Abstract
Glandular chemical defence relying on the action of salicylaldehyde is characteristic for Chrysomela leaf beetle larvae. The salicylaldehyde precursor salicin, sequestered from salicaceous host plants, is deglucosylated and the aglycon further oxidized by a salicyl alcohol oxidase (SAO) to the respective aldehyde. SAOs, key enzymes in salicin-based glandular chemical defence, were previously identified and shown to be of a single evolutionary origin in Chrysomela species. We here identified and characterized SAO of Phratora vitellinae, the only species outside the genus Chrysomela that produce salicylaldehyde as a defensive compound. Although Chrysomela and Phratora are not closest relatives, their SAOs share glucose–methanol–choline oxidoreductase (GMC) affiliation, a specific GMCi subfamily ancestor, glandular tissue-specific expression and almost identical gene architectures. Together, this strongly supports a single origin of SAOs of both Chrysomela and Phratora. Closely related species of Chrysomela and P. vitellinae use iridoids as defensive compounds, which are like salicylaldehyde synthesized by the consecutive action of glucosidase and oxidase. However, we elucidated SAO-like sequences but no SAO proteins in the glandular secretion of iridoid producers. These findings support a different evolutionary history of SAO, related genes and other oxidases involved in chemical defence in the glandular system of salicylaldehyde and iridoid-producing leaf beetle larvae.
Keywords: Phratora vitellinae, salicyl alcohol oxidase, chemical defence, Chrysomelidae
1. Introduction
Leaf beetle larvae of the subtribe Chrysomelina are efficiently protected against generalist predators (e.g. ants, wasps, ladybirds, spiders) and microbial infestation by the use of a glandular chemical defence [1–9]. When attacked by predators, the larvae release droplets of deterrent secretion through dorsal openings of eight pairs of defensive glands. The defensive secretion is partly biosynthesized and stored inside the gland reservoirs. Although a huge variety of defensive compounds in different Chrysomelina species exist, the consecutive action of a glucosidase and an oxidase is widespread to modify selectively ingested alcohol glucosides to bioactive compounds inside the glandular reservoirs [10].
Chrysomelina larvae most frequently use either iridoids or salicylaldehyde as chemical defensive compounds, the former being derived from 8-hydroxygeraniol-8-glucoside and the latter from salicin (figure 1) (reviewed in [13,14]). Taking the Chrysomelina phylogeny into account, the predominant de novo biosynthesis of iridoids (e.g. in the genera Gastrophysa, Phaedon, Phratora) is seen as the ancestral state in the evolution of deterrent compound production [12]. In comparison to this, the salicylaldehyde biosynthesis of Chrysomela larvae is a derived and more economic defence strategy, because the precursor salicin is sequestered from their salicaceous host plants [6]. Whether the salicylaldehyde biosynthesis is derived from the iridoid biosynthetic route (by shift of substrate specificities of the glandular reservoir oxidases) or different chemical defences in Chrysomelina evolved independently is not known so far. Furthermore, the origin of salicylaldehyde biosynthesis is not clear as beside the genus Chrysomela, also the more distant relative Phratora vitellinae produce salicylaldehyde [10,15,16] (figure 1). The salicylaldehyde precursor and biosynthesis, namely deglucosylation and oxidation, has been shown to be the same in Chrysomela and P. vitellinae larvae [6,10], but nevertheless a convergent origin of salicin-based chemical defence in both genera is discussed owing to phylogenetic analyses [12]. Therein P. vitellinae is placed isolated within iridoid producers without a close affiliation to the salicylaldehyde-producing genus Chrysomela. Moreover, P. vitellinae is the only species of the genus Phratora synthesizing salicylaldehyde instead of iridoids [15]. Therefore, to clarify the origin of salicylaldehyde-based chemical defence, the identification, characterization and comparison of the enzymes involved in its biosynthesis are required. In case of several Chrysomela ssp., salicyl alcohol oxidase (SAO), which catalyses the final step in salicylaldehyde formation, has been elucidated and previous studies indicate a similar enzymatic activity and the presence of an SAO-related protein in the glandular secretion of P. vitellinae [17–20]. However, the enzyme itself is not known.
