Synopsis
Selenium has been known for many years to have a role in boosting immune function, but the manner in which this element acts at the molecular level in host defense and inflammatory diseases is poorly understood. To elucidate the role of selenium-containing proteins in immune function, we knocked out the expression of this protein class in T cells or macrophages of mice by targeting the removal of the selenocysteine tRNA gene using loxP-Cre technology. Mice with selenoprotein-less T cells manifested reduced pools of mature and functional T cells in lymphoid tissues and an impairment in T cell-dependent antibody responses. Furthermore, selenoprotein deficiency in T cells led to an inability of these cells to suppress reactive oxygen species (ROS) production, which in turn affected their ability to proliferate in response to T cell receptor stimulation. Selenoprotein-less macrophages, on the other hand, manifested mostly normal inflammatory responses, but this deficiency resulted in an altered regulation in extracellular matrix-related gene expression and a diminished migration of macrophages in a protein gel matrix. These observations provided novel insights into the role of selenoproteins in immune function and tissue homeostasis.
Keywords: Macrophage, Selenium, Selenocysteine, T cells
Roles of selenium in immune function
Selenium is an important micronutrient in the diet of humans and other mammals. It is known to serve a wide variety of functions in health and development including roles in cancer and heart disease prevention, viral inhibition, male reproduction and immune function, and roles in delaying the aging process and the onset of AIDS in HIV positive patients(1). Interestingly, of all these health benefits attributed to selenium, a role of this element in boosting immune function was amongst the first indications that selenium, which incidentally was long thought to be a toxin(2), served as an advantageous agent in health. That is, only two years after selenium was shown to prevent liver necrosis in rats, which provided the first demonstration that this element was essential in the diet of mammals(3), McConnell injected 75Se into dogs and found that the isotope was incorporated into leucocytes(4). Since this early finding, numerous studies have suggested roles of selenium in both the adaptive and innate immune systems(5–15). The mechanism of how selenium acts at the molecular level in boosting the immune system is only beginning to be understood, which is primarily through the action of selenoproteins(16,17). The possible action of small molecular weight selenocompounds in enhancing immune activity has not been examined. The focus of this paper is on the role of selenoproteins in the immune system. Several excellent reviews on selenium in the immune system have recently been published that provide an in-depth analysis of wide range benefits that selenium offers in immune function(16–18).
Selenium metabolism in mammals
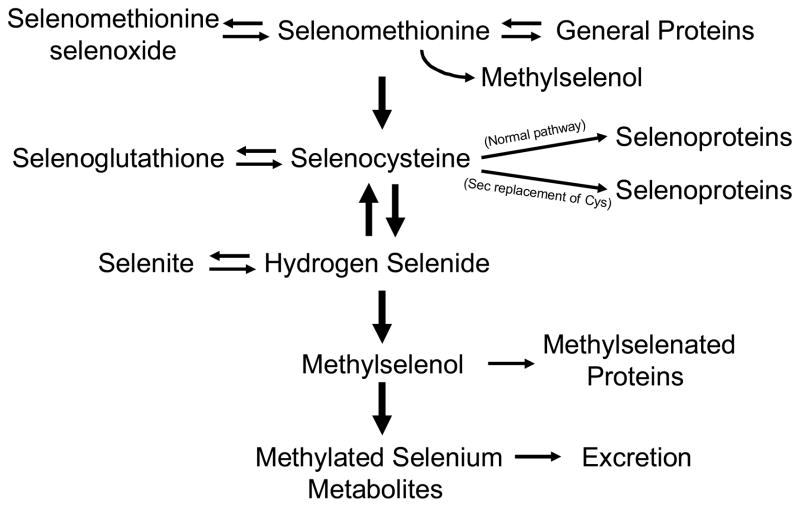
The possible means of how selenium is metabolized in mammalian systems is shown in Fig. 1. Selenium can potentially make its way into protein by multiple pathways, but only one of these pathways is used to specifically incorporate this element into selenoproteins: through the selenium-containing amino acid, selenocysteine (Sec), the 21st amino acid in the genetic code(19). Sec is programmed to be inserted into protein by the stop codon, UGA, and thus, this codeword serves a dual function in mammals. Its biosynthesis occurs by a novel pathway in eukaryotes in that the synthesis is carried out on Sec tRNA, designated Sec tRNA[Ser]Sec, and proceeds as follows: 1) serine is attached to tRNA[Ser]Sec in the presence of seryl-tRNA synthetase (SerS) and ATP to form Ser-tRNA[Ser]Sec; 2) then the seryl moiety is phosphorylated in the presence of O-phosphoseryl-tRNA[Ser]Sec kinase (PSTK) and ATP to form O-phosphoseryl-tRNA[Ser]Sec; and 3) O-phosphoseryl-tRNA[Ser]Sec in turn serves as a substrate for Sec synthase (SecS). In this reaction, monoselenophosphate (SeP), which is the active donor of selenium, donates selenium to the acceptor intermediate generated by SecS to yield selenocysteyl-tRNA[Ser]Sec. SeP is synthesized by selenophosphate synthetase 2 (SPS2) from selenide (or other selenium compounds) and ATP. The details of Sec biosynthesis have been reviewed elsewhere(20). The other pathways wherein selenium can be non-specifically incorporated into protein, namely by selenomethionine replacing methionine and selenocysteine replacing cysteine, may result in deleterious effects to the cell, especially if such insertion occurs to any appreciable extent.
Fig. 1. Selenium and selenomethionine metabolism in mammals.
