. Physical interaction between Rdh54 and Rad51 is functionally important because Rdh54 does not enhance the recombinase activity of the Escherichia coli RecA protein. Robust dsDNA-activated ATPase activity in Rdh54 generates unconstrained negative and positive supercoils in DNA. Efficient D-loop formation occurs with even topologically relaxed DNA, suggesting that via specific protein–protein interactions, the negative supercoils produced by Rdh54 are used by Rad51 for making DNA joints.
Keywords: Homologous recombination, DNA double-strand break repair, meiosis, DNA supercoiling
Homologous recombination is indispensable for faithful segregation of chromosomes in the first meiotic division, for DNA double-strand break repair, and for telomere maintenance when telomerase is inactivated. These and other homologous recombination processes are mediated by genes of the RAD52 epistasis group, which were first defined in the budding yeast Saccharomyces cerevisiae (for review, see Paques and Haber 1999; Sung et al. 2000). Emerging evidence strongly suggests that recombinational DNA repair by the RAD52 group of proteins is important for tumor suppression in mammals (for review, see Dasika et al. 1999).
In most homologous recombination events, a DNA double-strand break is formed and processed nucleolytically to yield ssDNA tails of considerable length. The DNA tails thus generated serve as substrate for recruitment of a number of recombination factors, which function in concert to conduct a DNA homology search to locate a chromosomal homolog and then catalyze the formation of a heteroduplex joint, called D-loop, between the initiating ssDNA tails and the chromosomal homolog (Paques and Haber 1999; Sung et al. 2000). The enzymatic process that leads to D-loop formation is called homologous DNA pairing and strand exchange (for review, see Kowalczykowski et al. 1994; Roca and Cox 1997). Ability to form a D-loop has been demonstrated in the Escherichia coli general recombinase RecA, which shows extensive structural and functional similarity to the S. cerevisiae Rad51 protein, a key member of the RAD52 epistasis group. However, Rad51 by itself forms a D-loop poorly, suggesting that other recombination proteins are needed for efficient D-loop formation. Consistent with this hypothesis, Rad54 protein has been shown to promote D-loop formation with Rad51 (Petukhova et al. 1998).
The S. cerevisiae RDH54/TID1 gene, also a member of the RAD52 epistasis group, affects the efficiency of DNA double-strand break repair and mitotic and meiotic recombination (Klein 1997; Shinohara et al. 1997; Arbel et al. 1999). Rdh54/Tid1 is homologous to Rad54 and possesses features characteristic of the Swi2/Snf2 family of proteins found to be essential for a variety of chromosomal transactions (Eisen et al. 1995). In two hybrid studies, Rdh54/Tid1 was shown to interact with Rad51 and the Dmc1 protein (Dresser et al. 1997), a homolog of Rad51 specifically involved in meiotic recombination (Bishop et al. 1992). We have now overexpressed and purified the RDH54/TID1-encoded product from yeast cells. Here we describe our biochemical studies revealing that Rdh54/Tid1 directly interacts with and cooperates with Rad51 to promote highly efficient D-loop formation. We also show an ability of Rdh54/Tid1 to produce unconstrained negative and positive supercoils in duplex DNA, an activity likely to be indispensable for the invasion of the homologous duplex molecule by the initiating ssDNA substrate during the D-loop reaction.
Results
Rdh54 has DNA-dependent ATPase activity
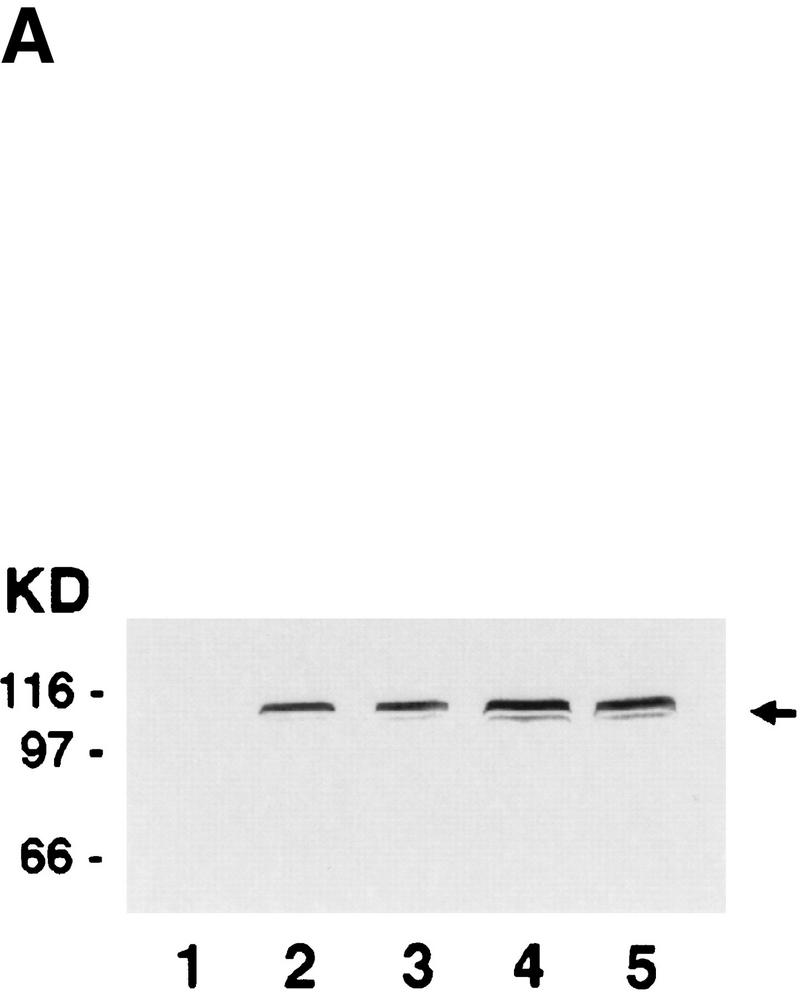
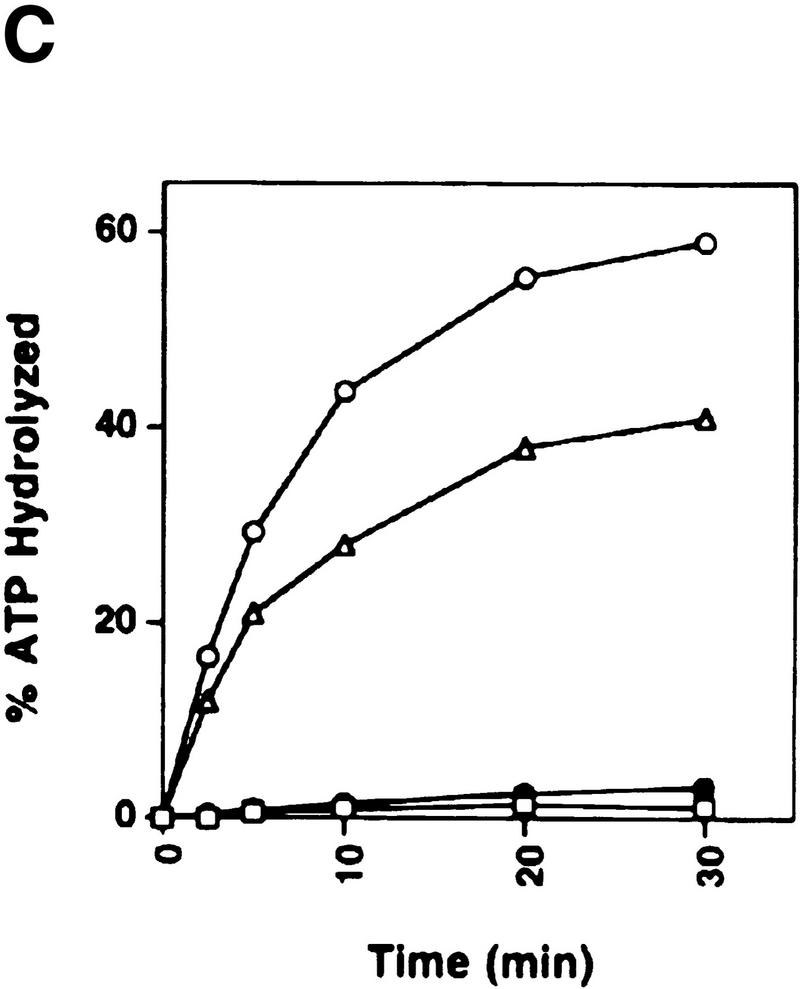
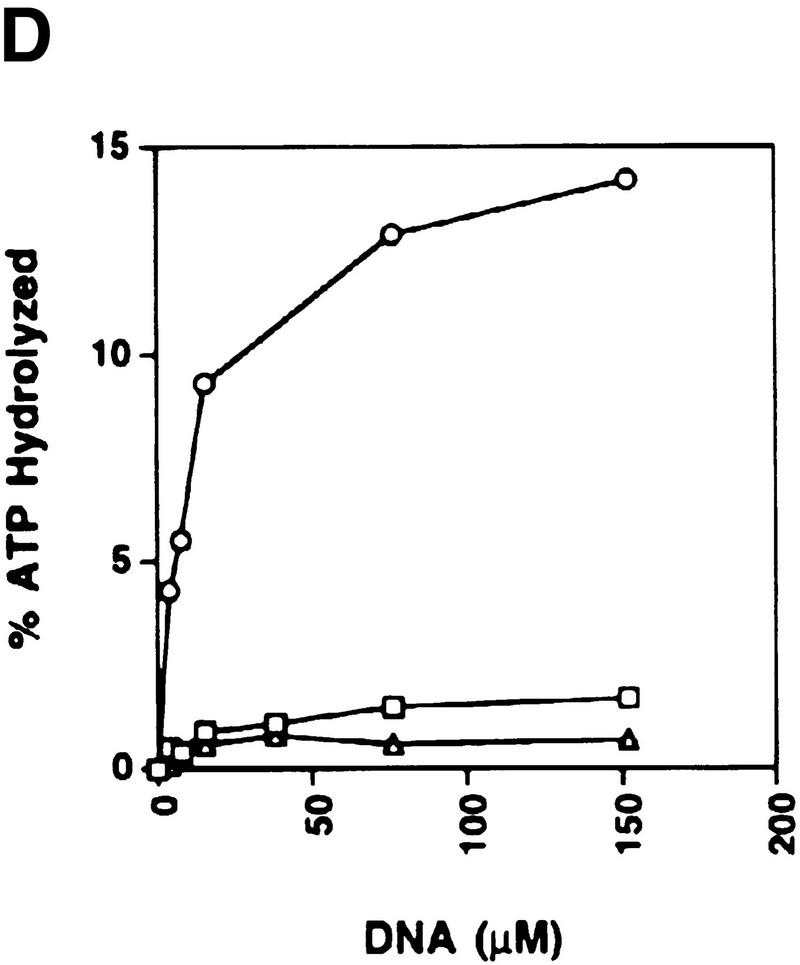
The RDH54/TID1-encoded product, henceforth referred to as Rdh54, was overexpressed in yeast cells (Fig. 1A) and purified to near homogeneity (Fig. 1B, lane 2), using a six-column procedure. We asked whether Rdh54 has ATPase activity because Walker type nucleotide binding motifs are present in this protein (Eisen et al. 1995). Whereas no ATP hydrolysis occurred in the absence of DNA, robust ATPase activity was seen in the presence of φX174 dsDNA and viral (+) strand DNA (Fig. 1C). Because dsDNA (kcat = 2.2 × 103 min−1) was a better cofactor than the viral (+) strand (kcat = 1.5 × 103 min−1) in activating ATP hydrolysis, we considered the possibility that ATP hydrolysis with the viral (+) strand was the result of secondary structure in the DNA. Indeed, an approximately 10-fold increase in ATPase activity was seen with the duplex obtained by hybridizing two complementary oligonucleotides (Fig. 1D), indicating that duplex DNA is the preferred cofactor in ATP hydrolysis.
