Abstract
Neuroblasts undergo asymmetric stem cell divisions to generate a series of ganglion mother cells (GMCs). During these divisions, the cell fate determinant Prospero is asymmetrically partitioned to the GMC by Miranda protein, which tethers it to the basal cortex of the dividing neuroblast. Interestingly, prospero mRNA is similarly segregated by the dsRNA binding protein, Staufen. Here we show that Staufen interacts in vivo with a segment of the prospero 3′ UTR. Staufen protein and prospero RNA colocalize to the apical side of the neuroblast at interphase, but move to the basal side during prophase. Both the apical and basal localization of Staufen are abolished by the removal of a conserved domain from the carboxyl terminus of the protein, which interacts in a yeast two-hybrid screen with Miranda protein. Furthermore, Miranda colocalizes with Staufen protein and prospero mRNA during neuroblast divisions, and neither Staufen nor prospero RNA are localized in miranda mutants. Thus Miranda, which localizes Prospero protein, also localizes prospero RNA through its interaction with Staufen protein.
Keywords: RNA localization, asymmetric segregation, Miranda, Staufen, Prospero, nervous system
As the nervous system develops, cellular diversity increases dramatically as thousands of neurons are born, and each takes on its own identity. An efficient method for generating diversity is to ensure that when a cell divides, each of its daughters assumes a distinct identity. This is most simply achieved by the unequal partitioning of cell fate determinants at each cell division. Such a mechanism is used in yeast to differentiate mother cells from their daughters (Bobola et al. 1996; Sil and Herskowitz 1996; Long et al. 1997; Takizawa et al. 1997), in early Caenorhabditis elegans development to distinguish the sister cells arising from the first embryonic divisions (for review, see Nelson and Grindstaff 1997), and in the CNS of both vertebrates (Chenn and McConnell 1995; Zhong et al. 1996) and Drosophila (Rhyu et al. 1994; Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995; Spana et al. 1995).
In the Drosophila embryonic CNS, neural precursors (or neuroblasts) divide in a stem cell lineage, giving rise to a series of smaller daughter cells called ganglion mother cells (GMCs). At least two cell fate determinants, the homeodomain protein Prospero (Doe et al. 1991; Vaessin et al. 1991) and the membrane-associated protein Numb (Uemura et al. 1989; Rhyu et al. 1994), are preferentially segregated to the GMC at cell division. To achieve this, the subcellular distribution of both proteins is regulated during the cell cycle (Rhyu et al. 1994; Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995; Spana et al. 1995). At prophase, Numb and Prospero form a tight crescent on the basal side of the neuroblast such that, as the GMC buds off, the two proteins are asymmetrically segregated to the daughter cell. Numb remains at the cortex in the GMC, whereas Prospero is released and enters the nucleus. Prospero specifies the GMC fate by repressing neuroblast-specific genes and activating GMC-specific genes (Doe et al. 1991; Vaessin et al. 1991; Matsuzaki et al. 1992). Although the role of Numb in directing GMC fates is unclear, it has been shown to specify the fate of one of the two daughters of the MP2 precursor (Spana et al. 1995). Numb segregates to the dMP2 daughter in which it inhibits the Notch signal transduction pathway (Spana and Doe 1996).
Prospero interacts with a protein called Miranda, which anchors it to the cell membrane (Ikeshima-Kataoka et al. 1997; Shen et al. 1997). In the absence of Miranda, Prospero is never localized at the cortex of the neuroblast and, as a result, enters the nucleus in both neuroblasts and GMCs. Miranda also forms a basal crescent in neuroblasts at prophase and is segregated with Prospero and Numb into the GMC at cell division. Once in the GMC, Miranda is rapidly degraded (Ikeshima-Kataoka et al. 1997; Shen et al. 1997) and Prospero is released.
The prospero mRNA is also asymmetrically localized in mitotic neuroblasts, and segregates into the GMC at cell division (Li et al. 1997; Broadus et al. 1998). This process has been shown to require a protein, Staufen, which contains five repeats of a putative dsRNA binding domain (dRBD; St Johnston et al. 1992). Staufen was first identified through its role in establishing the anterior–posterior asymmetry of the Drosophila oocyte (St Johnston 1995). Staufen associates with oskar mRNA to mediate its transport to the posterior of the oocyte, where the mRNA is anchored and translated (Ephrussi et al. 1991; Kim-Ha et al. 1991; St Johnston et al. 1991). Staufen also anchors bicoid mRNA at the anterior of the embryo (Ferrandon et al. 1994). In staufen mutants, the loss of asymmetry causes head defects and elimination of the abdomen (Schüpbach and Wieschaus 1989).
Of the five potential dsRNA binding domains in Staufen, dRBD2 and dRBD5 do not bind dsRNA in vitro and dRBD3 binds, but without sequence specificity (Bycroft et al. 1995; S. Grunert and D. St Johnston, unpubl.). As specific RNA binding has not been demonstrated in vitro by use of the full-length protein, an in vivo RNA injection assay was developed to demonstrate that Staufen mediates the localization of bcd through interaction with its 3′ UTR (Ferrandon et al. 1994). When the bcd 3′ UTR is injected into early embryos, it recruits the endogenous Staufen into ribonucleoprotein particles that localize to the astral microtubules, supporting the observation that RNA transport in the oocyte depends on microtubules (Pokrywka and Stephenson 1991; Clark et al. 1994; Pokrywka and Stephenson 1995).
Although Staufen mediates RNA localization both in the oocyte and in the nervous system, there are several obvious differences between the two processes. RNA localization in neuroblasts appears to depend on actin microfilaments rather than on microtubules (Broadus et al. 1998). Whereas RNA localization is essential in establishing polarity in the oocyte, it appears to be expendable in the nervous system and no obvious phenotype is observed in a staufen mutant (Li et al. 1997; Broadus et al. 1998; A.J. Schuldt, C.M. Davidson, and A.H. Brand, unpubl.). In the oocyte, mRNA translation relies on localization (Kim-Ha et al. 1995; Webster et al. 1997), whereas the translation and distribution of Prospero and Numb proteins is unaffected in staufen mutant embryos (Li et al. 1997; Broadus et al. 1998; A.J. Schuldt, C.M. Davidson, and A.H. Brand, unpubl.). Rather than acting as a primary mechanism for differentiating neuroblast and GMC cell fates, asymmetric RNA localization appears to support protein localization, ensuring that sufficient Prospero is either transported to, or can be translated by, the GMC (Broadus et al. 1998). This may be particularly important as GMCs do not transcribe prospero themselves. In support of this, the alteration in GMC cell fates that occurs when Prospero protein, or its activity, is reduced, is exacerbated by the loss of Staufen (Broadus et al. 1998). The combination of protein and RNA localization ensures the rapid and efficient segregation of Prospero to the GMC.
Staufen is not the only protein involved in RNA localization in the nervous system. Inscuteable, a novel membrane-associated protein with a putative SH3 binding site (Kraut and Campos-Ortega 1996), is required for prospero mRNA to move from the apical side of the neuroblast to the basal side at mitosis (Li et al. 1997). Inscuteable orients the mitotic spindle in neuroblasts and ensures that Numb and Prospero protein crescents localize on the basal side of the cell, rather than randomly (Kraut et al. 1996).