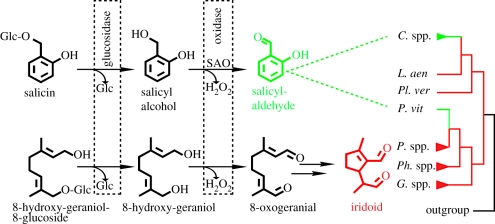
Figure 1.
Schematic of salicylaldehyde and iridoid biosynthesis in glandular reservoirs highlighting common enzymatic activities with boxes. The occurrence of those defensive compounds in various chrysomeline leaf beetle genera is plotted onto their phylogeny. Therein iridoid-based chemical defence (red) is the primary state. The derived salicylaldehyde-based defence (green) is present in the genus Chrysomela and P. vitellinae. This figure is a combination and modification of Pasteels et al. [10], Köpf et al. [11] and Termonia et al. [12]. C. spp., Chrysomela species; L. aen, Linaeidea aenea; Pl. ver, Plagiodera versicolora; P. vit, P. vitellinae; P. spp., Phratora species; Ph. spp., Phaedon species; G. spp.; Gastrophysa species, SAO, salicyl alcohol oxidase; Glc, glucose.
In the present study, we focus on the SAO of P. vitellinae to get insights into the evolution of salicin metabolism in conjunction with chemical defence and the origin of the SAO in chrysomelines. Here, we identified the SAO protein in the glandular secretion and used the corresponding cDNA for cloning and heterologous expression in an Sf9 insect cell line followed by enzyme assays. The SAO of P. vitellinae belongs to the glucose–methanol–choline oxidoreductase (GMC) family and shows high sequence similarity to Chrysomela ssp. SAOs. Enzymatic activity and Re specificity could be verified. Phylogenetic analyses indicate a common ancestor of all known SAOs, which is additionally supported by both common expression profiles and highly conserved gene architecture. The origin of chemical defence based on salicin sequestration in chrysomelines is discussed in consideration of these data and previous findings. This includes for the first time an elucidation of SAO-like sequences from transcriptome and genome screenings in iridoid-producing leaf beetle larvae.
2. Material and methods
(a). Leaf beetle larvae
Larvae of P. vitellinae and Phratora laticollis were collected from their respective host plant in the field near Bruxelles and reared in the laboratory (20°C, long-day conditions: 16 L : 8 D period) on S. caprea and Populus × canadensis for collecting glandular secretion. Larval tissues were stored in RNAlater (Qiagen) at −20°C until needed.
(b). Identification of the salicyl alcohol oxidase in the glandular secretion
Larval glandular secretion was collected in the laboratory using glass capillaries (Hirschmann Laborgeraete, ID: 0.28 mm, L: 100 mm) by gently squeezing the larvae with forceps until they protruded their glands. The emerging droplets are then collected with the capillaries and stored at −20°C. The secretion was directly used for one-dimensional sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gel runs with the help of the Criterion XT Precast Gel System (BioRad). Briefly, the separated protein bands were manually picked from deionized water using a 1–200 µl pipette and disposable tips cut to 3 mm ID blunt inlet. The gel plugs were transferred to 96-well microtitre plates (MTPs), reduced by 10 mM dithiothreitol, alkylated by 55 mM iodoacetamide and destained in 50 mM ammonium bicarbonate/50 per cent acetonitrile. Subsequently, the plugs were air-dried and overlayed with 50 mM ammonium bicarbonate containing 70 ng porcine trypsin (Sequencing grade, Promega). The