Figure 1.
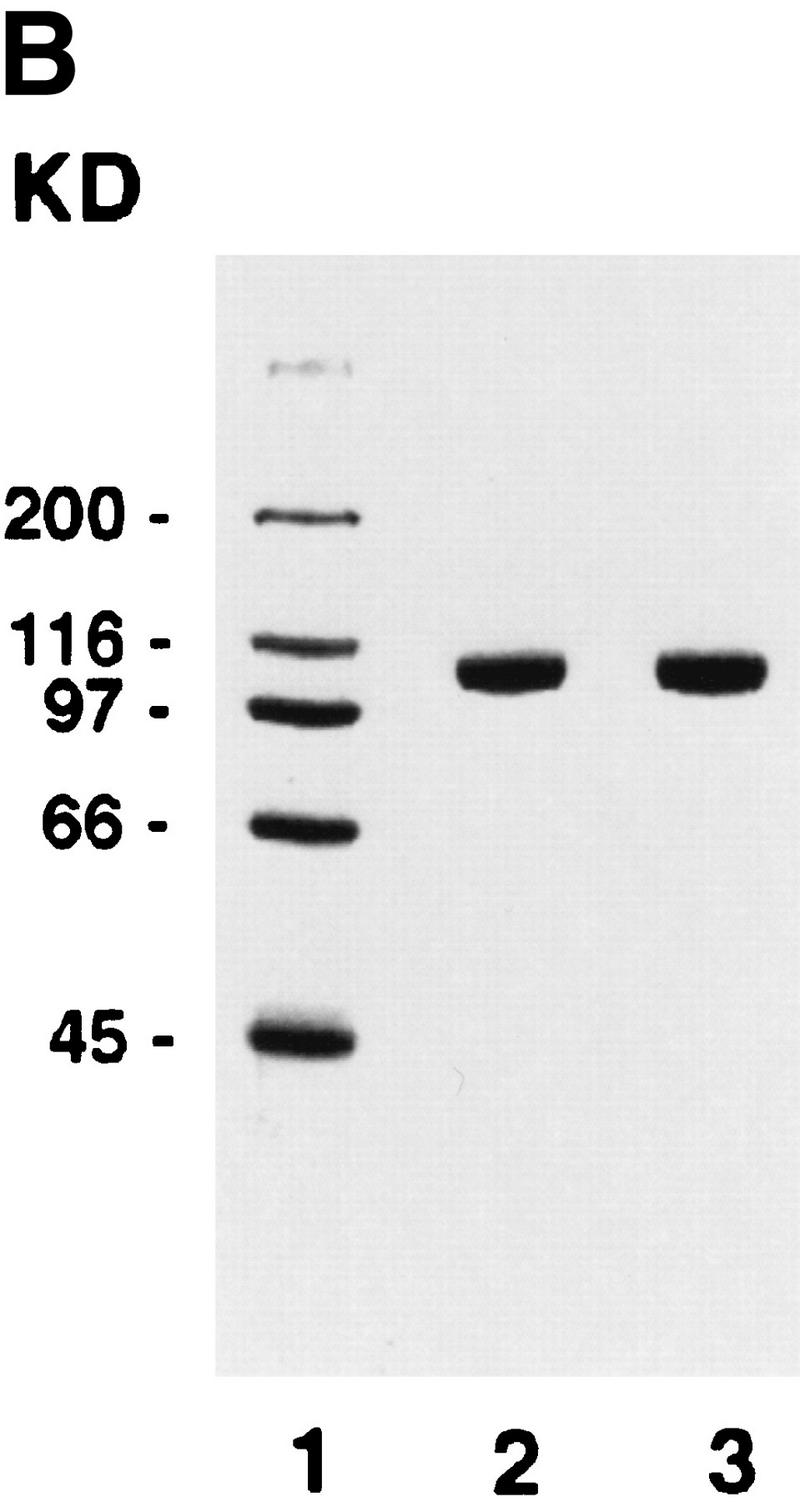
Purification and DNA-dependent ATPase activity of Rdh54 and rdh54 K351R. (A) Immunoblot analysis. Nitrocellulose blot containing extracts from strain BJ5464 harboring the empty vector pPM231 (2μ, GAL–PGK; lane 1) and from BJ5464 harboring plasmid pRdh54.1 (2μ, GAL–PGK–6His–RDH54; lane 2) and plasmid prdh54K/R.1 (2μ, GAL–PGK–6His–rdh54 K351R; lane 3), along with 50 ng of purified Rdh54 (lane 4) and rdh54 K351R (lane 5), were subjected to immunoblot analysis with anti-Rdh54 antibodies. The arrow marks the position of the Rdh54 and rdh54 K351R proteins. The level of Rdh54 in wild-type extract (lane 1) is too low to detect under the conditions used. (B) An 8% SDS–polyacrylamide gel containing 2 μg of purified Rdh54 (lane 2) and rdh54 K351R (lane 3) was stained with Coomassie Blue. (C) Purified Rdh54 (40 nm) was incubated with 1.5 mm [γ-32P]ATP in the absence of DNA (□) and in the presence of either φX viral (+) strand (▵; 30 μm nucleotides) or φX replicative form I DNA (○; 15 μm base pairs) for the indicated times at 37°C. The rdh54 K351R mutant protein (120 nm) was also incubated with φX replicative form I DNA (●; 15 μm base pairs); no ATP hydrolysis was detected with rdh54K351R when DNA was omitted (data not shown). (D) Double-stranded DNA is the preferred cofactor for ATP hydrolysis. Rdh54 (30 nm) was incubated with 1.5 mm [γ-32P]ATP with increasing concentrations of oligo 1 (□; in nucleotides), oligo 2 (▵; in nucleotides), or the duplex obtained by hybridizing oligo 1 and oligo 2 (○; in base pairs) for 8 min at 37°C.
To examine the role of ATP hydrolysis in Rdh54 functions, we changed the highly conserved Walker-type A lysine residue in Rdh54 to arginine and purified the rdh54 K351R mutant protein (Fig. 1B, lane 3). As shown in Figure 1C, rdh54 K351R has only 1%–2% of the ATPase activity of Rdh54. Genetic studies have revealed that the rdh54 K351 mutant is as defective in mitotic recombination as the rdh54 null mutant is and that meiotic viability is greatly reduced (H. Klein, unpubl.), indicating that ATP hydrolysis by Rdh54 is indispensable for its biological functions.