Inscuteable is able to bind to Staufen in a yeast two-hybrid screen, and may anchor Staufen at the apical cortex (Li et al. 1997). However, there is only a partial loss of Staufen localization in the absence of Inscuteable. Furthermore, although both Inscuteable and Staufen are required for the basal localization of prospero mRNA, both proteins were found only on the apical side of the cell (Li et al. 1997). Although there are now conflicting accounts of the subcellular distribution of Staufen (Broadus and Doe 1997; Li et al. 1997; Broadus et al. 1998), it is still unclear what mediates the transition of the prospero mRNA from the apical side of the neuroblast to the basal cortex.
We have investigated the factors required for the subcellular distribution of Staufen and prospero mRNA. We find that Staufen can interact, in vivo, with the prospero 3′ UTR. We have identified the region of Staufen required for its subcellular distribution, and show that this region interacts directly with Miranda. Miranda colocalizes with Staufen protein and prospero mRNA throughout the cell cycle, concentrating first on the apical side of the cell at interphase and then forming a basal crescent at mitosis. Furthermore, Miranda is required in vivo for the correct localization of Staufen protein and prospero mRNA. Thus, Miranda coordinates the subcellular distribution of both the Prospero protein and its mRNA, through a direct interaction with the prospero mRNA-binding protein, Staufen.
Results
Staufen binds to the 3′ UTR of the prospero mRNA in vivo
Previous reports suggested that Staufen can interact directly with the 3′ UTR of prospero mRNA in vitro (Li et al. 1997). However, the fragment of Staufen used in these experiments, dRBD3, can bind in vitro to any double-stranded RNA longer than 11 bp, including adenovirus VA1 RNA and the U1 and U2 snRNAs (St Johnston et al. 1992; S. Grunert and D. St Johnston, unpubl.). Furthermore, as full-length Staufen protein is extremely insoluble, specific RNA binding cannot be assayed in vitro. For these reasons, Ferrandon et al. (1994) developed an in vivo assay for RNA binding, which is based on the observation that Staufen forms ribonucleoprotein particles (RNPs) with RNAs with which it interacts. When bcd mRNA, but not a control dsRNA, is injected into the early embryo, Staufen is specifically recruited into particles at the site of injection.
To assay RNA binding in vivo, we injected the prospero 3′ UTR into embryos expressing a green fluorescent protein (GFP)–Staufen protein fusion and monitored the formation of Staufen RNP particles. The full-length prospero 3′ UTR forms particles (Fig. 1a), as does the bicoid 3′ UTR, but not the coding region of the prospero mRNA, nor VA1 RNA, even though it is able to form an extended secondary structure (data not shown). These RNPs are associated with the nuclei of the precellular embryo, and move with them to the cortex at stage 4 (Fig. 1d). However, unlike the RNP particles formed between Staufen and the bcd 3′ UTR (Ferrandon et al. 1994), the Staufen/prospero 3′ UTR particles do not associate with the astral microtubules (Fig. 1e). Similar results are observed when the prospero 3′ UTR is injected into embryos expressing wild-type Staufen (detected with anti-Staufen antibodies), rather than a GFP fusion, whereas VA1 RNA does not recruit Staufen (A.J. Schuldt and A.H. Brand, unpubl.).
Figure 1.
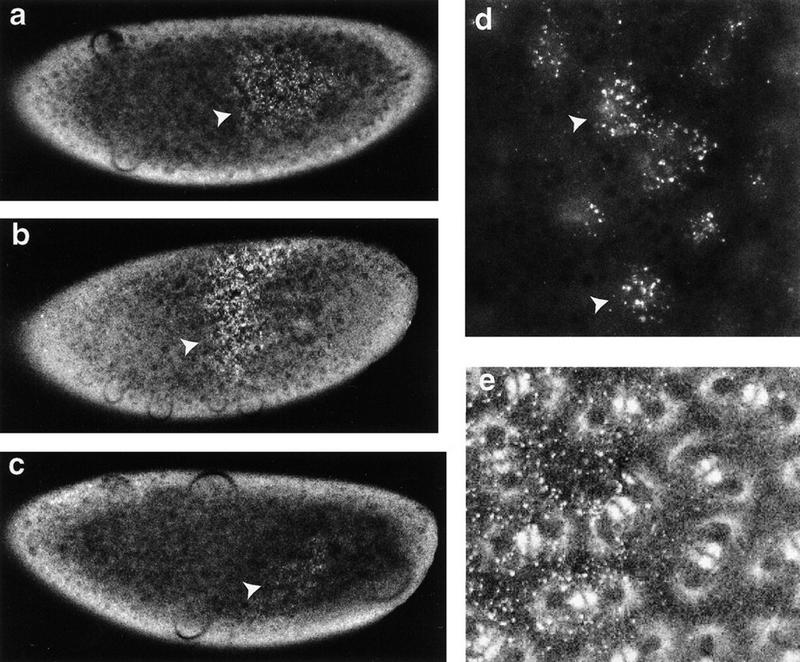
Staufen binds to the prospero 3′ UTR in vivo. Injection of the bcd 3′ UTR into the precellularization embryo has been shown to recruit Staufen into ribonucleoprotein particles at the site of injection (Ferrandon et al. 1994). We tested whether the prospero 3′ UTR also interacts with Staufen in this assay. In vitro-transcribed RNA was injected into living embryos expressing two different GFP fusion proteins: GFP–Staufen and Tau–GFP (Brand 1995; Micklem et al. 1997). (a) Full-length prospero 3′ UTR RNA (1.5 kb) recruits Staufen to the site of injection within 5–10 min (arrowhead). (b) A similar result is obtained with a 650-nucleotide segment from the 3′ end of the prospero 3′ UTR (arrowhead). (c) Conversely, an 800-nucleotide fragment from the 5′ end of the prospero 3′ UTR recruits almost no Staufen to the site of injection, even after a 20–30 min incubation (arrowhead). (d) The ribonucleoprotein particles containing GFP–Staufen and the prospero 3′ UTR cluster on the basal side of the nuclei and migrate with them as they move to the cortex of the embryo (arrowheads; 650-nucleotide fragment of the prospero 3′ UTR was injected). (e) Although the RNP particles are distributed over the nuclei, there is no apparent association with astral microtubules (labeled with Tau–GFP).
To further map the region of the prospero 3′ UTR with which Staufen interacts, we injected either the 3′ half of the UTR (Fig. 1b), or the 5′ half into embryos (Fig. 1c). Whereas the 3′ segment recruits Staufen into RNPs within 5–10 min of injection, the 5′ segment does so only slightly, if at all, after 20–30 min. Therefore, the region of the prospero mRNA recognized by Staufen lies in the terminal 650 bases of the mRNA.
Staufen colocalizes with Prospero throughout the cell cycle
Previous reports describing the subcellular distribution of Staufen have been conflicting. Li et al. (1997) reported that Staufen remains on the apical side of the neuroblast throughout the cell cycle, suggesting that it cannot directly mediate formation of the basal crescent of prospero mRNA. Conversely, Staufen is only observed on the basal side of neuroblasts cultured in vitro (Broadus and Doe 1997). Our results concur with those recently reported by Broadus et al. (1998) and demonstrate that Staufen colocalizes with Prospero protein at all stages of the cell cycle. In embryos, we observe that Staufen is concentrated on the apical side of the neuroblast at interphase, then forms a crescent on the basal side of the cell in prophase, where it remains through mitosis before partitioning to the GMC at division (Fig. 2a,c,e,g). A similar subcellular distribution is seen in living embryos (A.H. Brand, unpubl.). This dynamic pattern of localization shows Staufen to be correctly placed to bind the prospero mRNA throughout the cell cycle, and to mediate its segregation into the GMC (see Fig. 6a,b, below).