Pathways of selenium utilization in mammals are shown. Further discussion of selenium and selenomethionine metabolism may be found elsewhere(40,41). Sec is defined in the text and cys designates cysteine.
Roles of selenoproteins in immune function
In our studies on examining the role of selenoproteins in T cell development, we have taken advantage of the fact that selenoprotein synthesis is dependent on Sec tRNA which is responsible for inserting this selenium-containing amino acid into protein. By targeting the removal of the floxed Sec tRNA gene, designated Trsp, in T cells or macrophage of mice by Cre-recombinase under control of the Lck promoter or the lysozyme M (LysM) promoter, we generated selenoprotein-deficient T cells(21) or selenoprotein-deficient macrophage(22), respectively. The expression of selenoproteins in these two cell types and the consequences of the loss of selenoprotein expression in these cell types on immunity are discussed below. Another study involving the targeted removal of Trsp in macrophages and simultaneously Nrf2, a transcription factor that regulates antioxidant enzyme expression, demonstrated that the knockout mice manifested a reduction in viability and an increase in oxidative stress and susceptibility to hydrogen peroxide compared to mice in which either of the two genes was knocked out(23). Mice in which only Trsp was targeted for removal in macrophage had an increase in gene expression involving oxidative stress and detoxification enzymes. These results, along with the several lines of clinical evidence mentioned above, prompted us to examine whether selenoproteins link redox regulation to immunity and inflammation in mammals.
Selenoproteins in T cells
mRNA expression and T cell development
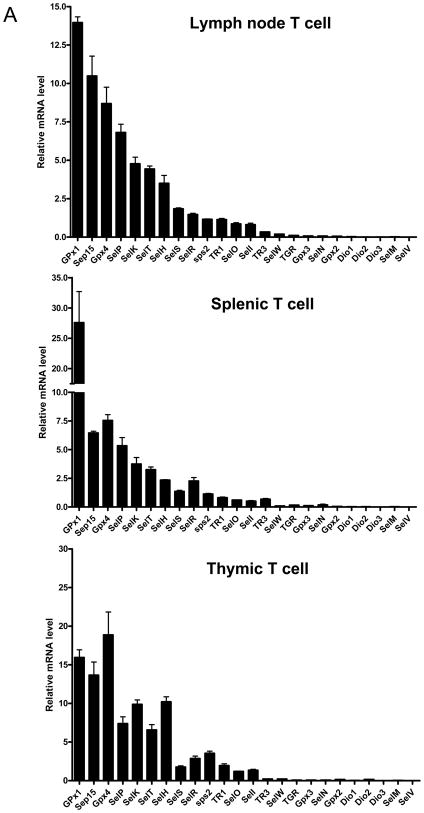
There are 24 selenoprotein genes in mice(24) and individual selenoproteins and selenoprotein mRNAs manifest tissue specificity in their patterns of expression and in the manner in which their expression is regulated(25). We examined the expression of the mRNA levels transcribed from each of the 24 selenoprotein genes by isolating total RNA from the three lymphoid organs, lymph nodes, spleen, and thymus, of normal mice and analyzing the expression by real-time PCR. As shown in Fig. 2A, GPx1 mRNA was highly expressed in all three organs. Several other selenoprotein mRNAs, namely Sep15, GPx4, SelP, SelK, SelT and SelH, were moderately to highly expressed in each organ. Several additional selenoprotein mRNAs were also made, while others were only slightly, or not at all, detectable. It is not clear why there is some variation in the levels of selenoprotein mRNA expression between the three tissues.
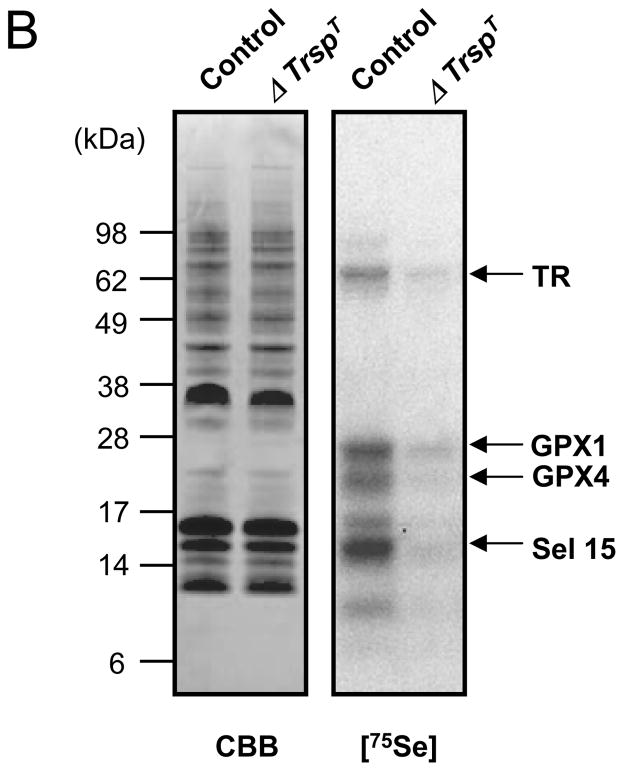
Fig. 2. Selenoprotein expression in T cells.
In (A), expression of selenoprotein mRNA was assessed by quantitative real-time PCR. Purified T cells were obtained from lymph nodes, spleen and thymus of mice as described(21). In (B), expression of selenoproteins was assessed by 75Se-labeling as described(27). The identities of major, labeled selenoproteins are designated on the right panel; and the left panel shows Coomassie-Blue stained gel which was used as a loading control.