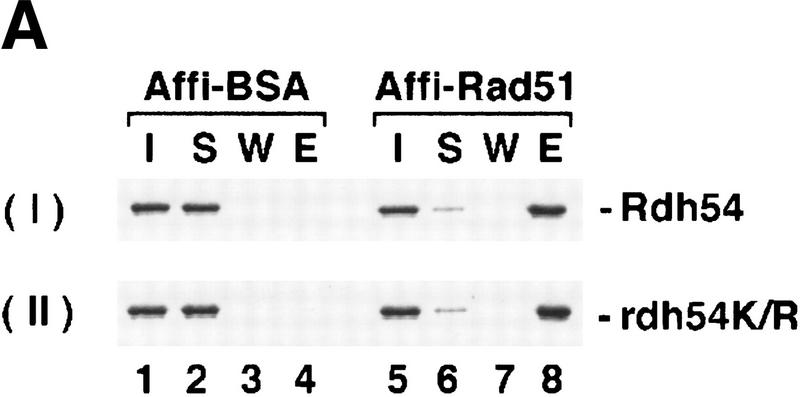
Rdh54 cooperates with Rad51 recombinase in D-loop formation
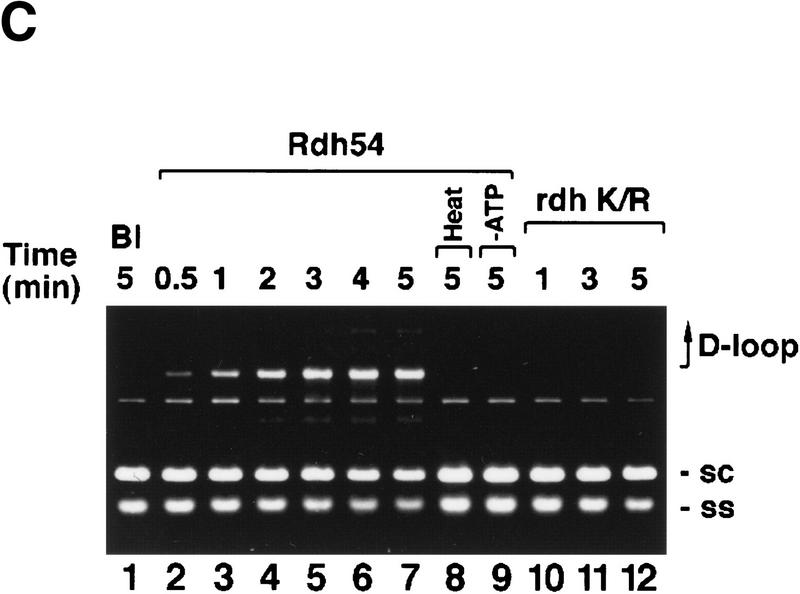
Rad51 forms a D-loop between a linear ssDNA molecule and a covalently closed duplex only poorly, even though it can make DNA joints in other model systems (for review, see Sung et al. 2000). It was therefore of considerable interest to examine whether Rdh54 would confer D-loop-forming ability to Rad51, especially given that these two proteins interact in the yeast two-hybrid system (Dresser et al. 1997) and in vitro, as shown in Figure 2A. Rdh54 was found to bind specifically to Affi-Gel 15 beads containing immobilized Rad51. To examine D-loop formation, we incubated Rad51, Rdh54, and the ssDNA-binding protein RPA, an important cofactor for the Rad51 recombinase activity (Sung 1994; Sugiyama et al. 1997), with linear φX (+) strand; the resulting nucleoprotein complex was reacted with φX replicative form I DNA (schematic shown in Fig. 2B). When we performed these experiments at 23°, 30°, and 37°C, the results were qualitatively the same at these reaction temperatures (data not shown). Here, we show the results from experiments conducted at 23°C because the relatively slow kinetics at this temperature enabled us to monitor the progress of the reaction more easily.
Figure 2.
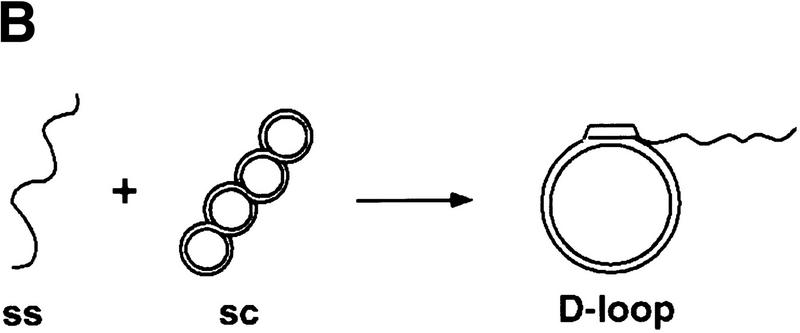
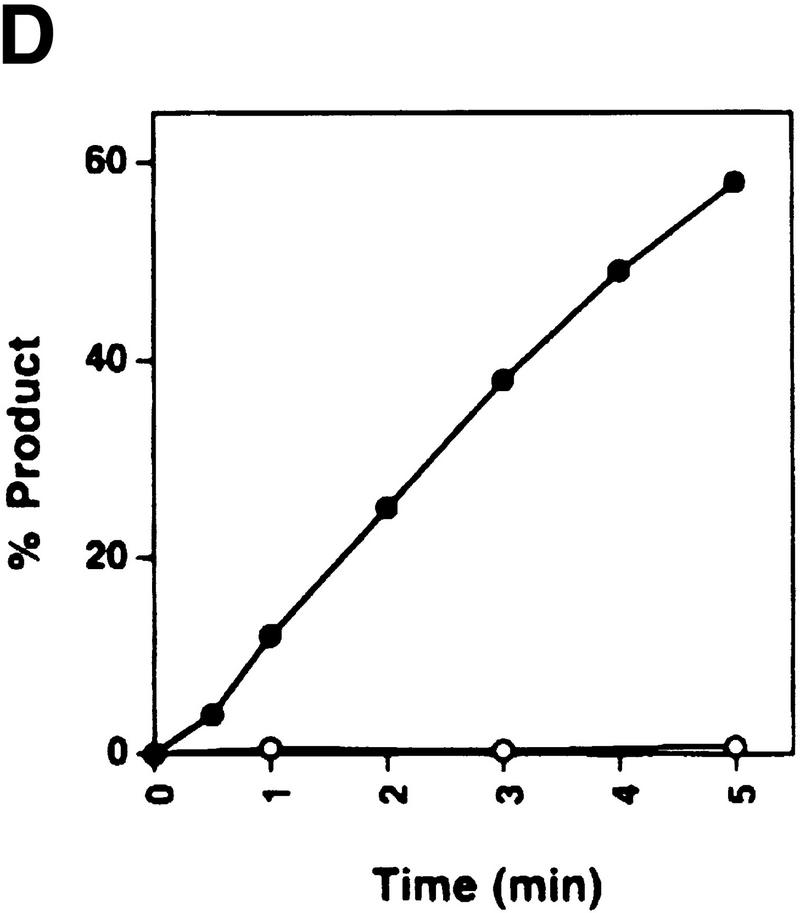
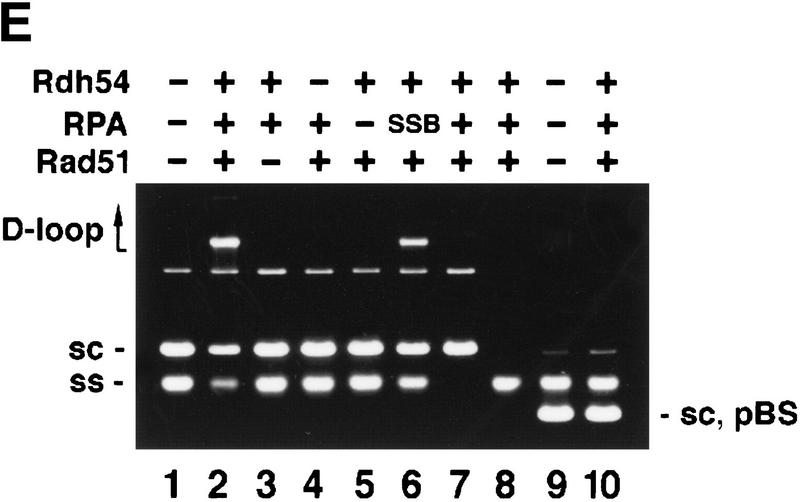
Rdh54 interacts with Rad51 and promotes D-loop formation. (A)Rad51 and Rdh54 interact physically. Rdh54 (panel I) or rdh54 K351R (panel II) was incubated with Affi-Gel 15 beads bearing bovine serum albumin (Affi-BSA) or Rad51 (Affi-Rad51). The beads were then washed with 100 mm KCl and eluted with 3% SDS. The input material (I), the supernatant containing unbound Rdh54 or rdh54 K351R (S), the KCl wash (W), and the SDS eluate (E) were run in an 8% polyacrylamide gel and stained with Coomassie Blue. Densitometric scanning of the gels revealed that > 80% of Rdh54 was retained on the Affi-Rad51 beads, whereas the Affi-BSA control beads bound < 3% of the Rdh54 protein. (B) Schematic of the D-loop reaction. Linear viral (+) strand (designated ss) is paired with the replicative form I DNA (designated sc) to yield a D-loop. (C) ATP hydrolysis-dependent promotion of D-loop formation by Rdh54. Linear φX viral (+) strand (ss) was incubated with Rad51, Rdh54, and RPA, and the nucleoprotein complex thus formed was reacted with φX replicative form I DNA (sc) at 23°C for the indicated times (lanes 2–9). The reaction mixture in lane 8 was heated at 92°C for 2 min before electrophoresis; ATP was omitted from the reaction mixture in lane 9. In lanes 10–12, Rad51, rdh54 K351R (rdh K/R), and RPA were incubated with the DNA substrates at 23°C for the indicated times. In lane 1, the DNA substrates were incubated in buffer without any recombination protein (Bl). In addition to the main D-loop species, more complex D-loop species are seen in the reaction involving wild-type Rdh54, especially in the later time points (lanes 2–7). (D) The gel in C was subjected to image analysis, and the data points were plotted. (●) results in lanes 2–7 of C involving wild-type Rdh54; (○) results in lanes 10–12 of C involving rdh54 K351R. (E) Requirements for D-loop formation. Linear φX viral (+) strand (ss) and replicative form I DNA (sc) were incubated in buffer without any recombination protein (lane 1), with Rad51, RPA, and Rdh54 (lane 2), with RPA and Rdh54 (lane 3), with Rad51 and RPA (lane 4), with Rad51 and Rdh54 (lane 5), and with Rad51, E. coli SSB, and Rdh54 (lane 6). In lane 7, φX replicative form I alone, and in lane 8, linear φX (+) strand alone, was incubated with Rad51, RPA, and Rdh54. In lanes 9 and 10, linear φX (+) strand and the unrelated pBlueScript replicative form I DNA (sc, pBS) were incubated in buffer without any recombination protein (lane 9) or with the combination of Rad51, RPA, and Rdh54 (lane 10). The reaction temperature was 23°C.