Figure 2.
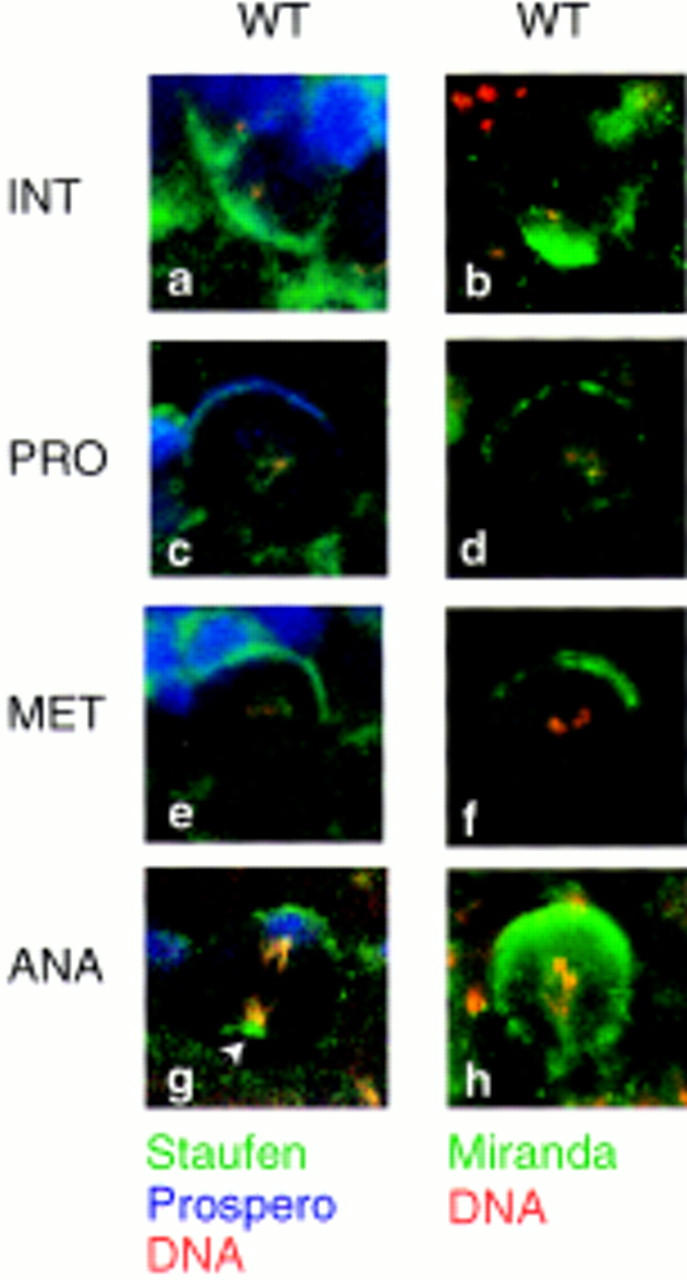
Staufen, Prospero, and Miranda colocalize throughout the cell cycle. Staufen (in green; a,c,e,g), Prospero (in blue; a,c,e,g), and Miranda (in green; b,d,f,h) colocalize in neuroblasts. At interphase, all three proteins are found on the apical side of the cell (a,b); at prophase, they form a crescent at the basal cortex (c,d), which is maintained at metaphase (e,f) and anaphase (g,h) before segregation into the GMC. Interestingly, Staufen is often also associated with the centrosome on the apical side of the neuroblast at anaphase (arrowhead in g), as is Miranda (see Fig. 5j). DNA is labeled in red in all panels; basal is up, apical down.
Figure 6.
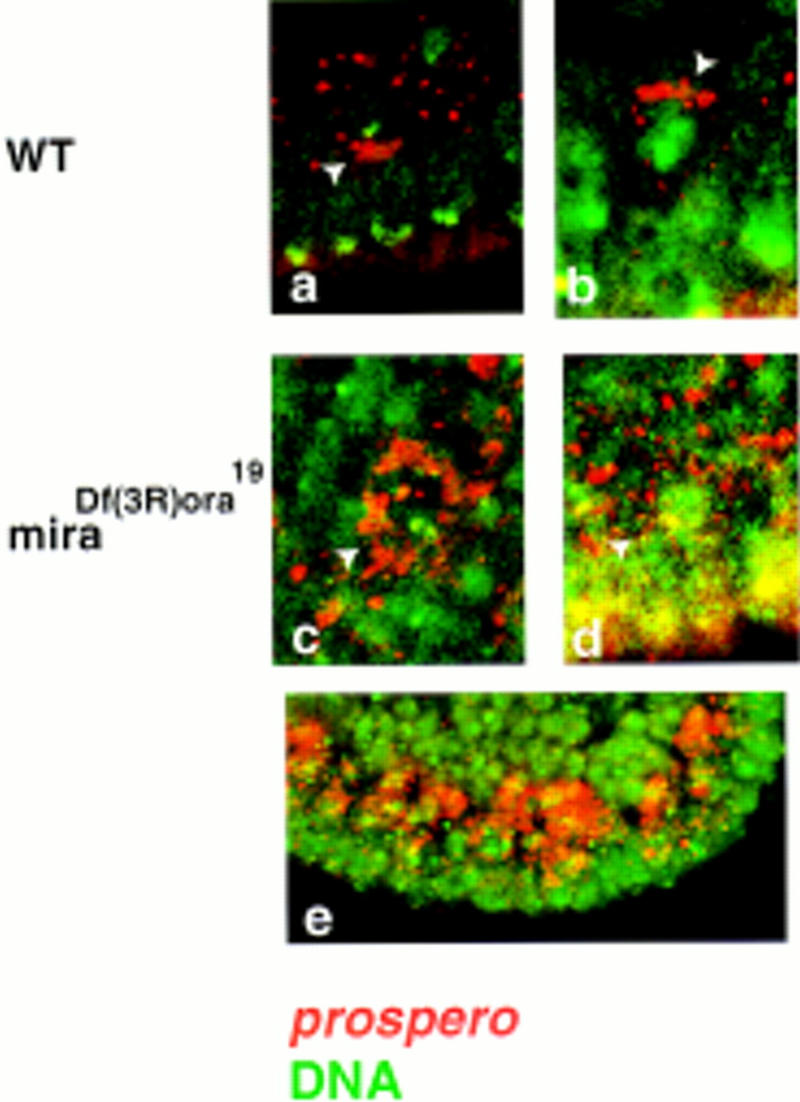
Miranda is required to localize prospero mRNA. prospero mRNA localizes on the apical side of neuroblasts at interphase (arrow in a) and moves to the basal cortex during mitosis (arrowhead in b). In the absence of Miranda, the RNA is evenly distributed in all neuroblasts (c,d,e). The arrowheads in c and d indicate individual neuroblasts. prospero mRNA is labeled in red, DNA in green.
Staufen localization requires dRBD5
Experiments in the oocyte have identified two regions of Staufen that are required for its function during oogenesis: a 99-amino-acid region in the middle of dRBD2, and the carboxy-terminal 157 amino acids that include dRBD5 (Micklem 1998). As neither the dRBD2 insert nor the carboxy-terminal domain binds dsRNA in vitro (S. Grunert and D. St Johnston, unpubl.), these two regions may be involved in some other aspect of Staufen function. When Staufen protein that lacks either the dRBD2 insert or the carboxy-terminal domain is expressed maternally, it can only partially rescue the abdominal defects caused by Staufen null mutations (Micklem 1998; C.M. Davidson and A.H. Brand, unpubl.).