We also examined selenoprotein expression in T cells by labeling control and selenoprotein deficient (ΔTrspT) mice with 75Se, isolating the T cells from thymus and examining the labeled selenoproteins by gel electrophoresis (Fig. 2B). Several selenoproteins were labeled in control mice, some of which have been identified in earlier studies(26). Virtually no selenoproteins were labeled in the ΔTrspT cells and the faint labeling is most likely due to contamination from other cell types(27).
The thymus, spleen and lymph nodes of affected mice manifested moderate to severe atrophy wherein their mass and cellularity were decreased to varying degrees ranging between 50 and 80% of those of control mice. The T cell population in lymphoid organs, which was analyzed by flow cytometry, was altered significantly. For example, the CD3+ population in splenocytes of ΔTrspT mice was about 50% of that of control splenocytes, while the CD8+ T cell subpopulation was more substantially reduced (>50%). Although the consequences of the partial loss of functional T cells, and in particular CD8+ T cells, was not clearly defined in our study, other studies have shown that selenium deficiency in the diet, which is known to cause a loss or reduction in selenoprotein function, impairs antiviral immunity and exacerbates viral pathogenesis(28,29).
T cell activation
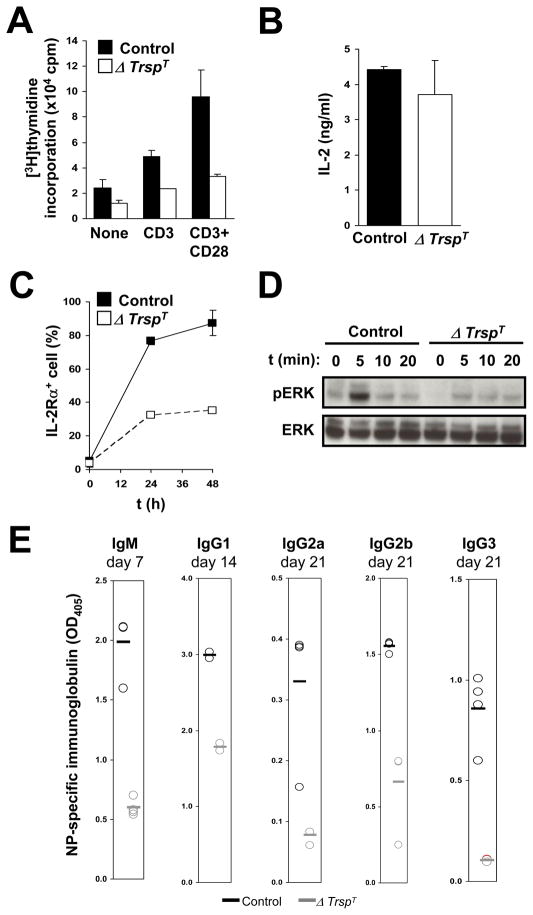
We next tested the ability of selenoprotein-deficient T cells to be activated by T cell receptor (TCR) signaling. Purified control T cells and ΔTrspT cells were cultured in the presence or absence of anti-CD28 and anti-CD3 antibodies wherein the presence of these antibodies mimicked TCR activation and stimulated T cells to proliferate(21). The amount of 3H-thymidine incorporated into both cell lines was measured after 60 hrs as an indication of the level of proliferation. Control cells incubated with both antibodies manifested a far more pronounced level of proliferation compared to unstimulated cells whereas the level of proliferation of ΔTrspT cells was dramatically reduced under the same conditions (Fig. 3A). The data suggested that selenoprotein expression appeared to be essential for the ability of T cells to proliferate in response to TCR stimulation.
Fig. 3. Activation of control and selenoprotein-deficient cells by TCR stimulation.
T cells were isolated from lymph nodes of control and ΔTrspT mice. In (A), the level of proliferation was assessed by incorporation of [3H]-thymidine in absence or presence of anti-CD3/CD28 or anti-CD3 alone. The results of the averages of triplicate determinations from six control and ΔTrspT mice, respectively, are shown. In (B and C), the levels of IL-2 production (B) and cell surface expression of IL-2Rα (C) in CD3/CD28-stimulated T cells are shown. In (D), Erk activation in T cells stimulated with anti-CD3 and anti-CD28 was analyzed at the time intervals indicated. Both phosphorylated (pERK) and total (ERK) proteins are shown. In (E), serum levels of major Igs at indicated time points were determined by ELISA in control and ΔTrspT mice following immunization with NP-OVA. Circles represent Ig levels of each animal. Bars represent the mean values. Experimental details are given in Shrimali et al.(21)
Since TCR activation is known to produce IL-2 in T cells and generate T cell IL-2 receptor (IL-2R) expression, we initially examined IL-2 production in control and ΔTrspT cells (Fig. 3B)(21). Both cell lines produced similar amounts of IL-2 in response to TCR activation. However, ΔTrspT cells manifested a much reduced IL-2R induction compared to control cells (Fig. 3C).
Since TCR is coupled to multiple signaling pathways and it is known that mitogen-activated protein kinase ERK links TCR activation to T cell proliferation(30), we examined ERK activation in T control and selenoprotein-deficient cells in response to stimulation with CD3/CD28 antibodies(21). As shown in Fig. 3D, T cells from ΔTrspT mice manifested a very slight appearance of the phosphorylated or active forms of ERK following treatment with anti-CD3 and anti-CD28 antibodies, while T cells from control mice manifested a rapid appearance of the active forms of ERK.