Joint molecules were formed in reactions that contained ATP, Rad51, Rdh54, and RPA (Fig. 2C, lanes 2–7, and 2D). The joint molecules resulted from pairing between the linear (+) strand and the replicative form I DNA because they were not seen when either DNA species was omitted (Fig. 2E, lanes 7 and 8) or when φX replicative form I DNA was replaced with the unrelated pBlueScript replicative form I DNA (Fig. 2E, lanes 9 and 10). Omission of Rad51, Rdh54, or RPA abolished the formation of joint molecules (Fig. 2E, lanes 3–5). Significantly, the E. coli ssDNA-binding protein SSB can partially substitute for RPA (Fig. 2E, lane 6). The joint molecules were stable to deproteinization treatment, but heating the reaction mixture to 92°C for 2 min greatly decreased its level and released replicative form I DNA and linear (+) strand (Fig. 2C, lane 8). In the experiments shown, the φX circular viral (+) strand was linearized at the PstI site, but the same results were obtained when linearization of the circular (+) strand occurred at the AvaII site (data not shown).Therefore, we concluded that a D-loop is formed efficiently by the combination of Rad51, Rdh54, and RPA. As shown in Figure 2C, rdh54 K351R was ineffective in D-loop formation (lanes 10–12), indicating that ATP hydrolysis by Rdh54 is indispensable. The slower-migrating joint molecules (Fig. 2C) likely corresponded to more complex D-loop structures consisting of multiple molecules of ssDNA molecules and/or dsDNA molecules. In these experiments, a ratio of ssDNA to Rad51 of 30 nucleotides per protein monomer was used, although in other model systems, a ratio of three nucleotides per Rad51 monomer is optimal (Sung and Robberson 1995; Sugiyama et al. 1997); increasing Rad51 to this level resulted in a three- to fourfold reduction of D-loop formation (data not shown).
Rdh54 modifies DNA topology
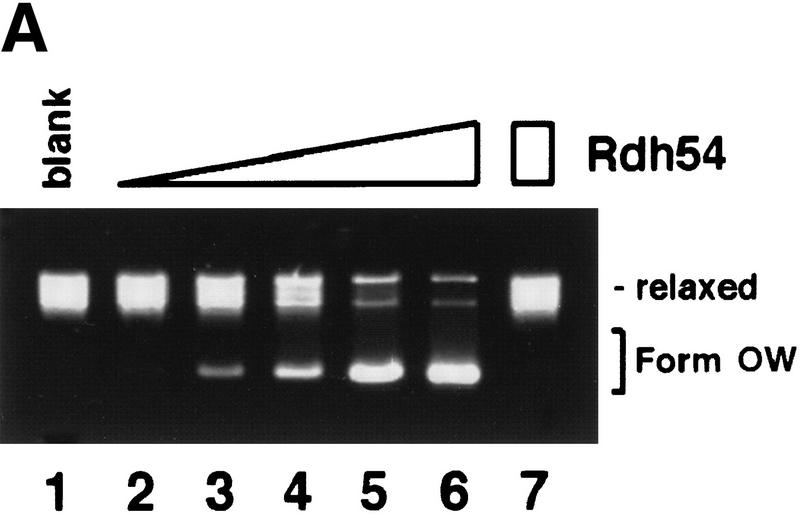
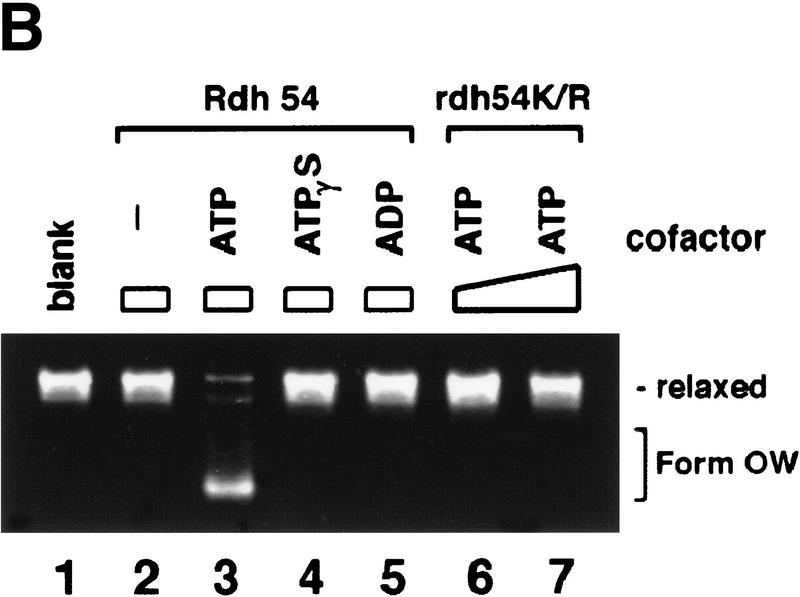
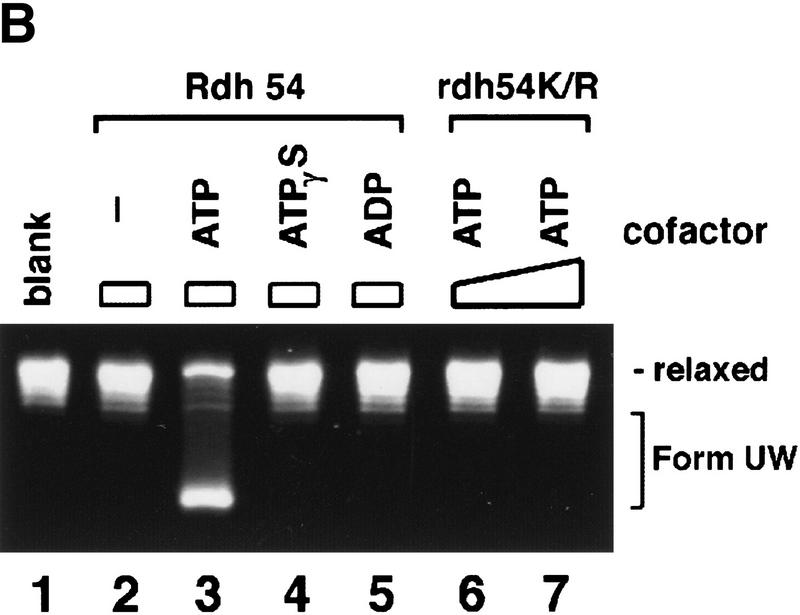
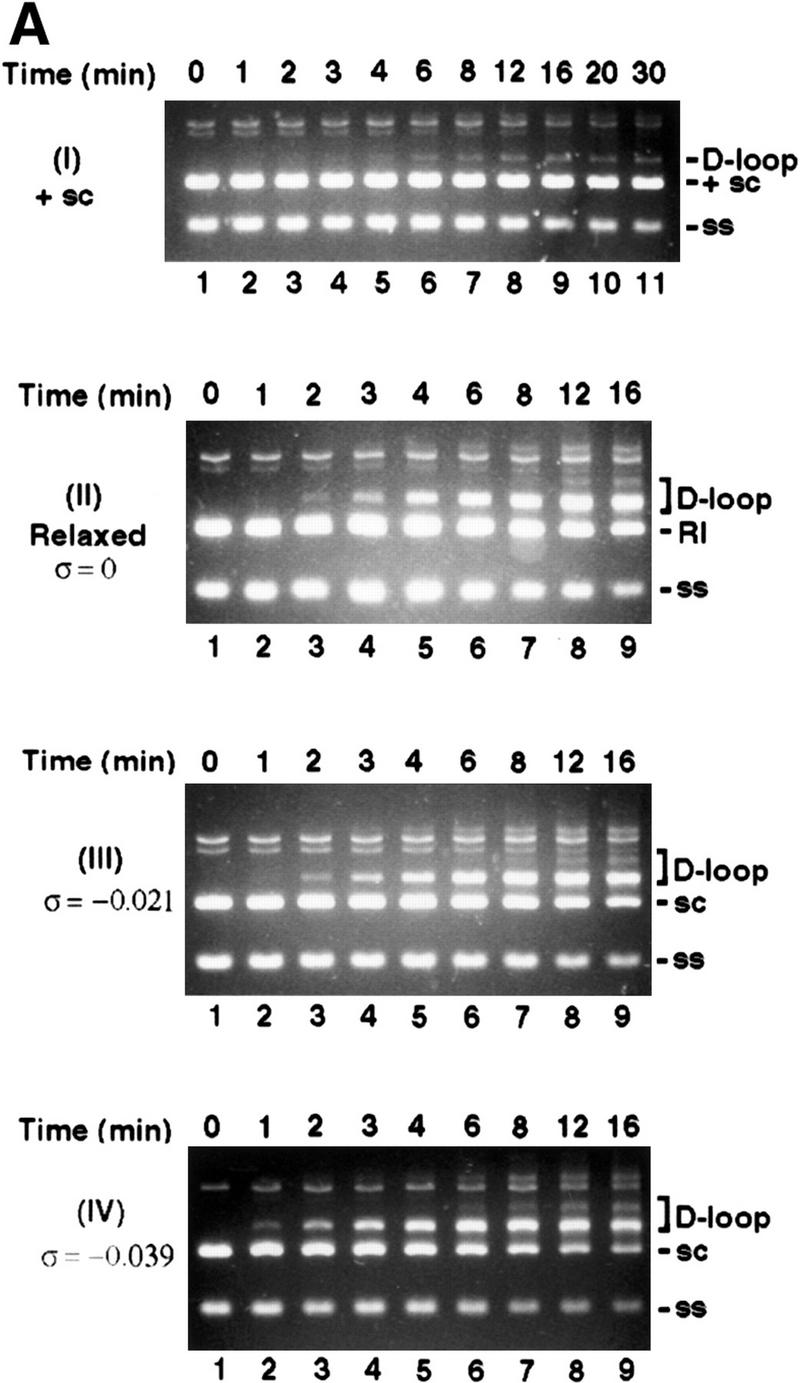
Although Rdh54 contains DNA helicase-like motifs (Eisen et al. 1995) and has dsDNA-activated ATPase activity, it is devoid of DNA helicase activity (data not shown). However, if ATP hydrolysis drives a change in the DNA topology that does not entail extensive separation of DNA base pairs, then such a change may not be revealed by the conventional helicase assay. To investigate this possibility, we incubated topologically relaxed DNA with ATP, topoisomerase I, and Rdh54, followed by analysis in an agarose gel to look for a linking number change (ΔLk) in the DNA. These experiments were first conducted with E. coli topoisomerase I, which removes negative supercoils but is inactive toward positive supercoils. As shown in Figure 3A, incubation of the relaxed DNA with Rdh54 and topoisomerase I resulted in a change in the ΔLk, as indicated by formation of a DNA species that had a higher electrophoretic mobility. Because subsequent analyses indicated that this novel DNA species was positively supercoiled, it was designated Form OW (overwound) DNA. The omission of ATP or its substitution with ADP or the nonhydrolyzable analog ATP-γ-S abolished the formation of Form OW DNA (Fig. 3B), and rdh54 K351R did not produce Form OW DNA in the presence of ATP (Fig. 3B). That Form OW DNA was positively supercoiled was revealed by using two-dimensional gel analysis (Fig. 3C, III) and by the observation that it was relaxed by calf thymus topoisomerase I (data not shown), which acts on both negatively and positively supercoiled DNA templates, but not by using E. coli topoisomerase I (Fig. 3C, IV). Thus, the results indicated that Rdh54 generates both negatively and positively supercoiled domains in the DNA template on ATP hydrolysis. Preferential removal of the negative supercoils by E. coli topoisomerase I resulted in accumulation of positive supercoils and the formation of Form OW DNA (Fig. 3). Maximal level of Form OW DNA occurred at 220 nm Rdh54 (DNA to Rdh54 ratio of 84 base pairs per protein monomer; Fig. 3A, lane 5), although even at 22 nm Rdh54 (840 bp per protein monomer), some Form OW DNA was seen (Fig. 3A, lane 2).