To test whether either of these domains is required for Staufen localization in neuroblasts, we assayed the subcellular distribution of Staufen mutants that lack the dRBD2 insert (ΔdRBD2) or the carboxy-terminal domain of Staufen (ΔdRBD5). The removal of the dRBD2 insert has no effect on Staufen distribution in neuroblasts at any stage of the cell cycle (Fig. 3a,c,e,g). However, the loss of the carboxy-terminal 157 amino acids of Staufen eliminates both apical and basal localization. StaufenΔdRBD5 is distributed throughout the cytoplasm from interphase through mitosis (Figure 3b,d,f,h). Therefore, the carboxyl terminus of Staufen is necessary to direct asymmetric distribution of the protein in neuroblasts. If the normal subcellular distribution of Staufen is mediated by a specific protein–protein interaction, then the site of the interaction may reside within the 157-amino-acid domain removed in StaufenΔdRBD5.
Figure 3.
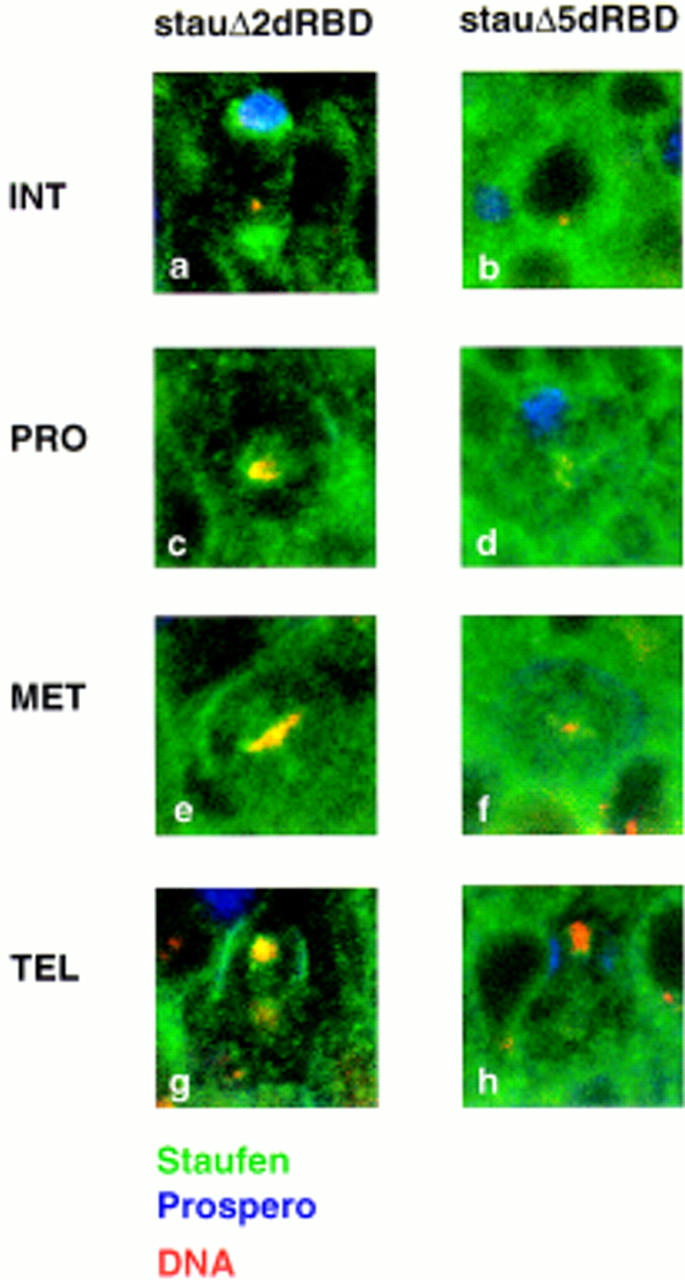
Deletion of dRBD5 eliminates Staufen localization. Deletion of the loop in dRBD2 of Staufen has little effect on protein localization throughout the cell cycle (a,c,e,g). At interphase, StaufenΔdRBD2 concentrates on the apical side of the cell (a) before moving to the basal cortex during mitosis (c,e,g). Deletion of dRBD5, however, prevents Staufen localization to either the apical side of the neuroblast at interphase (b), or the basal side at prophase (d), metaphase (f), or anaphase (h). Staufen is labeled in green, Prospero in blue, and DNA in red in all panels; basal is up, apical down.
Staufen binds directly to Miranda through dRBD5
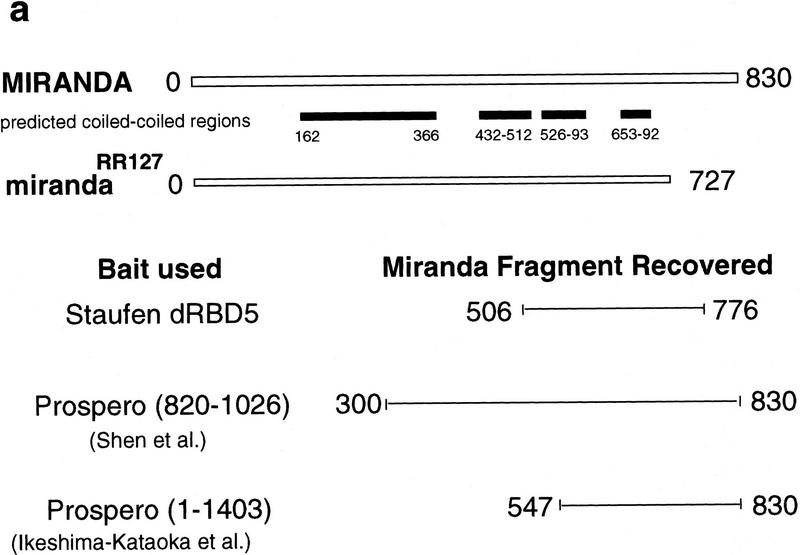
Staufen dRBD5 does not interact with RNA in vitro (S. Grunert and D. St Johnston, unpubl.) but is required in vivo for Staufen protein mRNA localization, suggesting that it may interact with other proteins that anchor Staufen at the apical and basal cortex, or mediate its transport from one side of the neuroblast to the other. To identify proteins that might interact with dRBD5 to direct Staufen crescent formation, we carried out a yeast two-hybrid screen on a random-primed embryonic cDNA library using a LexA–dRBD5 fusion protein as bait (Fields and Song 1989; Bartel et al. 1993; Poortinga et al. 1998). From 4,000,000 transformants, we isolated 10 positive clones, and were able to recover the library plasmid from 6 of these. All six clones contained the same insert, which encodes amino acids 506–776 of the Miranda protein.
To test the specificity of the dRBD5/Miranda interaction, we retransformed the Miranda clone into yeast that contained either the original bait (lexA–dRBD5) or the control baits (lexA–lamin or lexA–BRCA2). Only yeast containing both lexA–dRBD5 and Miranda–VP16 express high levels of β-galactosidase (Fig. 4b). Furthermore, this region of Miranda does not interact with all dRBDs, because no β-galactosidase activity is observed when Miranda–VP16 is cotransformed with LexA–Staufen dRBD1 (Fig. 4b). Thus, Miranda binds specifically to Staufen dRBD5, and does not interact with a closely related domain from the same protein.
Figure 4.