MTPs were covered with aluminium foil and the proteins were digested overnight at 37°C. The resulting peptides were extracted from the gel plugs by adding twice 50 µl of 50 per cent acetonitrile in 0.1 per cent trifluoroacetic acid for 20 min and the extracts were collected in an extraction MTP and vacuum-dried to remove any remaining liquid and ammonium bicarbonate. The tryptic peptides were reconstituted in 6 µl aqueous 0.1 per cent formic acid (FA). The selected volumes of samples (ca 4.5 µl) were injected on a nanoAcquity nanoUPLC system (Waters, Milford, MA, USA). Mobile phase A (0.1% aqueous FA, 15 µl min−1 for 1 min) was used to concentrate and desalt the samples on a 20 × 0.180 mm Symmetry C18, 5 µm particle precolumn. The samples were then eluted on a 100 mm × 75 µm ID, 1.7 µm BEH nanoAcquity C18 column (Waters). Phases A and B (100% MeCN in 0.1% FA) were linearly mixed in a gradient to 5 per cent phase B in 0.33 min, increased to 40 per cent B in 10 min and finally increased to 85 per cent B in 10.5 min, holding 85 per cent B to 11 min and decreasing to 1 per cent B in 11.1 min of the run. The eluted peptides were transferred to the nanoelectrospray source of a Synapt HDMS tandem mass spectrometer (Waters) equipped with metal-coated nanoelectrospray tips (Picotip, 50 × 0.36 mm, 10 µm I.D, Waters). The source temperature was set to 80°C, cone gas flow at 20 l h–1 and the nanoelectrospray voltage was 3.2 kV. The TOF analyzer was used in a reflectron mode. The MS/MS spectra were collected at 1 s intervals (50–1700 m/z). A 650 fmol µl–1 human Glu-fibrinopeptide B in 0.1 per cent FA/acetonitrile (1 : 1 v/v) was infused at a flow rate of 0.5 µl min–1 through the reference NanoLockSpray source every 30th scan to compensate for mass shifts in the MS and MS/MS fragmentation mode. The data were collected by MassLynx v. 4.1 software. ProteinLynx Global Server Browser v. 2.3 software (both Waters) was used for baseline subtraction and smoothing, deisotoping, de novo peptide sequence identification and database searches. The peptide fragment spectra were searched against the Uniprot ‘Chrysomelidae’ taxonomy-defined subdatabase downloaded on 18 March 2010 from http://www.uniprot.org/. The protein identification from MS/MS fragment spectra used peptide mass tolerance 15 ppm and minimum three peptides found, estimated calibration error 0.002 Da, 0.03 Da mass deviation of de novo-sequenced peptides, one possible missed cleavage, and carbamidomethylation of cysteins, possible oxidation of methionines, and possible deamidation of asparagines and glutamines, respectively.
(c). Amplification of full-length salicyl alcohol oxidase encoding cDNA and genes
An expressed tag sequence (EST) encoding the P. vitellinae SAO identified by using Chrysomela SAO primers were used for RACE PCR with the SMART RACE cDNA Amplification Kit (Clontech) according to the manufacturers' guidelines. Genes of P. vitellinae SAO and P. laticollis SAO-like protein were amplified with the LA Taq Polymerase (Takara), following the recommended instructions for long-distance PCR. Gel-purified bands were prepared for shearing on a HydroShear DNA Shearing Device (GeneMachines) and then cloned into SmaI-digested pUC19 vector (Fermentas) for shotgun sequencing. Positive clones were picked manually and grown overnight in DYT medium.