Figure 3.
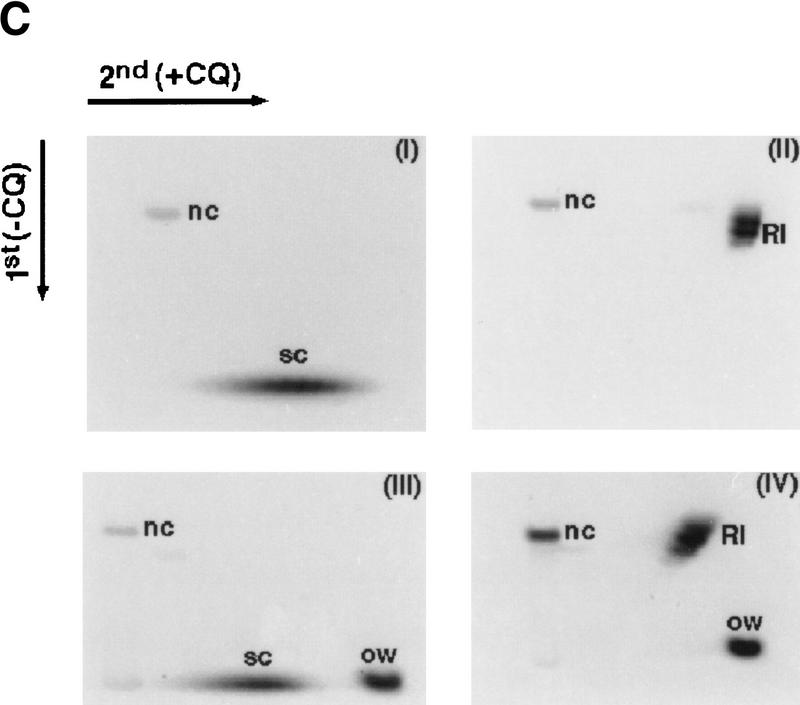
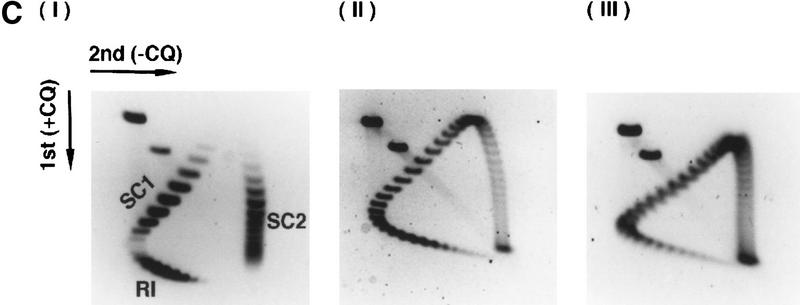
DNA topology modification as revealed by treatment with E. coli topoisomerase I. (A) Increasing concentrations of Rdh54 (22, 44, 88, 220, and 440 nm in lanes 2–6, respectively) were incubated with relaxed φX DNA (18.5 μm base pairs) in the presence of ATP and E. coli topoisomerase I; the DNA species were resolved in a 0.9% agarose gel and visualized by staining with ethidium bromide. In lane 1, the DNA was incubated in buffer with topoisomerase but no Rdh54; in lane 7, the DNA was incubated with 440 nm Rdh54 in the absence of topoisomerase. The product of DNA topology modification is designated Form OW. (B) ATP hydrolysis is required for DNA topology modification. In lanes 2–5, 200 nm Rdh54 was incubated with relaxed DNA and E. coli topoisomerase I in the absence of a nucleotide or in the presence of ATP, ATP-γ-S, or ADP as indicated, and the DNA product was analyzed as described in A. The rdh54 K351R mutant protein (200 and 400 nm in lanes 6 and 7, respectively) was also examined for the ability to remodel DNA. In lane 1, relaxed DNA was incubated in buffer with topoisomerase and ATP but no Rdh54 or rdh54 K351R. The DNA concentration was 18.5 μm base pairs in all of the reaction mixtures. (C) Two-dimensional gel analysis of Form OW DNA. In panels I and II, negatively supercoiled φX DNA isolated from cells (ς = −0.06) without (I) or with (II) prior treatment with E. coli topoisomerase I was subject to two-dimensional gel analysis. In panels III and IV, a mixture of negatively supercoiled φX DNA and purified Form OW DNA without (III) or with (IV) prior treatment with E. coli topoisomerase I was subject to two-dimensional gel analysis. Note that the negatively supercoiled DNA, but not Form OW DNA, was relaxed by E. coli topoisomerase I (IV). In these gel analyses, the first dimension was conducted in the absence of chloroquine (−CQ) and the second dimension in the presence of chloroquine (+CQ). (nc) nicked circular DNA;(ow) Form OW DNA; (Rl) relaxed DNA; (sc) negatively supercoiled DNA.
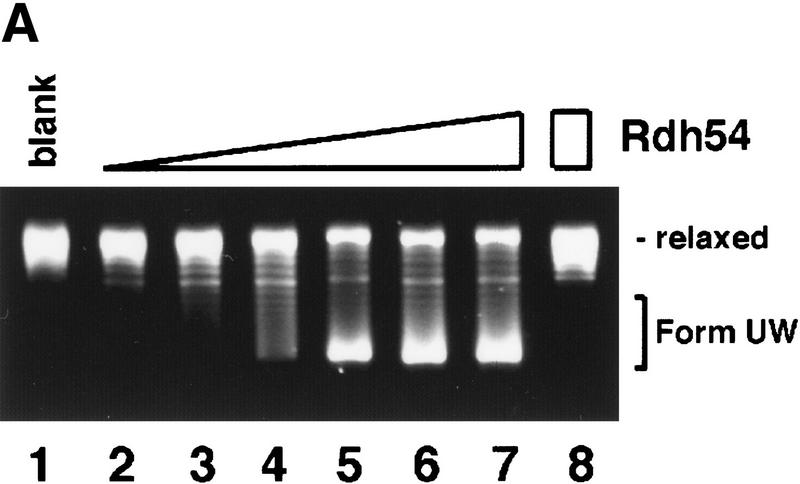
We next used calf thymus topoisomerase I in the DNA topology modification reaction. At the concentration of Rdh54 (220 nm, corresponding to 84 base pairs of duplex DNA per protein monomer) that gave maximal amount of Form OW DNA in the experiment with E. coli topoisomerase I (Fig. 3A, lane 5), there was no change in the linking number of the DNA substrate when calf thymus topoisomerase I was used (Fig. 4A, lane 3). This result allowed us to deduce that the negative and positive supercoils produced by up to 220 nm Rdh54 are largely unconstrained because both are apparently subject to complete removal by calf thymus topoisomerase I with no net linking number change in the DNA (Fig. 4A, lanes 2 and 3). At much higher concentrations of Rdh54 (440 nm to 1.76 μm), heterogeneous DNA species with linking numbers different than that of the relaxed substrate were seen (Fig. 4A, lanes 4–7). The formation of these novel DNA species was again completely dependent on ATP hydrolysis (Fig. 4B). These DNA species could be relaxed by E. coli topoisomerase I, indicating that they were negatively supercoiled (data not shown); we have designated these species collectively as Form UW (underwound). This conclusion was also confirmed by two-dimensional gel analysis, which revealed that Form UW DNA contained upward of one negative superhelical turn to >40 such turns (Fig. 4C, II and III). These results indicated that some of the negative supercoils produced become constrained at high concentrations of Rdh54 (Fig. 4A, lanes 4–7). This deduction is consistent with the observation that at high concentrations of Rdh54 at which production of Form UW DNA with calf thymus topoisomerase I was maximal, the level of Form OW DNA produced by treatment with E. coli topoisomerase I was in fact reduced (data not shown).
Figure 4.