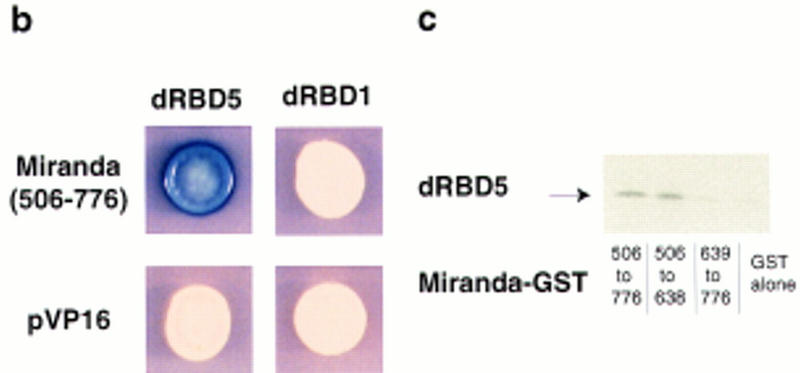
Staufen interacts with Miranda. (a) miranda encodes a protein of 830 amino acids (open rectangle in a, marked 0–830 amino acids), within which are four potential coiled–coil domains (solid boxes). The carboxy-terminal 103 amino acids are deleted in mirandaRR127 (open rectangle, 0–727 amino acids). By use of Staufen dRBD5 as bait in a yeast two-hybrid screen, we isolated six clones, all of which encode amino acids 506–776 of Miranda (black line). When amino acids 820–1026 of Prospero were used as bait in a similar screen, Shen et al. (1997) isolated amino acids 300–830 of Miranda. Ikeshima-Kataoka et al. (1997) used amino acids 1–1403 of Prospero as bait and recovered a fragment of Miranda encoded by amino acids 547–830. (b) Only yeast expressing both Staufen dRBD5 and Miranda amino acids 506–776 activate expression of lacZ (blue colony). Coexpression of either Staufen dRBD1 with Miranda, Staufen dRBD5 with the pVP16 vector, or Staufen dRBD1 with the pVP16 vector, does not activate transcription (white colonies). (c) 35S-Labeled Staufen dRBD5 interacts in vitro with GST–Miranda amino acids 506–776 (first lane) and GST–Miranda amino acids 506–638 (second lane) but not with GST–Miranda amino acids 639–776 (third lane) or GST alone (fourth lane).
To more precisely map the region of Miranda protein that interacts with Staufen-dRBD5, we divided the fragment of Miranda identified in the yeast two-hybrid screen into two parts (amino acids 506–638 and amino acids 639–776), and examined their interaction with dRBD5 in a GST pull-down assay (Hagemeier et al. 1991). 35S-Labeled Staufen–dRBD5 coprecipitates with both the full-length fragment, GST–Miranda amino acids 506–776, and the amino-terminal segment of this region, GST–Miranda amino acids 506–638, but shows no interaction above background with the carboxy-terminal segment, GST–Miranda amino acids 639–776 (Fig. 4c). This suggests that the Staufen binding site in Miranda corresponds to the predicted coiled–coil domain that extends from amino acids 526–593.
Miranda colocalizes with Staufen throughout the cell cycle
We have shown that Miranda and Staufen can interact in a yeast two-hybrid screen and in vitro. If the two proteins also interact in Drosophila embryos, they should colocalize in neuroblasts at specific stages of the cell cycle. Miranda has been reported to be evenly distributed throughout the cytoplasm and around the cell cortex at interphase, and to form a basal crescent during mitosis (Ikeshima-Kataoka et al. 1997; Shen et al. 1997). Therefore, Staufen might bind to one protein on the apical side of the neuroblast, for example, Inscuteable, and to a second on the basal side of the cell, Miranda. However, when we assay the subcellular distribution of Miranda we find that, like Staufen, Miranda concentrates predominantly on the apical side of the cell at interphase (Fig. 2b). Interestingly, miranda mRNA is also localized predominantly on the apical side of the neuroblast (Fig. 5m). Miranda protein then forms a crescent on the basal side of the neuroblast at prophase, where it remains until after cell division (Fig. 2d,f,h; Ikeshima-Kataoka et al. 1997; Shen et al. 1997). Therefore, the subcellular distribution of Miranda suggests that it might interact with Staufen at all stages of the cell cycle.
Figure 5.
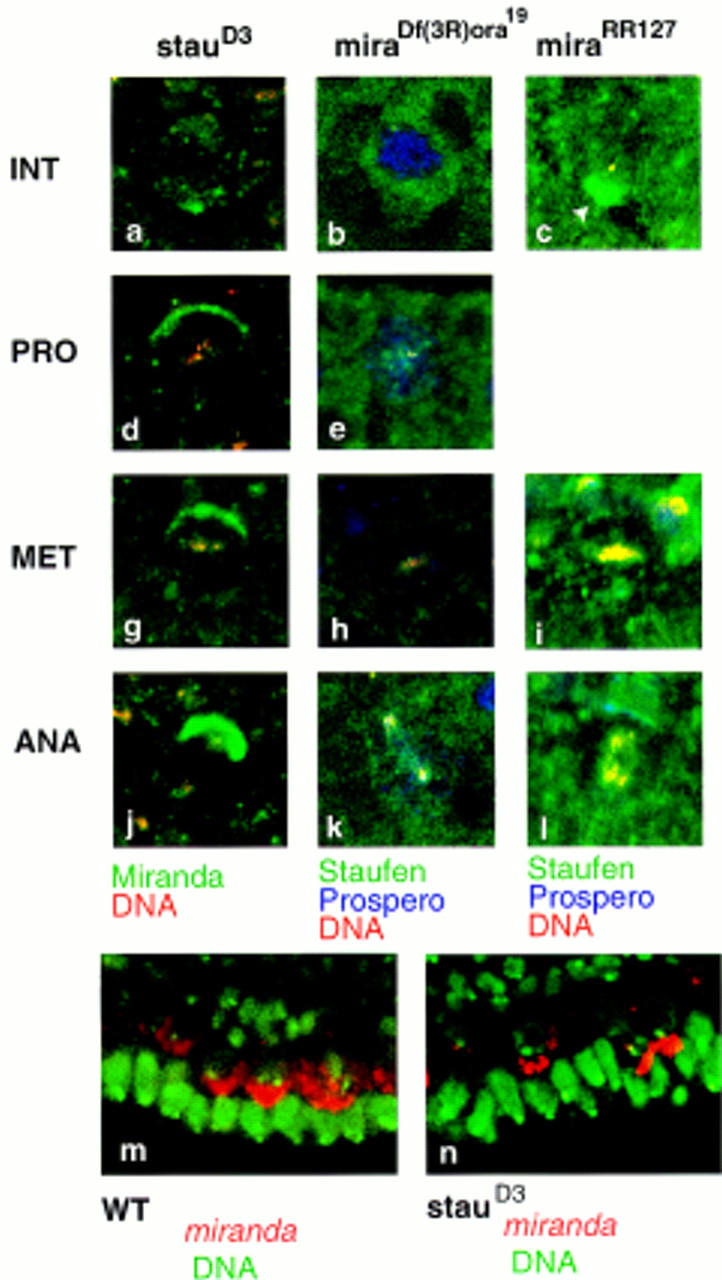
The subcellular localization of Staufen requires Miranda. Miranda localizes normally throughout the cell cycle in staufenD3 mutants (a,d,g,j; Miranda labeled in green, DNA in red). Staufen subcellular localization, however, is absolutely dependent on Miranda (b,e,h,k; Staufen labeled in green, Prospero in blue, DNA in red). In mirandaRR127, Staufen is appropriately localized, but concentrates more strongly at the apical side of the neuroblast at interphase (arrowhead in c; Staufen labeled in green, Prospero in blue, DNA in red), and forms a weaker basal crescent at mitosis (i,l). miranda mRNA (labeled in red, DNA in green) localizes predominantly on the apical side of the neuroblast throughout the cell cycle in both wild-type (m) and staufenD3 embryos (n).