(d). Western blot and enzyme assays
Forty-eight hours after transfection, the culture medium of Sf9 cells was harvested, concentrated 20-fold with iCON concentrators (20 ml/9 K, PIERCE) and the crude protein extract was then used for Western blot analysis and enzyme assays. For detection of the heterologously expressed proteins in Western blots, Anti-V5-HRP antibody (Invitrogen) and SuperSignal West His Probe Kit (Pierce) were used. The enzyme assay was performed in 0.5 ml plastic tubes in 50 mM potassium phosphate buffer (pH 6.0) at 30°C for 10 min to 2 h with a final salicyl alcohol (Sigma) or 8-hydroxygeraniol [21] concentration of 350 µM to 3 mM in a total volume of 100 µl. Assays were stopped by freezing in liquid nitrogen. After extraction with 100 µl ethyl acetate (Roth) and centrifugation for 5 min at 5000g, the organic phase was directly used for GCMS analysis (ThermoQuest Finnigan Trace GC-MS 2000 (Quadrupole) equipped with Alltech EC 5-column, 15 m × 0.25 mm, film thickness 0.25 µm). Compounds were eluted under programmed conditions: 45°C for 1 min, ramped at 10°C min–1 to 200°C, followed by a 30°C min–1 ramp to 280°C for 3 min. Helium carrier gas was maintained at a flow rate of 1.5 ml min–1. Eluting compounds were detected by mass spectrometry and compared with authentic references. The Re specificity of the enzyme was determined using chiral 1R-[1-2H1]-salicyl alcohol [20]. The resulting salicyl aldehyde showed no deuterium labelling, demonstrating the Re specificity of the oxidation.
(e). Analysis of glucose–methanol–choline oxidoreductase expression
Third larval instars were dissected. Fat bodies, defensive glands, gut and Malpighian tubules were stabilized in RNAlater (Qiagen) and stored at −20°C for further applications. For qPCR, 500 ng of total RNA pooled from the tissue of 20 individuals was reverse transcribed with a mix of random and oligo-dT20 primers. Real-time PCR was done in optical 96-well plates on an MX 3000P (Stratagene). All steps were performed with Verso SYBR Green 2-Step QRT-PCR Kit Plus ROX Vial (Thermo Scientific) following the manufacturer's instructions. Specific amplification of transcripts was verified by melting curve analysis. All primers were designed by the help of Primer 3 (v. 0.4.0.). We used eukaryotic initiation factor-4A (eIF4A) and elongation factor-1α (EF1α) as reference genes to normalize quantities of our genes of interest. For the analysis of raw data, we used qBase, choosing a logarithmic view of the relative expression level on the y-axis of the graphs, where the lowest transcript abundance was set to 1.
(f). Heterologous expression in Sf9 cells
The cDNA encoding the SAO protein was amplified by PCR using gene-specific primers, including a 5′ Kozak sequence and lacking a stop codon for epitope and His-tag fusion expression after ligation into pIBV5-His-TOPO TA vector (Invitrogen) used for Sf9 insect cell expression. The correct sequence and direction of cloning were verified by sequencing. Sf9 cells were cultivated in Sf-900 II SFM (GIBCO) on tissue culture dishes (100 × 20 mm, FALCON) at 27°C until 60 per cent confluence. Transfection was performed with Insect Gene Juice (Novagen) following the manufacturer's protocol.
(g). Phylogenetic analyses
Multiple alignments of protein sequences were carried out using ClustalW [22]. Phylogenetic relationships were inferred using a neighbour-joining algorithm [23] implemented in MAFFT taking insertions and deletions into account. The bootstrap re-sampling analysis with 1000 replicates was performed to evaluate the tree topology. A model-based phylogenetic analysis using Bayesian Markov Chain Monte Carlo inference was also carried out as implemented in MrBayes v. 3.1.2 consisting of four Markov chains. The analysis was run for 1 000 000 generations, sampling from the trees every 100 generations, and the first 1000 generations were discarded as the ‘burn-in’. Trees were combined into a single summary tree.