DNA topology modification as revealed by treatment with calf thymus topoisomerase I. (A) Increasing concentrations of Rdh54 (0.088, 0.22, 0.44, 0.88, 1.32, and 1.76 μm in lanes 2–7, respectively) were incubated with relaxed φX DNA (18.5 μm base pairs) in the presence of 2 mm ATP and 4 units of calf thymus topoisomerase I; the DNA products were analyzed in a 0.9% agarose gel and visualized by staining with ethidium bromide. In lane 1, the DNA was incubated in buffer with topoisomerase but no Rdh54; in lane 8, the DNA was incubated with 1.76 μm Rdh54 in the absence of topoisomerase. The product of DNA topology modification is designated Form UW. (B) ATP hydrolysis is required for DNA topology modification. Rdh54 (0.88 μm in lanes 2–5) was incubated with relaxed DNA and calf thymus topoisomerase I in the absence of a nucleotide or in the presence of ATP, ATP-γ-S, or ADP as indicated; the DNA product was analyzed by electrophoresis as described in A. In lane 1, relaxed DNA was incubated in buffer with topoisomerase but no Rdh54. The rdh54 K351R mutant protein (1 and 2 μm, in lanes 6 and 7, respectively) was also analyzed for the ability to remodel DNA. In lane 1, relaxed DNA was incubated in buffer with topoisomerase but no Rdh54 or rdh54 K351R. The DNA concentration was 18.5 μm base pairs in all the reaction mixtures. (C) Two-dimensional gel analysis of Form UW DNA. In panel I, a mixture of relaxed φX DNA (Rl), slightly negatively supercoiled φX DNA (SC1; average ς = −0.017), and negatively supercoiled φX DNA isolated from cells (SC2; average ς = −0.06) was subjected to two-dimensional gel analysis in which the first dimension was conducted in the presence of chloroquine diphosphate (+CQ) and the second dimension was conducted without it (−CQ). Analysis of Form UW DNA made by treating relaxed φX DNA (18.5 μm base pairs) with 0.6 μm (panel II) and 0.8 μm (panel III) Rdh54 and topoisomerase was also performed.
Relevance of DNA topology in D-loop formation
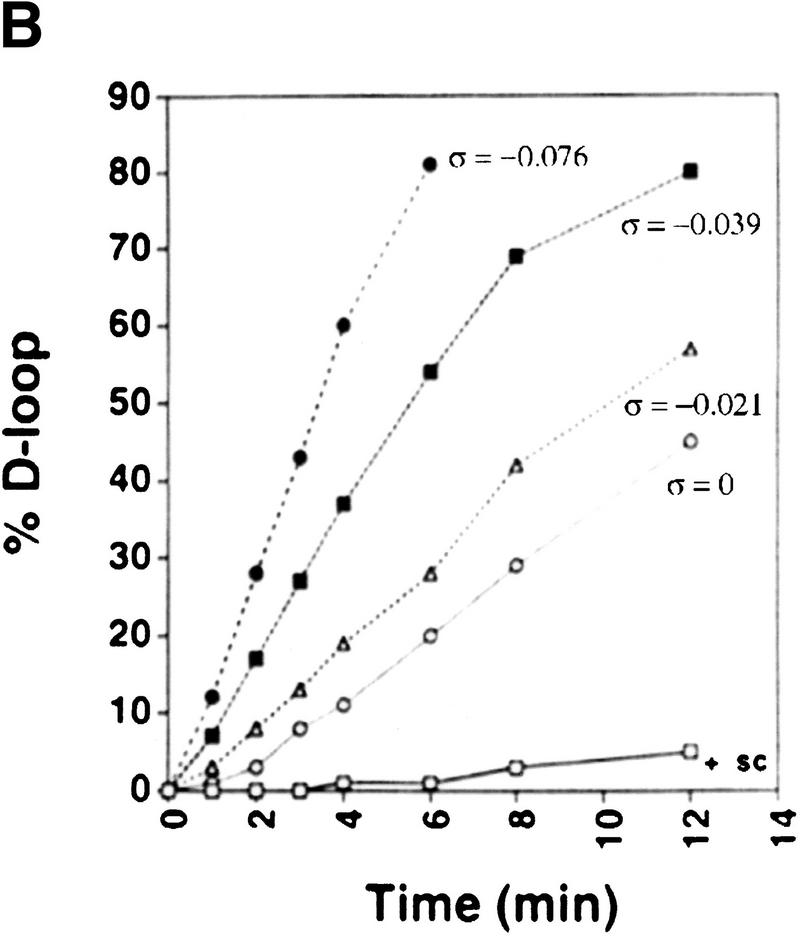
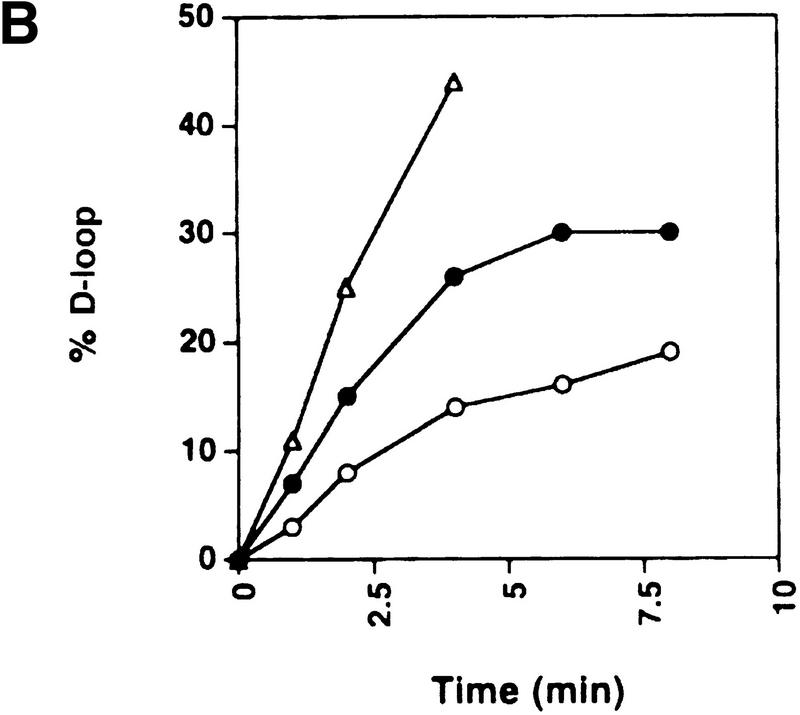
To address whether the positive or negative supercoils produced by Rdh54 are important for the D-loop reaction, we examined D-loop formation with positively supercoiled DNA, relaxed DNA, and a series of negatively supercoiled DNA substrates. As shown in Figure 5, the level of D-loop formation increased with the degree of negative superhelicity in the substrate, implying that the negative supercoils generated in the DNA topology modification reaction are germane for D-loop formation. Surprisingly, robust D-loop formation was seen even with topologically relaxed DNA, suggesting that the ability to supercoil DNA negatively can overcome the topological constraints encountered in DNA joint formation involving a topologically relaxed DNA substrate. In these experiments (Fig. 5), ethidium bromide was included in the gel analysis to improve the resolution of the joint molecules formed with the relaxed (ς = 0) and slightly supercoiled (ς = −0.021) DNA substrates, but omission of ethidium bromide from the gel analysis did not affect the levels of joint molecules formed with these substrates. With the entire range of DNA substrates, DNA joint formation required Rad51, Rdh54, and RPA; rad54 K351R was inactive (data not shown). For the E. coli RecA recombinase and its cognate single-strand binding factor SSB, no D-loop was formed with positively supercoiled DNA or relaxed DNA, and a much more pronounced dependence of D-loop formation on the degree of negative superhelicity in the DNA template was seen than occurred in the case of Rad51-Rdh54-RPA, regardless of whether ethidium bromide was included in the gel analysis (data not shown). With all the DNA substrates tested, the combination of Rad51-Rdh54-RPA appeared to be considerably more adept in D-loop formation than was RecA-SSB (data not shown).
Figure 5.
Dependence of D-loop formation on negative supercoiling. (A) Linear φX viral (+) strand (ss) was incubated with Rad51, Rdh54, and RPA, and the nucleoprotein complex thus formed was reacted with positively supercoiled DNA (+ sc;, panel I), topologically relaxed DNA (Rl; ς = 0; panel II), or negatively supercoiled DNA (sc) substrates with ς values of −0.021 (panel III) and −0.039 (panel IV) at 23°C for the indicated times. To effect separation of the D-loop from other DNA species, we analyzed the reaction mixtures in agarose gels containing 10 μm ethidium bromide. (B) The data points from image analyses of the gels in A and from another reaction performed under the same conditions but with duplex substrate having a ς value of −0.076 are plotted.
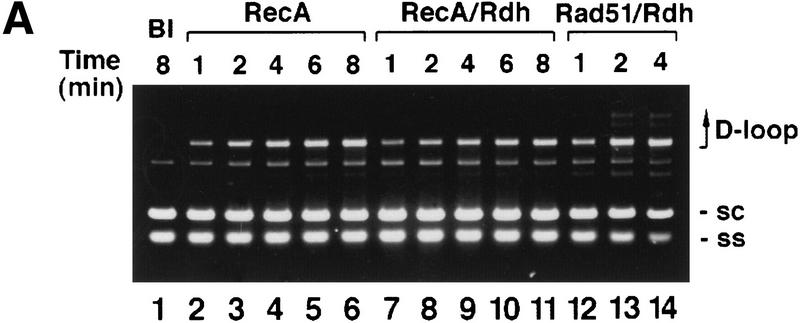
Next, we addressed whether D-loop formation was dependent only on the ability of Rdh54 to modify DNA topology or whether it also required physical interaction of Rdh54 with Rad51. We reasoned that if change in DNA topology alone is critical for promoting D-loop formation, then Rdh54 should also stimulate the ability of RecA to make D-loop. However, with negatively supercoiled DNA (ς = −0.06) isolated from cells as substrate, Rdh54 actually suppressed the D-loop reaction mediated by RecA (Fig. 6). The same inhibitory effect by Rdh54 was seen over a wide range of RecA concentrations, using other negatively supercoiled DNA templates (ς values of −0.021, −0.039, and −0.076), when RPA was used instead of SSB and with buffer conditions optimal for the RecA-mediated reaction or those we used for the Rad51–Rdh54–RPA reaction (Fig. 6; data not shown). Rdh54 also did not enable RecA/SSB to make a D-loop with relaxed DNA or positively supercoiled DNA (data not shown). Thus, the ability of Rdh54 to promote D-loop formation is highly specific for Rad51, suggesting that the interaction between these two proteins is indispensable for the D-loop reaction.