The subcellular distribution of Staufen requires Miranda
Miranda interacts with both Prospero and Numb in vitro, but is required to tether only Prospero to the cell membrane in vivo (Ikeshima-Kataoka et al. 1997; Shen et al. 1997). Numb is itself a membrane-associated protein and although it may form a complex with Miranda, the interaction is not necessary to maintain Numb at the cortex. Prospero, in contrast, is a homeodomain protein that moves into the nucleus unless tethered by Miranda at the membrane. Staufen is not itself membrane associated, suggesting that it too may be tethered to the cortex by a specific protein–protein interaction. As Staufen binds to Miranda in a yeast two-hybrid screen, and the two proteins colocalize in neuroblasts, it seemed possible that Miranda might anchor Staufen to the cortex.
To test if Miranda is required to localize Staufen in vivo, we followed the subcellular distribution of Staufen in miranda null mutant embryos [Df(3R)ora19]. In the absence of Miranda, Staufen is found throughout the neuroblast cytoplasm at all stages of the cell cycle (Fig. 5b,e,h,k). Similar results are seen in an EMSinduced allele, mirandaYY227 (data not shown). Prospero protein is also evenly distributed (Fig. 5b,e,h,k). We assayed the distribution of Miranda in a staufen mutant, to determine whether the localization of the two proteins is mutually dependent (Fig. 5a,d,g,j). Miranda localizes normally in the absence of Staufen, suggesting that it acts upstream of both Staufen and Prospero in the process of asymmetric protein localization. miranda mRNA also remains apical in staufen mutant embryos (Fig. 3n).
Although Prospero is never localized at the cortex in most miranda mutants, one mutant, mirandaRR127, does localize Prospero but fails to release it from the cortex even after the two proteins are partitioned to the GMC (Ikeshima-Kataoka et al. 1997). The mutation removes the carboxy-terminal 103 amino acids of Miranda, within which lie sites for phosphorylation by protein kinase C (PKC). If phosphorylation regulates the release of Prospero from Miranda, then the Staufen–Miranda interaction might be similarly regulated.
We assayed the subcellular distribution of Staufen in mirandaRR127 mutant embryos. In these embryos, Prospero remains at the cortex and cannot enter the GMC nucleus. We cannot determine whether Staufen localization in the GMC is affected, because Staufen is normally cytoplasmic or cortical, but we do observe an increased accumulation of Staufen at the apical side of the neuroblast at interphase (Fig. 5c). At metaphase, less Staufen is concentrated in a basal crescent (cf. Fig. 5i with Fig. 2e). Therefore, Miranda may anchor Staufen at the apical side of the cell, and regulate the release of both proteins to allow formation of a basal crescent at prophase.
prospero mRNA localization requires Miranda
We have shown that Miranda mediates both the apical and basal localization of Staufen and may regulate its transition from apical to basal during the cell cycle. If prospero mRNA localization is Staufen dependent then, by extension, prospero mRNA should be mislocalized in the absence of Miranda. As predicted, prospero mRNA is present throughout the cytoplasm in miranda mutant embryos (Fig. 6c–e). Miranda, therefore, mediates the asymmetric distribution not only of the Prospero protein, but also of its mRNA.
Discussion
Miranda colocalizes with Staufen and prospero mRNA throughout the cell cycle
Here we demonstrate that the subcellular distribution of Miranda and Staufen is sufficient to explain all phases of prospero mRNA localization in neuroblasts. First, Staufen interacts in vivo with the prospero mRNA through a 650-nucleotide region of the 3′ UTR. Second, Staufen colocalizes with prospero mRNA in neuroblasts throughout the cell cycle. Staufen first concentrates on the apical side of the neuroblast at interphase, then moves to the basal side of the cell at prophase, and segregates preferentially into the GMC at cytokinesis. Third, both the apical and basal localization of Staufen are eliminated when dRBD5 is deleted. This region is sufficient to bind to Miranda in a yeast two-hybrid screen and in a GST pull-down assay. Fourth, we show that Miranda colocalizes with Staufen protein and prospero mRNA throughout cell cycle, concentrating first on the apical side of the cell and then forming a basal crescent at prophase. Finally, Miranda is required in vivo to localize Staufen, both apically and basally, and for prospero mRNA localization. Thus, Miranda, which localizes Prospero protein, also localizes prospero mRNA through its interaction with the RNA binding protein, Staufen (Fig. 7).
Figure 7.
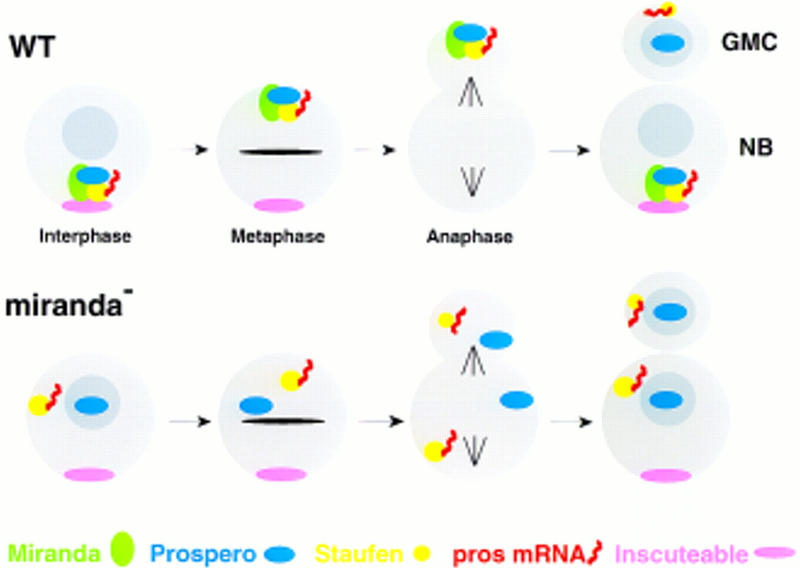
Miranda mediates the asymmetric segregation of Prospero, Staufen, and prospero mRNA. (Top) In wild-type neuroblasts, Miranda forms a complex with Prospero protein, Staufen and prospero mRNA, on the apical side of the cell at interphase. The complex moves to the basal side of the cell during mitosis, where it is anchored at the cortex prior to segregation into the GMC. Miranda is then degraded and Prospero enters the GMC nucleus. (Bottom) In the absence of Miranda, Staufen and prospero mRNA are randomly distributed in the neuroblast cytoplasm at interphase, whereas Prospero enters the nucleus. During mitosis, all three factors are evenly partitioned between the neuroblast and the budding GMC. After cytokinesis, Prospero enters the nucleus of both the neuroblast and the GMC, whereas Staufen and prospero mRNA remain randomly distributed in the cytoplasm of both cells.