The above data demonstrated that the loss of selenoprotein expression in T cells resulted in defects in the development of functionally mature T cells and their TCR-dependent activation. It is highly likely that such anomalies result in an impairment in T cell-mediated immune responses. To further examine this possibility, control and ΔTrspT mice were immunized with NP-OVA, an antigen that stimulates T cell antibody production. Serum was taken from the immunized mice at different time periods after immunization and the levels of antigen-specific immunoglobulins were assessed by enzyme-linked immunosorbent assay. Of the several immunoglobulin classes examined, IgM, IgG1, IgG2a, IgG2b and IgG3, the serum levels of antigen-specific antibodies were poorly raised in ΔTrspT mice (Fig. 3E). Thus, these data suggest that selenoproteins play an important role in T cell immunity.
Reactive oxygen species in ΔTrspT cells
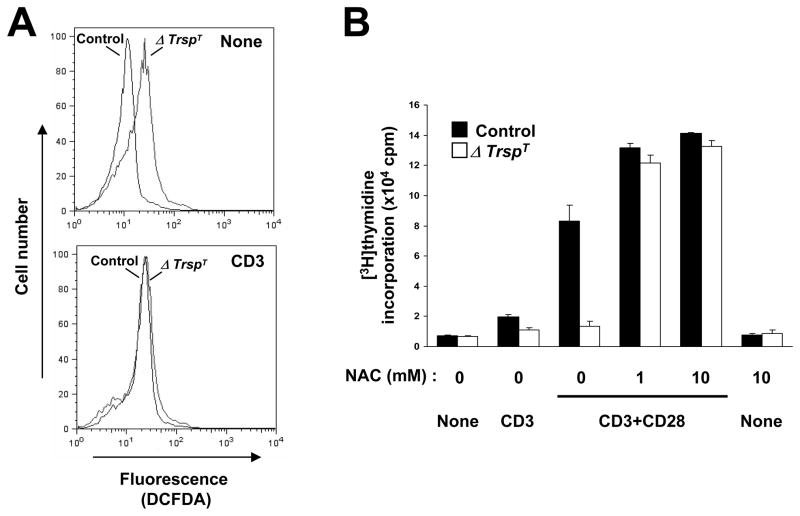
Since several selenoproteins are known to act as antioxidants and therefore have roles in protecting cells against oxidative stress, we examined whether selenoprotein-deficient T cells produce higher levels of ROS than the corresponding control cells and how this may relate to T cell defects in ΔTrspT mice. To elucidate the basal and TCR stimulated ROS generation in ΔTrspT and control cells, we labeled the cells with cell-permeable, oxidation sensitive DCFDA dye. Interestingly, as shown in Fig. 4A, the oxidation of DCFDA due to the generation of ROS was present to a greater extent in T cells of ΔTrspT mice than in control mice even without TCR stimulation. TCR stimulation enhanced ROS production in control cells, but had little effect on ROS levels in ΔTrspT cells. The data suggest that selenoproteins have a major role in suppressing ROS production in T cells and that the loss of this protein class results in T cells being defective in TCR-induced and ROS-sensitive responses.
Fig. 4. ROS production in TCR-stimulated T cells.
In (A), control and ΔTrspT T cells isolated and purified from lymph nodes and either unstimulated or stimulated with anti-CD3 antibody were used. ROS production was analyzed in by flow cytometry using DCFDA. In (B), control and ΔTrspT T cells were either unstimulated (None) or stimulated as indicated in the presence of various concentrations of NAC. T cell proliferation and all other determinations used in the figure are described in Shrimali et al.(21)
Finally, ΔTrspT cells were incubated in the presence of N-acetyl cysteine (NAC), which is known to block endogenously produced ROS, to assess whether this reagent can restore the ability of these cells to proliferate in response to TCR stimulation (Fig. 4B). The ability of the selenoprotein deficient T cells to proliferate in response to TCR stimulation was restored by NAC treatment. NAC alone did not induce T cell proliferation. These observations suggest that selenoproteins are essential for reducing ROS generation in T cells and that ROS production prevents T cell activation.
Selenoproteins in macrophages
Selenoprotein mRNA and selenoprotein expression and ROS levels
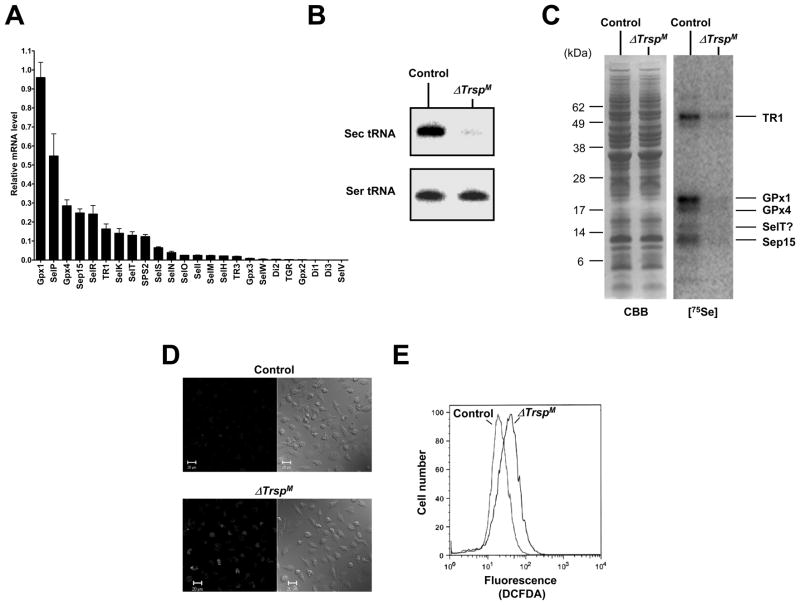
To determine the relative expression levels of selenoproteins mRNAs in mouse macrophages, total RNA was isolated from bone marrow-derived, normal macrophages and the expression of mRNA levels transcribed from all 24 mouse selenoprotein genes were analyzed by real-time PCR. As shown in Fig. 5A, relatively high levels of selenoprotein mRNAs corresponding to GPx1, GPx4, TR1, Sep15, and selenoproteins P, R, K and T and SPS2 were expressed. Interestingly, the most highly expressed selenoprotein mRNA in macrophage, like that in T cells (see above), was GPx1 and a major role of this selenoenzyme is as an antioxidant. Similarly, Sep15, GPx4 and selenoproteins P, K and T were also highly expressed in macrophages.