Figure 6.
Effect of Rdh54 is specific for Rad51. (A)Linear φX viral (+) strand (ss) and replicative form I DNA (sc) were incubated with RecA and SSB (lanes 2–11) in the absence (RecA; lanes 2–6) or presence (RecA/Rdh; lanes 7–11) of Rdh54 for the times indicated. In lanes 12–14, the DNA substrates were incubated with Rad51, RPA, and Rdh54 (Rad51/Rdh) for the times indicated. The reaction temperature was 23°C, and the reaction mixtures were analyzed in an agarose gel without ethidium bromide. In lane 1, the DNA substrates were incubated in buffer without any recombination protein (Bl). (B) The data points from image analysis of the gel in A are plotted. (●) results in lanes 2–6 of A involving RecA and SSB; (○) results in lanes 7–11 of A involving RecA, SSB, and Rdh54; (▵) results in lanes 12–14 of A involving Rad51, RPA, and Rdh54.
Discussion
In homologous recombination processes, DNA double-strand breaks are processed nucleolytically to yield ssDNA tails of a considerable length. These ssDNA tails are then used for the invasion of an intact DNA homolog (sister chromatid or homologous chromosome) to form a D-loop. In addition to establishing stable linkage among the recombining DNA molecules, D-loop formation is critical for priming DNA synthesis to replace the genetic information eliminated during end-processing of DNA double-strand breaks (Paques and Haber 1999; Sung et al. 2000). The results reported here indicate that Rdh54 promotes D-loop formation by the Rad51 recombinase. Although the two-hybrid analysis of Dresser et al. (1997) has revealed that Rad51 and Rdh54 interact in vivo, our in vitro experiment involving purified Rad51 and Rdh54 further indicates that interaction between these two factors occurs directly. In addition, we have shown that Rdh54 modifies DNA topology in a fashion that requires ATP hydrolysis and generates extensive positive and negative supercoils. Because the level of D-loop is stimulated by negative supercoiling, it seems reasonable to suggest that the negative supercoils produced by Rdh54 are important for D-loop formation. The unconstrained negative supercoils generated by Rdh54 could render strand invasion by the Rad51-ssDNA nucleoprotein complex more facile because DNA strand separation in duplex DNA is favored by negative supercoiling. In fact, with the ability to supercoil DNA, even a topologically relaxed DNA template is efficiently used for D-loop formation.
Tracking of transcribing RNA polymerases along the DNA template creates a negatively supercoiled domain behind the path of polymerase movement and a positively supercoiled domain ahead of it (Wu et al. 1988). A similar tracking mechanism has been invoked to explain the generation of both negative and positive supercoils by DNA helicases and type I restriction enzymes (Yuan et al. 1980; Yang et al. 1989). We suggest that the free energy from ATP hydrolysis also fuels the tracking of Rdh54 on duplex DNA. We speculate that such a tracking mechanism not only produces the unconstrained supercoils observed experimentally in this study but may also enhance the efficiency at which the incoming duplex DNA molecule is sampled by the Rad51–Rdh54–RPA–ssDNA complex for homology.
Yeast Rad54, a homolog of Rdh54 and member of the RAD52 epistasis group, also promotes D-loop formation by Rad51 and RPA (Sung et al. 2000). Published work has indicated that yeast Rad54 and human Rad54 also modify DNA topology (Petukhova et al. 1999; Tan et al. 1999), and our unpublished studies have indicated a similar ability of yeast Rad54 to induce both negative and positive supercoils in DNA. Despite these similarities between Rdh54 and Rad54, the latter is notably effective in promoting DNA joint formation in model systems that involve linear duplex DNA molecules (Petukhova et al. 1998, 1999), whereas the stimulatory effect of Rdh54 is much more specific for the D-loop reaction (data not shown). Genetic studies have also indicated that mitotic interchromosomal recombination and meiotic recombination are more dependent on Rdh54, whereas Rad54 appears to be more specifically involved in sister chromatid-based recombination and intrachromosomal recombination in the mitotic phase (Klein 1997; Arbel et al. 1999). Aside from the differences in biochemical properties between Rad54 and Rdh54, specific interactions of Rad54 and Rdh54 with other recombination factors may also confer specificities in various recombination processes. Consistent with this conjecture, Rdh54 has been found to bind the meiosis-specific recombinase Dmc1 in yeast two-hybrid studies (Dresser et al. 1997). Our biochemical results suggest that an interaction of Rdh54 with Dmc1 may enable the latter to promote formation of DNA intermediates during meiotic recombination.
In mouse embryonic stem cells, chicken DT40 cells, and Drosophila, deletion of the RAD54 gene engenders defects in recombination and repair of DNA damage induced by ionizing radiation, methyl methanesulfonate, and the DNA cross-linking agent mitomycin C (Esser et al. 1997; Kooistra et al. 1999; Takata et al. 1999). Recently, another Rad54/Rdh54-like factor, Rad54B, has been identified in humans (Hiramoto et al. 1999; Matsuda et al. 1999). Thus, similar to the situation in yeast, human cells appear to possess multiple Rad54/Rdh54-like proteins. The results reported in this article provide a biochemical model for examining the roles of Rad54/Rdh54-like factors in recombination and DNA repair in multicellular organisms.
Materials and methods
Plasmids for protein expression
A portion of Rdh54 (amino acid residues 226–472) was expressed in E. coli with the T7 promoter and was used as antigen for raising polyclonal antisera. For expression in yeast, RDH54 gene and rdh54 K351R mutant gene containing a six-histidine sequence at the carboxyl terminus was fused to the GAL–PGK promoter to yield plasmids pRdh54.1 (2μ, LEU2-d, GAL–PGK–6His–RDH54) and prdh54K/R.1 (2μ, LEU2-d, GAL–PGK–6His–rdh54 K351R), which were introduced into the yeast strain BJ5464 (Matα, ura3-52, trp-1, leu2Δ1, his3Δ200, pep4::HIS3, prbΔ1.6R). Cells were cultured in leucine dropout medium, and protein expression was induced by adding galactose, as described by Petukhova et al. (1999).
Recombination proteins
All these steps were performed at 4°C. For purification of Rdh54 protein, we prepared extract from 1 kg of yeast paste (strain BJ5464 harboring pRdh54.1) in 1 liter of cell breakage buffer (50 mm Tris-HCl at pH 7.5, 10% sucrose, 2 mm EDTA, 2 mm DTT, 150 mm KCl, and protease inhibitors) by using a French press, as described (Petukhova et al. 1999), and clarified it by ultracentrifugation (100,000g for 90 min). The supernatant from the centrifugation step (Fraction I) was passed through a Q Sepharose column (2.5 × 24 cm; 120 ml total; Amersham Pharmacia Biotech). The Q column flow (Fraction II) was applied onto an SP Sepharose column (2.5 × 24 cm; 120 ml total), which was washed with K buffer (20 mm KH2PO4 at pH 7.4, 10% glycerol, 0.5 mm EDTA, and 0.5 mm DTT) containing 150 mm KCl and then developed with a 600-ml gradient of 150–600 mm KCl in K buffer. Rdh54 eluted from SP Sepharose at ∼380 mm KCl as determined by immunoblotting, and the peak fractions were pooled (Fraction III; 50 ml total) and fractionated in a column of Bio-Gel HTP hydroxyapatite (1.5 × 4 cm; 7 ml total; Bio-Rad) by using a 150-ml gradient of 20–280 mm KH2PO4 in K buffer. Hydroxyapatite fractions containing the peak of Rdh54, eluting at ∼180 mm KH2PO4, were pooled (Fraction IV; 10 ml total) and mixed with 0.5 ml of nickel NTA-agarose for 2 hr. The nickel matrix was poured into a glass column with an internal diameter of 0.6 cm; it was washed with 8 ml of 10 and 20 mm imidazole and then with 3 ml of 150 mm imidazole in K buffer containing 150 mm KCl. The 150-mm imidazole eluate (Fraction V) was further fractionated in a 1-ml Mono S column (Amersham Pharmacia Biotech), using a 20-ml gradiant of 150–500 mm KCl in K buffer. Rdh54 was eluted from Mono S at ∼360 mm KCl, and ∼200 μg of nearly homogeneous protein (Fraction VI) was obtained from the starting 1 kg of yeast paste. The rdh54 K351R mutant was purified from strain BJ5464 harboring prdh54K/R.1 by using the same fractionation procedure. Purified Rdh54 and rdh54 K351R were concentrated to 2 mg/ml in Centricon-30 microconcentrators (Amicon) and stored in small portions at −70°C. Three independent preparations of Rdh54 and two independent preparations of rdh54 K351R gave the same results in all the biochemical experiments. Rad51 and RPA were purified from yeast as described previously (Sung 1997). RecA was a gift from Michael Cox (Department of Biochemistry, University of Wisconsin, Madison), and SSB was purchased from Pharmacia.