Miranda and Staufen interact in vivo
If RNA localization is an integral part of the process of asymmetric protein segregation, then something must coordinate the two processes. Here we show that Miranda performs this function. Miranda is a novel protein containing four coiled–coil structures, two leucine zippers, four putative destruction boxes, and eight potential PKC phosphorylation sites (Ikeshima-Kataoka et al. 1997; Shen et al. 1997). The carboxy-terminal 103 amino acids, which encode the PKC phosphorylation sites and are removed in mirandaRR127, regulate the release of Prospero from the cortex. The cell cycle control of Prospero release, and Miranda turnover, may, therefore, be regulated by phosphorylation.
We show that Staufen binds to amino acids 506–638 of Miranda, and that Staufen is still localized in mirandaRR127 mutants. The Staufen binding site is contained within a domain of Miranda that has been shown also to interact with Prospero (amino acids 445–727; Ikeshima-Kataoka et al. 1997). This region encodes the third and fourth coiled–coil domains (Fig. 4a). Staufen binds to the 132 amino acids between 506–638, which encode the third coiled–coil domain.
Staufen dRBD5 is an unusual example of the dRBD protein motif, which contains the conserved amino acids that form the hydrophobic core of the domain, but lacks the basic amino acids required for RNA binding (St Johnston et al. 1992; Bycroft et al. 1995). Because dRBD5 does not bind dsRNA in vitro (S. Grunert, D.R. Micklem, and D. St Johnston, unpubl.), its conservation suggests that it might be involved in some other aspect of Staufen function. We show that the carboxy-terminal 157 amino acids of Staufen mediate its interaction with Miranda. The removal of this domain, in StaufenΔdRBD5, eliminates all aspects of Staufen subcellular localization. The only conserved region within this 157 amino acids is dRBD5. The 78 amino acids encompassing dRBD5 are sufficient to bind to Miranda in a yeast two-hybrid screen and in vitro, further defining the interaction domain.
Miranda coordinates the asymmetric distribution of Prospero and its mRNA
Miranda has been shown to anchor Prospero protein to the basal cortex of the neuroblast, and to regulate its release from the cortex once segregated to the GMC. Here we demonstrate that Miranda also directs the subcellular distribution of Staufen and, in so doing, localizes the prospero mRNA. In the absence of Miranda, Prospero enters the neuroblast nucleus, and Staufen and prospero mRNA are evenly distributed in the cytoplasm.
Miranda binds to Prospero protein and to Staufen, which in turn binds prospero mRNA, to form a complex on the apical side of the neuroblast. The complex may be anchored by Inscuteable at interphase, and then released as the cell cycle progresses. In mirandaRR127, Staufen accumulates on the apical side of the cell, suggesting that Miranda may regulate release from the apical cortex. Miranda, Prospero, Staufen, and prospero mRNA then move as a group to the basal side of the cell during mitosis, a process that appears to require actin microfilaments. Staufen and Miranda also associate with the apical centrosome, although the importance of this interaction is unclear. Once at the basal cortex, the complex is anchored by factors that have not, as yet, been identified. However, as Miranda acts as the adapter between protein and RNA localization, these factors may be isolated in screens for other Miranda binding proteins.
After cytokinesis, Miranda is rapidly degraded in the GMC, and Prospero is released and enters the nucleus. It may be important, therefore, to minimize translation of new Miranda protein in the GMC. Whereas prospero mRNA is specifically segregated to the GMC, miranda mRNA remains tightly anchored on the apical side of the neuroblast. By tethering miranda mRNA in this way, Miranda protein, but not miranda mRNA, is partitioned to the GMC at cell division.
Several interesting questions remain to be answered. What regulates the release of Miranda from the apical side of the cell? How are Miranda, Prospero, Staufen, and prospero mRNA transported to the basal side of the neuroblast? Do they move as a complex, and how are they anchored at the basal cortex? Prospero and Staufen bind to the same region of Miranda, but it is not known whether they bind to the same molecule simultaneously. The answers to these questions may help to elucidate the mechanism of asymmetric protein and RNA localization not only in the nervous system, but also in other tissues, and in other organisms.
Materials and methods
Drosophila mutants and transgenic lines
Flies carrying deficiency Df(3R)ora19, which deletes miranda (Shen et al. 1997), were a kind gift of Yuh Nung Jan (University of California, San Francisco). The mirandaRR127 and mirandaYY227 mutants (Ikeshima-Kataoka et al. 1997) were kindly provided by Fumio Matsuzaki (National Institute of Neuroscience, Tokyo, Japan). staufenD3 is a protein null (St Johnston et al. 1991).
Expression of Myc-tagged derivatives of Staufen was driven in the germ line of homozygous staufenD3 females from the α4 tubulin promoter. The Staufen fusion proteins consist of the first 9 amino acids of α4 tubulin, a 16-amino-acid myc epitope, followed by amino acids 18–1026 of Staufen. To express wild-type myc-tagged Staufen, an FspI–ClaI fragment (324–1140, numbered as in GenBank M69111) from staufen cDNA E3 (St Johnston et al. 1991) was cloned into the BamHI site of D277 (a modified pCaTub67MatpolyA vector; Micklem et al. 1997) to generate D287. The CelII–NotI fragment of cDNA E3 (483–end) was then inserted between the CelII site and the NotI site in the polylinker to produce D288.
The StaufenΔdRBD5 derivative was generated in a similar fashion, but lacks the carboxy-terminal 157 amino acids of Staufen, downstream of the PvuII site at position 2879. StaufenΔdRBD2 is identical to D288, except that the 99-amino-acid insertion in the middle of dRBD2 has been replaced with the 8-amino-acid loop that occurs in the equivalent position in dRBD3. Further information on the sequence of these constructs is available on request. DNA constructs were introduced into the germ line of w; stauD3, sp/CyO flies by P-element-mediated transfomation (Rubin and Spradling 1982). Multiple transformant lines were obtained for each construct and lines giving the highest levels of expression were analyzed further.
Expression of GFP–Staufen was driven from the α4 tubulin promoter in vector D277M. We used the GFP variant, mGFP6, to enhance the fluorescence and solubility of the fusion protein. Staufen fusions with other GFP variants give no fluorescence, possibly because of the production of an insoluble fusion protein [data not shown; see Siemering et al. (1996) for a discussion of solubility and fluorescence]. mGFP6 carries mutations that improve the maturation and spectral properties of the protein, F64L, S65T (Heim et al. 1995; Cormack et al. 1996) and V163A, I167T, S175G (Heim et al. 1994; Siemering et al. 1996), and codon usage changes that remove a cryptic intron and optimize expression (Haseloff et al. 1997; data not shown). The sequence of mGFP6 is available on request. mGFP6 was PCR amplified as a PstI–BglII fragment and cloned into D277M, replacing the myc epitope tag. Staufen was then cloned as a BglII–NotI fragment from D288 downstream of mGFP6. The transgenic line used in this study, α4 tubulin–mGP6–Staufen2.1, is an insert on 3R that rescues the maternal effect lethality of a staufen null mutation.
RNA injection
prospero RNA was transcribed with an Ambion transcription kit. RNA was injected at a concentration of 1 μg/ml (in water) into 0–1 hr embryos (Ferrandon et al. 1994) expressing a GFP–Staufen fusion protein, driven by the α4 tubulin promoter (described above), and a Tau–GFP fusion protein (Brand 1995), also driven by the α4 tubulin promoter (Micklem et al. 1997). Time lapse images of injected embryos were collected by confocal microscopy with a BioRad MRC1024 and a Nikon E800 microscope. Images were imported into Adobe Photoshop 4.0, and assembled in Adobe Illustrator 6.0.