Fig. 5. Selenoprotein gene expression in macrophages.
In (A), selenoprotein gene expression in macrophages was analyzed by real-time PCR and is shown as the relative mRNA level to that of Gusb which was used as the internal control. In (B), northern blot analysis showing levels of Sec tRNA in control and ΔTrsp macrophage. In (C), 75Se-labeled selenoproteins were visualized by autoradiography after SDS electrophoresis in control and ΔTrsp macrophage as shown in the right panel. Left panel shows Coomassie Blue stained gel which served as a loading control. The identities of the major, labeled selenoproteins are designated on the right of the panel. In (D), macrophages were stained with DCFDA and ROS production analyzed by confocal microscopy. Fluorescence (left panels) and phase contrast (right panels) images are shown. In (E), macrophages were stained with DCFDA and ROS production analyzed by flow cytometry. Experimental details of the studies shown in this figure are given in Carlson et al.(22)
We deleted Trsp in macrophage (ΔTrspM) and examined the effects of Trsp loss on selenoprotein expression and function. Initially, we demonstrated that Trsp was no longer expressed in this cell type (Fig. 5B). Selenoprotein expression was also examined in macrophages by incubating the bone marrow-derived macrophages from control and ΔTrspM mice in the presence of 75Se and electrophoresing a protein extract on a polyacrylamide gel (Fig. 5C. left panel). Several selenoproteins whose mRNAs were also detected by real-time PCR were visualized on the gel, while the expression of selenoproteins in ΔTrspM cells were almost completely abolished (Fig. 5C, right panel). The low levels of Sec tRNA and selenoproteins observed in ΔTrspM macrophages in Figs. 5B and C are likely due to contamination of the bone marrow-derived macrophage preparations with non-macrophage cells as has been observed in studies involving other cell types(27).
The functions of only about half of selenoproteins have been characterized and most are oxidoreductases and/or antioxidants(31). Since selenoprotein-less T cells accumulate higher ROS levels than the corresponding control cells (see Fig. 4A), ROS levels in ΔTrspM macrophages were also assessed by staining (Figs. 5D and E). Resting selenoprotein-deficient macrophages manifested higher steady state ROS levels than control macrophages.
In vitro and in vivo inflammatory responses
Due to the highly significant role that macrophages have in initiating inflammation, it was important to assess the ability of the corresponding selenoprotein-deficient cells in the inflammation process. Macrophages generate cytokines in response to tissue injury and microbial infection. The fact that several inflammatory stimuli such as lipopolysaccharide (LPS) cause transient ROS accumulation as part of cellular signaling events(32,33), and several selenoproteins, e.g., GPx isoforms, are known to regulate inflammatory responses(34,35) prompted us to examine the effect of selenoprotein deficiency and the resulting redox imbalance on macrophage inflammatory response. We initially treated control and ΔTrspM macrophages with LPS and examined the degradation and replenishment of I Bα. It should be noted that the transcription factor NF-κB, and mitogen-activated protein kinase (ERK, JNK and p38) signaling pathways are essential for cellular responses to inflammatory stimuli and that both the degradation and replenishment of I Bα are indicative of NF-κB activation. However, in LPS-treated control and ΔTrspM macrophages, the degradation and replenishment of IκBα occurred independently of selenoprotein status and induction of the active forms of the protein kinases (i.e., phosphorylated ERK, JNK and p38) was similar and therefore considered normal(22). Furthermore, comparison of LPS-induced inflammatory gene expression in the two bone marrow-derived macrophage groups and analyses of gene expression by real-time PCR revealed that the magnitude and kinetics of expression of the genes encoding the chemokines KC (Cxcl1), macrophage inflammatory protein-2 (Cxcl2), and the cytokines tumor necrosis factor-α (Tnf-α) and interleukin-1β (Il1b) were similar(22).
We next turned our attention to examining the effects of selenoprotein deficiency in macrophages on the pathology of mice by exposing control and ΔTrspM mice to various models of inflammatory response. The models used included zymosan-induced peritonitis, LPS endotoxemia and chemical irritant (12-O-tetradecanoylphorbol-13-acetate [TPA]) dermatitis. In these models, control and ΔTrspM mice had comparable rates of neutrophil infiltration, rates of mortality and levels of cytokine production and local edema formation. Since in vitro and in vivo inflammatory responses in both control and ΔTrspM macrophages were similar, we have not shown the data herein, but refer the reader to the original work(22). Interestingly, the loss of selenoprotein expression and the deregulated ROS generation in ΔTrspM macrophages did not appear to cause any detectable detrimental effects concerning inflammatory responses.