DNA substrates
DNA substrates with different degrees of negative supercoiling were made by treating replicative form I φX 174 DNA (GIBCO BRL) with calf thymus topoisomerase I (GIBCO BRL) and increasing concentrations of ethidium bromide (Sung et al. 1993). The superhelical densities of the DNA preparations were determined by one-dimensional and two-dimensional gel analyses. Topologically relaxed DNA was obtained by treatment of form I DNA with calf thymus topoisomerase I without ethidium bromide. To make positively supercoiled DNA, we treated topologically relaxed DNA (38 μm base pairs) with Rdh54 (450 nm) and 5 μg of E. coli topoisomerase I in 400 μl of buffer R (35 mm potassium MOPS at pH 7.2, 2 mm ATP, 3 mm MgCl2, 1 mm DTT, 100 μg/ml BSA, and an ATP-regenerating system consisting of 20 mm creatine phosphate and 30 μg/ml creatine kinase) at 37°C for 15 min. To purify the DNA substrates, we extracted reaction mixtures once with buffered phenol, ran them in 0.8% agarose gels, and then isolated the desired DNA species from gel slices by using the Geneclean kit (Bio 101). The agarose gels used in the purification of the topologically relaxed DNA and slightly negatively supercoiled DNA (with ς = −0.021) contained 10 μm ethidium bromide to effect separation of these DNA species from the nicked and linear forms. All of the purified DNA species were filter-dialyzed in Centricon-30 microconcentrators into TE (10 mm Tris-HCl at pH 7.5, and 0.2 mm EDTA).
ATPase assay
The indicated amounts of Rdh54 or rdh54K351R were incubated with 1.5 mm [γ-32P]ATP in a final volume of 10 μl of reaction buffer (30 mm Tris-HCl at pH 7.5, 5mm MgCl2, 1 mm DTT, 50 μg/ml BSA, and 30 mm KCl) in the absence of DNA and with either 100 ng of φX 174 viral (+) strand (30 μm nucleotides) or replicative form I DNA (15 μm base pairs) at 37°C for the indicated times. The level of ATP hydrolysis was determined by thin layer chromatography in polyethylenimine cellulose sheet. The oligonucleotides used in Figure 1D were as follows: oligo 1, 5′-AAATGAACATAAAGTAAATAAGTATAAGGATAATACAAAATAAGTAAATGAATAAACATAGAAAATAAAGTAAAGGATATAAA-3′; and oligo 2, the exact complement of oligo 1.
Binding of Rdh54 to Affi-gel immobilized Rad51
Purified Rdh54 or rdh54 K351R, 2 μg in 100 μl of buffer T (20 mm Tris-HCl at pH 7.5, 10% glycerol, 0.5 mm EDTA, 0.5 mm DTT, 0.01% NP-40, 100 mm KCl and 200 μg/ml BSA), was mixed with 6 μl of Affi-Gel 15 beads containing covalently conjugated Rad51 (2 mg/ml) or BSA (5 mg/ml) for 15 min at 23°C. The beads were collected by a 5-sec centrifugation in a microcentrifuge and washed with 150 μl of buffer T, before being treated with 30 μl of 3% SDS at 37°C for 10 min to elute bound Rdh54. The input material (10 μl), the supernatant that contained unbound Rdh54 (10 μl), the wash (15 μl), and the SDS eluate (5 μl) were analyzed by SDS-PAGE to determine their content of Rdh54 or rdh54 K351R
D-loop assay
A 26-mer oligonucleotide was hybridized to φX viral (+) strand to create a PstI site and then treated with PstI for linearization. Alternatively, a 26-mer oligonucleotide was used to generate an AvaII site for linearization. Linearized viral (+) strands were banded in 0.9% agarose gels and purified from gel slices by using the Geneclean kit. Reaction mixtures (50 μl final volume) were assembled by mixing 2.2 μg of Rad51 (1 μm) added to 1 μl of storage buffer and 500 ng of linear φX viral (+) strand (30 μm nucleotides) added to 2 μl of TE in 40 μl of buffer R. After a 3-min incubation at 37°C, 12 μg of RPA (2 μm) in 2 μl of storage buffer was added, followed by a 3-min incubation at 37°C; then 1 μl of storage buffer with or without 1.6 μg of Rdh54 (290 nm) was incorporated, followed by a 2-min incubation at 23°C. To complete the reaction mixture, we incorporated 3 μl of TE containing 600 ng of φX174 replicative form I DNA (18.5 μm base pairs) with the indicated topological state and 4 μl of 50 mm spermidine. The reaction mixture was incubated at 23°C, and 5-μl portions were withdrawn at the indicated times, deproteinized with 0.5% SDS and 0.5 mg/ml proteinase K for 15 min at 37°C, and subjected to electrophoresis in 0.85% agarose gels in TAE buffer, as described (Petukhova et al. 1998). The gels were stained in ethidium bromide (2 μg/ml in water) for 1 hr, destained for 18 hr in water, and then subjected to image analysis in a NucleoTech gel documentation station equipped with a CCD camera. For better resolution of topologically relaxed DNA, the slightly supercoiled DNA (ς = −0.021), and the D-loop species formed with these DNA substrates from the nicked circular and linear forms present, we included 10 μm ethidium bromide in the agarose gels used in Figure 5A. The reaction mixtures in Figure 5A were otherwise deproteinized as usual without ethidium bromide. In some cases (e.g., Fig. 2E), the reaction mixtures were scaled down appropriately, but otherwise they were assembled and analyzed in the manner described.
DNA topology modification reaction
Increasing concentrations of Rdh54 were incubated with 120 ng of relaxed φX 174 DNA (18.5 μm base pairs) in 9.5 μl of buffer R for 3 min at 23°C, followed by the addition of 4 U calf thymus topoisomerase I (GIBCO BRL) or 100 ng of E. coli topoisomerase I in 0.5 μl of 50mm Tris-HCl (pH 7.5), 50% glycerol, 1 mm DTT, and 100 μg/ml BSA. Reaction mixtures were incubated at 37°C for 10 min, quenched by 0.5% SDS, and treated with 1 mg/ml proteinase K for 15 min at 37°C. Reaction products were resolved in 0.8% agarose gels run in TAE buffer. DNA species were visualized by staining with ethidium bromide (3 μg/ml in water) for 1 hr, destained for 16 hr in water, and then analyzed using the gel documentation station.
Two-dimensional gel analysis
All of the electrophoresis steps were performed using Kodak BioMax MP1015 agarose gel units. In Figure 3B, for the first dimension, the DNA samples were run in 0.9% gels in TAE buffer at 110 mA and 23°C for 6 hr. The gels were soaked for 18 hr at 4°C in 30 gel volumes of TAE buffer containing 30 μm chloroquine diphosphate. The second dimension of the electrophoresis was performed in TAE buffer containing 30 μm chloroquine diphosphate at 110 mA and 23°C for 6 hr. In Figure 4B, for the first dimension, the DNA samples were run at 110 mA and 23°C in 0.85% gels in TAE buffer containing 15 μm chloroquine diphosphate for 9 hr. Gels were soaked in 30 gel volumes of TAE buffer at 4°C for 18 hr. The second dimension of the electrophoresis was performed in TAE buffer at 110 mA and 23°C for 6 hr. After completion of the second dimension, we stained gels with ethidium bromide (3 μg/ml in water) for 90 min, destained them for 24 hr in water, and then analyzed them using the gel documentation station.
Acknowledgments
We thank Sabrina Stratton and Sefton Vergano for excellent assistance; Stephen Van Komen for help with art work; James Wang, Sue-Jane Chen, and Yuk-Ching Tse-Dinh for their gifts of E. coli topoisomerase I; and Michael Cox for the gift of RecA protein. This study was supported by National Institutes of Health research grants ES07061, GM57814, and GM53738 and training grant T32AG00165.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sung@uthscsa.edu; FAX (210) 567-7277.
E-MAIL hannah.klein@med.nyu.edu; FAX (212) 263-8166.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.826100.
References
- Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;8:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Domingues AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser JR, Hendriks W, Swagemakers SMA, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Hiramoto T, Nakanishi T, Sumiyoshim T, Fukudam T, Matsuura S, Tauchi H, Komatsu K, Shibasaki Y, Inui H, Watatani M, et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancers. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- Klein H. RDH54, a RAD54 homologue in Saccharomyces cerevisiae,is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra R, Vreeken K, Zonneveld JB, de Jong A, Eeken JC, Osgood CJ, Buerstedde JM, Lohman PH, Pastink A. The Drosophila melanogaster RAD54 homolog, DmRAD54, is involved in the repair of radiation damage and recombination. Mol Cell Biol. 1999;19:6269–6275. doi: 10.1128/mcb.17.10.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–65. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Miyagawa K, Takahashi M, Fukuda T, Kataoka T, Asahara T, Inui H, Watatani M, Yasutomi M, Kamada N, et al. Mutations in the RAD54 recombination gene in primary cancers. Oncogene. 1999;18:3427–3430. doi: 10.1038/sj.onc.1202692. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton SA, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. ATP hydrolysis-dependent duplex DNA unwinding and promotion of Rad51 catalyzed homologous DNA pairing by yeast Rad54 protein. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Roca IA, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP dependent homologous DNA pairing and strand exchange by the yeast Rad51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Sung P, Reynolds P, Prakash L, Prakash S. Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J Biol Chem. 1993;268:26391–26399. [PubMed] [Google Scholar]
- Sung P, Van Komen S, Trujillo K. Recombination factors of Saccharomyces cerevisiae. Mutat Res. 2000;451:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1999;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TL, Essers J, Citterio E, Swagemakers SM, de Wit J, Benson FE, Hoeijmakers JH, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yang L, Bret Jessee C, Lau K, Zhang H, Liu LF. Template supercoiling during ATP-dependent DNA helix tracking: Studies with simian virus 40 large tumor antigen. Proc Natl Acad Sci. 1989;86:6121–6125. doi: 10.1073/pnas.86.16.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R, Hamilton DL, Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980;20:237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]