Immunohistochemistry
Antibody staining was carried out according to Patel (1994) with the following modifications: To preserve the cytoskeleton, embryos were fixed for 5 min in a 1:1 mix of undiluted (37%) formaldehyde and heptane. PBS, 0.1% Triton X-100 replaces PEM (100 mm PIPES, 2 mm EGTA, 1 mm MgSO4) throughout. Rabbit anti-Staufen (St Johnston et al. 1991) was used at a dilution of 1:500; mouse anti-Prospero mAb MR2A (Spana and Doe 1995; a kind gift from Chris Doe) was used at a dilution of 1:2; rabbit anti-Miranda A96c (Shen et al. 1997; a kind gift from Yuh Nung Jan) was used at 1:1000. Secondary antibodies, directly conjugated to FITC, Texas Red, or Cy5 were used at a dilution of 1:200. Embryos were mounted in DNA stain solution (Lundell and Hirsh 1994; sonicated 2 mg/ml of 1,4-phenylenediamine in 4 mm Na2CO3, 90% glycerol) and visualized by confocal microscopy, as above.
In situ hybridization
A 1.5-kb BamHI fragment of the prospero cDNA (a kind gift from W. Chia, Institute of Molecular and Cell Biology, National University of Singapore) was used as a template to generate a random primed, digoxigenin-labeled, DNA probe (Boehringer Mannheim). In situ hybridization to whole mount embryos was carried out as described previously (Tautz and Pfeifle 1989), with embryos incubated overnight in a hybridization mix at 48°C. The embryos were washed, then incubated with alkaline phosphatase-conjugated anti-digoxigenin antibodies. The alkaline phosphatase reaction was carried out by use of the fluorescent substrate, HNPP/Fast Red TR (Boehringer Mannheim) as described by Goto and Hayashi (1997). Embryos were mounted in DNA stain solution and visualized by confocal microscopy, as above.
Yeast two-hybrid screen and GST pull downs
To construct the LexA-dRBD5 bait, the carboxy-terminal 78 amino acids of Staufen (bases 3116–3355) were amplified by the primers GGAATTCGCTGGAGTGCACATGAAGGAGCA and CGGGATCCTTACCCCAGCTTGCTGAGGAT, and cloned between the EcoRI and BamHI sites of PBTM116-nls (Bartel et al. 1993). The resulting plasmid was transformed into yeast strain L40 (Hollenberg et al. 1995). The same yeast were then transformed with a random-primed cDNA library in pVP16 made from 0- to 4-hr embryonic mRNA (a generous gift of Gretchen Poortinga and Susan Parkhurst; Poortinga et al. 1998). Of ∼4,000,000 transformants, 138 colonies grew on His− plates. Ten of these His+ colonies also showed β-galactosidase activity. cDNA library plasmids were successfully recovered from six of these. The specificity of the interaction between these positive clones and lexA–dRBD5 was examined further by testing for their ability to activate his3 and lacZ expression when cotransformed with the original bait. LexA–Lamin, LexA–BRCA2, and LexA–Staufen dRBD1 served as negative controls.
For GST pull down assays, three fragments of Miranda (amino acids 506–776, amino acids 506–638, or amino acids 639–776) were cloned in-frame with GST in pGEX2T (Pharmacia), and purified after expression in Escherichia coli C41 (Bannister and Kouzarides 1996; Miroux and Walker 1996). Staufen dRBD5 (amino acids 937–1026) was cloned into pING14 (Taunton et al. 1996; Brehm et al. 1998). In vitro translated [35S]methionine-dRBD5 was used in GST pull down assays, as described by Hagemeier et al. (1991). Briefly, 500 ng of GST fusion protein on beads was incubated with 2–5 ml of dRBD5 in Z buffer. The beads were then washed three times in NETN buffer and bound protein was resolved by SDS-PAGE.
Acknowledgments
We thank Fumio Matsuzaki for communicating results prior to publication. We are grateful to Gretchen Poortinga and Susan Parkhurst for kindly providing the yeast two-hybrid library prior to publication. For generously providing DNA constructs, antibodies, and Drosophila lines, we thank Bill Chia, Chris Doe, Yuh Nung Jan, and Fumio Matsuzaki. We thank Chris Phelps for comments on the manuscript. A.J.S. is supported by a BBSRC special studentship; J.A. is funded by a studentship from the Boehringer Ingelheim Fonds; J.H. is supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC). This work was funded by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ahb@mole.bio.cam.ac.uk; FAX 44-1223-334089.
References
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bartel PL, Chien C, Sternglanz R, Fields S. Using the two hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular interactions in development: A practical approach. Oxford, UK: IRL Press; 1993. pp. 153–179. [Google Scholar]
- Bobola N, Jansen R-P, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Brand A. GFP in Drosophila. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister A, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Broadus J, Doe CQ. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric localization in Drosophila neuroblasts. Curr Biol. 1997;7:827–835. doi: 10.1016/s0960-9822(06)00370-8. [DOI] [PubMed] [Google Scholar]
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- Bycroft M, Grünert S, Murzin AG, Proctor M, St Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila Staufen protein reveals homology to the N terminal domain of ribosomal protein S5. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Clark I, Giniger E, Ruohola-Baker H, Jan LY, Jan YN. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr Biol. 1994;4:289–300. doi: 10.1016/s0960-9822(00)00068-3. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. Facs optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–465. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nüsslein-Volhard C, St Johnston D. Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Goto S, Hayashi S. Cell migration within the embryonic limb primordium of Drosophila as revealed by a novel fluorescence method to visualize mRNA and protein. Dev Genes Evol. 1997;207:194–198. doi: 10.1007/s004270050107. [DOI] [PubMed] [Google Scholar]
- Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acid Res. 1991;21:160–162. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering DR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Prasher DC, Tsien RY. Wavelength mutations and post-translational autoxidation of green fluorescent protein. Proc Natl Acad Sci. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue specific basic helix-loop-helix proteins with a two hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar messenger RNA by Bruno, an ovarian RNA binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–630. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Kraut R, Campos-Ortega JA. inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeletal adapter protein. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Li P, Yang XH, Wasser M, Cai Y, Chia W. Inscuteable and staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. A new visible light DNA fluorochrome for confocal microscopy. BioTechniques. 1994;16:434–440. [PubMed] [Google Scholar]
- Matsuzaki F, Koisumi K, Hama C, Yoshioka T, Nabeshima T. Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem Biophys Res Comm. 1992;182:1326–1332. doi: 10.1016/0006-291x(92)91878-t. [DOI] [PubMed] [Google Scholar]
- Micklem DR. “Trans-acting factors required for the posterior localisation of oskar mRNA.” Ph.D. thesis. Cambridge, UK: Cambridge University; 1998. [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, Gonzalez-Reyes A, St Johnston D. mago nashi is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli—mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Grindstaff KK. Cell polarity: Par for the polar course. Curr Biol. 1997;7:R562–R564. doi: 10.1016/s0960-9822(06)00282-x. [DOI] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical uses in cell and molecular biology. San Diego, CA: Academic Press; 1994. pp. 445–487. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development. 1991;113:55–66. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- ————— Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–370. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Poortinga, G., M. Watanabe, and S.M. Parkhurst. 1998. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of Numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable elements vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- Sil A, Herskowitz I. Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The Prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- ————— Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Spana E, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. In situ hybridization to embryos with non-radioactive probes. In: Ashburner M, editor. Drosophila, a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 194–198. [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes & Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jiang M-M, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]