Macrophage microarray analysis
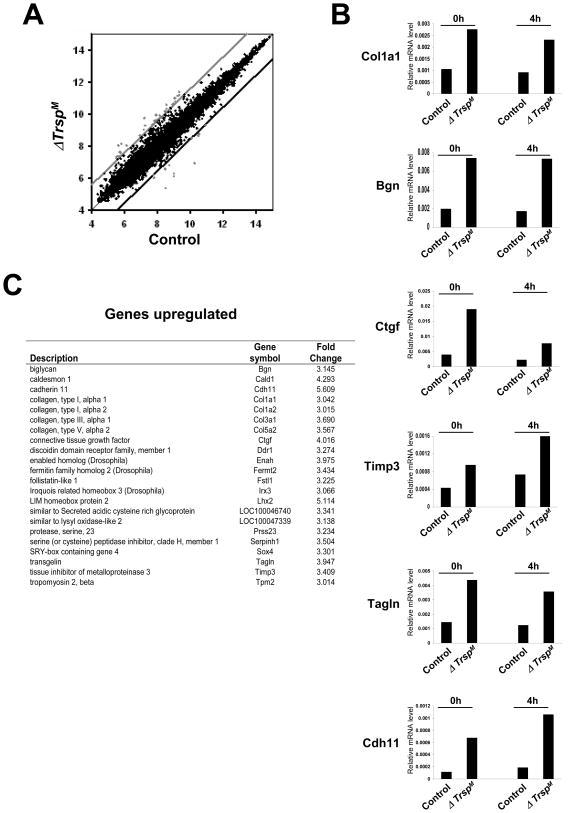
Since there were unexpectedly little or no distinguishable inflammatory phenotypes in ΔTrspM mice, it seemed reasonable that selenoproteins may have other functions in macrophages in addition to roles in inflammatory gene expression. To explore this possibility, an analysis of the global gene expression profiles was carried out in control and ΔTrspM macrophages. We used whole-genome DNA microarrays from mice to assess the expression levels of mRNAs in the two macrophage groups. Those genes that manifested a significantly higher or lower expression in ΔTrspM macrophages compared to control macrophages are shown in Fig. 6A and are designated by those dots (designating mRNAs) that fell outside the lines above and below normal expression levels indicating significant changes.
Fig. 6. Microarray analysis of ΔTrspM macrophages.
In (A), genes that were upregulated and downregulated in ΔTrspM macrophages are shown in the scatter plot. The ordinate and abscissa represent logarithmic values of the signal intensity of individual microarray spots. The upper and lower cut-off lines illustrate the margins of gene expression ratio wherein 3-fold or higher upregulated genes (spots above the light gray line) and 3-fold or lower downregulated genes (spots below the dark black line) are shown as outside the upper and lower cut-off lines, respectively. In (B), those genes whose expression levels were identified in DNA microarray analysis manifesting an alteration greater than 3-fold were validated by real-time PCR. The relative expression of genes in macrophages that were either untreated or treated with LPS is shown. In (C), expression of genes and protein products that were upregulated by selenoproteins is shown. Experimental details of the studies shown in this figure are given in Carlson et al.(22)
The expression levels of those mRNAs that showed significant changes in selenoprotein-deficient macrophages were verified by real-time PCR analysis (Fig. 6B). Interestingly, many genes that deviated significantly in their expression levels from normal in ΔTrspM macrophages were functionally related to the formation, remodeling cellular interaction with the extracellular matrix (ECM). Each of the ECM-related genes that we found to be significantly altered in ΔTrspM macrophages were upregulated compared to the corresponding control cells (Fig. 6C). These included genes that encode ECM components (collagen chains, extracellular proteoglycans, and secreted glycoproteins; Col1a1, Col5a2, Bgn, and Ctgf), inhibitors of ECM proteolysis (metalloendopeptidase and serine-type endopeptidase inhibitors; Timp3, and Serpinh1), and ECM-induced cytoskeletal remodeling (actin binding proteins; Tagln, Enah, and Cald1).
Selenoprotein deficiency and macrophage invasiveness
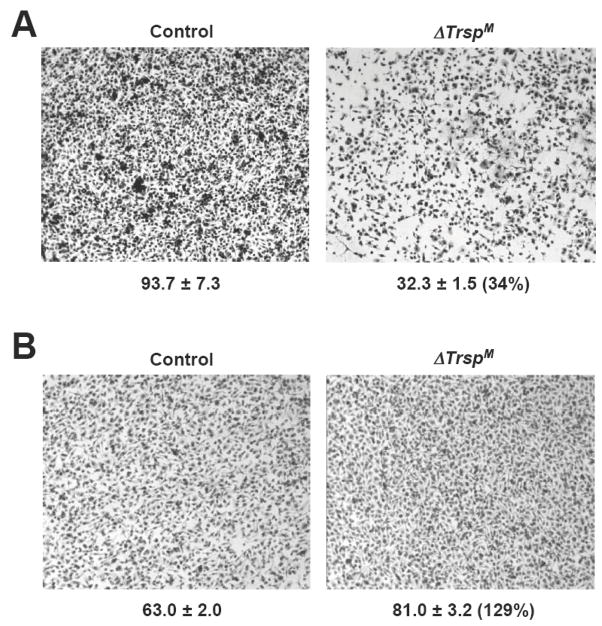
The microarray analysis of ΔTrspM macrophages suggested that macrophage-ECM interactions were altered by the lack of selenoprotein expression in such a manner that matrix remodeling was decreased and the surrounding ECM was reinforced. Such effects might be expected to impair the migration of macrophages through the ECM and basement membrane. To test this possibility, we compared the invasiveness of control and ΔTrspM macrophages in a protein gel matrix. The migration of selenoprotein-deficient macrophages was dramatically reduced in the gel-laden transwell chambers (Fig. 7A). We repeated this assay, but without the gel in the transwell chamber to assess whether the gel invasion phenotype arose from a cell-intrinsic motility defect. Surprisingly, the migration of ΔTrspM macrophages was slightly higher than control macrophages (Fig. 7B) suggesting that the changes observed in ECM-related gene expression in macrophages appeared to decrease the ability of the cell to migrate only in surroundings consisting of ECM components.
Fig. 7. Analysis of ΔTrspM macrophage invasion in a protein gel matrix.
In (A), macrophage migration was examined by a transwell assay with a protein gel matrix (Matrigel) layer in the upper chamber. Numbers below each panel indicate the relative migration of macrophages and they represent the mean ± standard deviation (n=3). Numbers in parenthesis indicate percentage of cell migration in the ΔTrspM sample relative to the control sample. In (B), macrophage migration was examined by a transwell assay in the absence of a Matrigel layer and the data presented as in A. Experimental details of the studies shown in this figure are given in Carlson et al.(22)
Conclusion
Our data show that selenoproteins have an essential role in T cell proliferation in response to TCR stimulation. Although there is substantial evidence that many selenoproteins function as antioxidants(31), the fact that NAC can compensate for the loss of selenoprotein expression in T cells and reverse their defect of TCR-induced proliferation provides further evidence that members of this selenium-containing protein class serve to maintain redox homeostasis. Not unexpectedly, we observed elevated ROS generation in T cells and macrophages. The fact that the ROS dysregulation in ΔTrspM macrophages did not result in altered inflammatory responses suggested that their increased levels and oxidative damage in selenoprotein-deficient cells were below the threshold to elicit effecting inflammatory responses. Selenoproteins were shown to also have a role in macrophage invasiveness.
Since GPx4 is one of the more highly expressed selenoproteins in both T cells and macrophages, we are currently examining the effect of targeting the removal of this essential selenoenzyme(36) in these two cell types. In addition, we are using mouse models to investigate the effects of deficient, adequate, and supplemental levels of selenium intake on the immune response mediated by T cells against infectious disease resistance involving influenza virus (collaboration with M. Beck) that do not express selenoproteins or express only essential, housekeeping selenoproteins, but not non-essential, stress-related selenoproteins(37). This study will also allow us to characterize the critical selenium metabolites (i.e., selenoproteins or low-molecular weight selenocompounds) involved in immune function and examine their roles in resistance to infectious disease. Recent epidemiological and molecular evidence suggests a strong link between metabolic regulatory mechanisms and immune homeostasis(38,39). Our study has just begun to reveal how selenium and selenoproteins function at the crossroads of metabolism and immunity. Future studies will seek to offer deeper insights into the effects of dietary selenium on the onset and progression of immune disorders, and help devise better-informed public health policies.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research, and in addition, a specific grant from the Office of Dietary Supplements, NCI, NIH and the Cutaneous Biology Research Center through the Massachusetts General Hospital/Shiseido Co. Ltd. agreement to JMP and NIH grants awarded to VNG. The authors declare that there are no conflicts of interest. BAC, M-HY, RKS and RI carried out the research and participated in the planning of experiments and the writing of the manuscript, and VNG, DLH and JMP planned the experiments and wrote the manuscript. We thank the Journal of Biological Chemistry for the use of Figures 1–4 and BMC Immunology for the use of Figures 5–7 that were originally published therein.
References
- 1.Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. New York, NY: Springer Science+Business Media, LLC; 2006. [Google Scholar]
- 2.Oldfield JE. Selenium: A historical perspective. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. New York, NY: Springer Science+Business Media, LLC; 2006. pp. 1–6. [Google Scholar]
- 3.Schwarz K, Foltz CM. Selenium as an integral part of factor 3 against dietary necrotic liver degeneration. J Am Chem Soc. 1957;79:3292–3293. [PubMed] [Google Scholar]
- 4.McConnell K. Selenium-75-binding in dog leucocytes. Tex Rep Biol Med. 1959;17:120–122. [PubMed] [Google Scholar]
- 5.Bainbridge DR. Use of (75Se)L-Selenomethionine as a label for lymphoid cells. Immunology. 1976;30:135–144. [PMC free article] [PubMed] [Google Scholar]
- 6.Beckett GJ, Arthur JR, Miller SM, et al. Selenium, immunity and disease. In: Hughes DA, Bendich A, Darlington G, editors. Dietary Enhancement of Human Immune Function. Totowa, NJ: Humana Press; 2003. [Google Scholar]
- 7.Behne D, Wolters W. Distribution of selenium and glutathione peroxidase in the rat. J Nutr. 1983;113:456–461. doi: 10.1093/jn/113.2.456. [DOI] [PubMed] [Google Scholar]
- 8.Bhaskaram P. Micronutrient malnutrition, infection, and immunity: an overview. Nutr Rev. 2002;60:S40–45. doi: 10.1301/00296640260130722. [DOI] [PubMed] [Google Scholar]
- 9.Brown KM, Arthur JR. Selenium, selenoproteins and human health: a review. Public Health Nutr. 2001;4:593–599. doi: 10.1079/phn2001143. [DOI] [PubMed] [Google Scholar]
- 10.Gromer S, Eubel JK, Lee BL, et al. Human selenoproteins at a glance. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiremidjian-Schumacher L, Roy M. Selenium and immune function. Z Ernahrungswiss. 1998;37(Suppl 1):50–56. [PubMed] [Google Scholar]
- 12.McKenzie RC, Arthur JR, Miller SM, et al. Selenium and the immune system. In: Calder PC, Field CJ, Gill NS, editors. Nutrition and Immune Function. Oxford, UK: CAB International; 2002. [Google Scholar]
- 13.McKenzie RC, Rafferty TS, Arthur JR, et al. Effects of selenium on immunity and aging. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. New York, NY: Springer Science+Business Media, LLC; 2006. pp. 311–322. [Google Scholar]
- 14.Spallholz JE. Selenium and glutathione peroxidase: essential nutrient and antioxidant component of the immune system. Adv Exp Med Biol. 1990;262:145–158. doi: 10.1007/978-1-4613-0553-8_12. [DOI] [PubMed] [Google Scholar]
- 15.Turner RJ, Finch JM. Selenium and the immune response. Proc Nutr Soc. 1991;50:275–285. doi: 10.1079/pns19910037. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann PR. Mechanisms by which selenium influences immune responses. Arch Immunol Ther Exp (Warsz) 2007;55:289–297. doi: 10.1007/s00005-007-0036-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann PR, Berry MJ. The influence of selenium on immune responses. Mol Nutr Food Res. 2008;52:1273–1280. doi: 10.1002/mnfr.200700330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arthur JR, McKenzie RC, Beckett GJ. Selenium in the immune system. J Nutr. 2003;133:1457S–1459S. doi: 10.1093/jn/133.5.1457S. [DOI] [PubMed] [Google Scholar]
- 19.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu XM, Carlson BA, Zhang Y, et al. New developments in selenium biochemistry: selenocysteine biosynthesis in eukaryotes and archaea. Biol Trace Elem Res. 2007;119:234–241. doi: 10.1007/s12011-007-8003-9. [DOI] [PubMed] [Google Scholar]
- 21.Shrimali RK, Irons RD, Carlson BA, et al. Selenoproteins mediate T cell immunity through an antioxidant mechanism. J Biol Chem. 2008;283:20181–20185. doi: 10.1074/jbc.M802559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson BA, Yoo MH, Sano Y, et al. Selenoproteins regulate macrophage invasiveness and extracellular matrix-related gene expression. BMC Immuno. 2009;10:57. doi: 10.1186/1471-2172-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T, Kelly VP, Motohashi H, et al. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem. 2008;283:2021–2030. doi: 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- 24.Kryukov GV, Castellano S, Novoselov SV, et al. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann PR, Hoge SC, Li PA, et al. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gladyshev VN, Stadtman TC, Hatfield DL, et al. Levels of major selenoproteins in T cells decrease during HIV infection and low molecular mass selenium compounds increase. Proc Natl Acad Sci U S A. 1999;96:835–839. doi: 10.1073/pnas.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson BA, Novoselov SV, Kumaraswamy E, et al. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 28.Beck MA. Selenium and vitamin E status: impact on viral pathogenicity. J Nutr. 2007;137:1338–1340. doi: 10.1093/jn/137.5.1338. [DOI] [PubMed] [Google Scholar]
- 29.Hurwitz BE, Klaus JR, Llabre MM, et al. Suppression of human immunodeficiency virus type 1 viral load with selenium supplementation: a randomized controlled trial. Arch Intern Med. 2007;167:148–154. doi: 10.1001/archinte.167.2.148. [DOI] [PubMed] [Google Scholar]
- 30.Pages G, Guerin S, Grall D, et al. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 31.Hatfield DL, Carlson BA, Xu XM, et al. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog Nucleic Acid Res Mol Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 32.Hsu HY, Wen MH. Lipopolysaccharide-mediated reactive oxygen species and signal transduction in the regulation of interleukin-1 gene expression. J Biol Chem. 2002;277:22131–22139. doi: 10.1074/jbc.M111883200. [DOI] [PubMed] [Google Scholar]
- 33.Park HS, Jung HY, Park EY, et al. Cutting edge: direct interaction of TLR4 with NAD(P)H oxidase 4 isozyme is essential for lipopolysaccharide-induced production of reactive oxygen species and activation of NF-kappa B. J Immunol. 2004;173:3589–3593. doi: 10.4049/jimmunol.173.6.3589. [DOI] [PubMed] [Google Scholar]
- 34.Brigelius-Flohe R, Friedrichs B, Maurer S, et al. Interleukin-1-induced nuclear factor kappa B activation is inhibited by overexpression of phospholipid hydroperoxide glutathione peroxidase in a human endothelial cell line. Biochem J. 1997;328 (Pt 1):199–203. doi: 10.1042/bj3280199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esworthy RS, Aranda R, Martin MG, et al. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 36.Conrad M, Schneider M, Seiler A, et al. Physiological role of phospholipid hydroperoxide glutathione peroxidase in mammals. Biol Chem. 2007;388:1019–1025. doi: 10.1515/BC.2007.130. [DOI] [PubMed] [Google Scholar]
- 37.Carlson BA, Moustafa ME, Sengupta A, et al. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. 2007;282:32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 38.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finley JW. Bioavailability of selenium in foods. Nutri Rev. 2006;64:146–151. doi: 10.1111/j.1753-4887.2006.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 41.Jackson MI, Combs GF., Jr Selenium and anticarcinogenesis: Underlying mechanisms. Curr Opin Clin Nutr Metab Care. 2008;11:718–726. doi: 10.1097/MCO.0b013e3283139674. [DOI] [PubMed] [Google Scholar]