3. Results
(a). Identification and sequence analysis of salicyl alcohol oxidase protein and cDNA
One-dimensional SDS–PAGE gels of P. vitellinae larval glandular secretion displayed a highly abundant protein at about 75 kDa (figure 2). Through de novo MS fingerprinting, we could identify this major glandular protein as a member of the GMC oxidoreductase family. In particular, de novo peptide sequences could be assigned to known Chrysomela SAOs [18,19]. The corresponding cDNA was obtained using internal SAO primers designed to amplify a partial core fragment followed by P. vitellinae sequence-specific 3′- and 5′-RACE. This led to a single full-length cDNA of 2040 bp with an ORF of 1881 bp that encodes a protein of 626 amino acids. Sequence comparison showed an identity of about 72 per cent to leaf beetle SAOs of Chrysomela lapponica, Chrysomela populi and Chrysomela tremulae at the amino acid level, including the N-terminal signal peptide for the secretory pathway (electronic supplementary material, figure S1 and table S1). In total, 17 peptides identified by nanoLC-MS/MS of the 75 kDa protein band extract matched the full-length sequence, allowing approximately 40 per cent of total sequence coverage (electronic supplementary material, figure S2 and table S2), thus verifying that the amplified transcript corresponds to the protein present in the glandular secretion. The calculated molecular mass of P. vitellinae SAO-like protein with/without the signal peptide (70/68 kDa) is lower than its actual mass (approx. 75 kDa). Comparable post-translational modifications as shown for Chrysomela SAOs are indicated by seven predicted N-glycosylation sites in P. vitellinae SAO-like protein. This predicted glycosylation was confirmed by a band shift assay treating the heterologously expressed protein with PNGase F (figure 3). Interestingly, not only the total number but also four positions of predicted N-glycosylation sites are conserved in Chrysomela ssp. and P. vitellinae SAOs.
Figure 2.
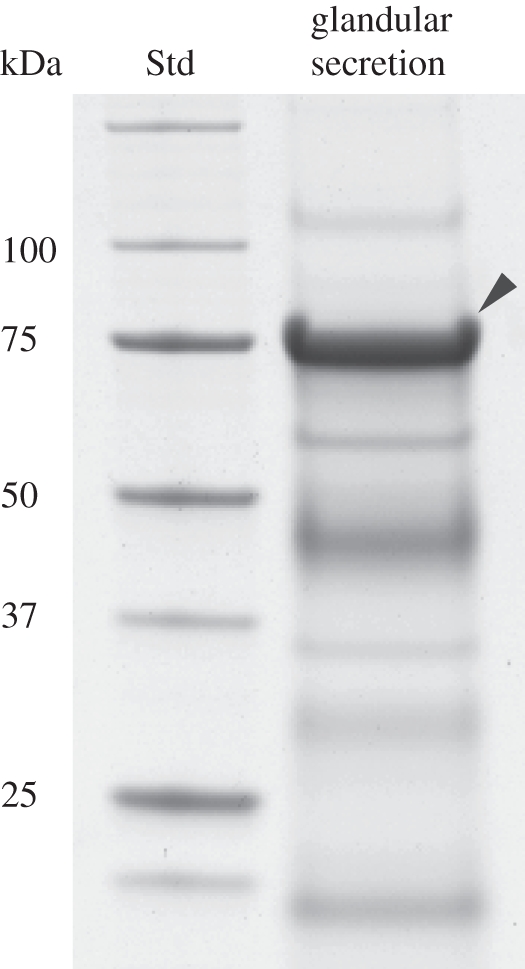
Coomassie Brilliant Blue-stained one-dimensional SDS protein gel of the larval glandular secretion of P. vitellinae. The most prominent band marked with an arrow corresponds to SAO. The size of the protein standard on the left is indicated in 103 Da.
Figure 3.
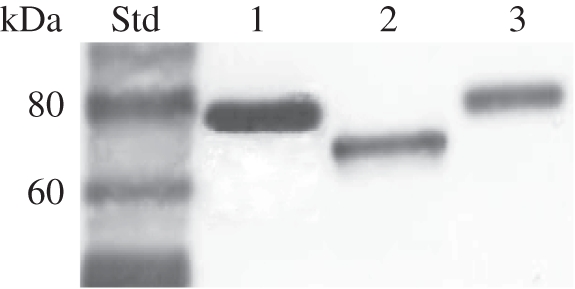
Western blot of heterologously expressed SAOs in Sf9 insect cells. Lanes 1 and 2 correspond to the P. vitellinae SAO. Size differences are due to a PNGase F treatment of probe 2 (lane 2), suggesting post-translational N-glycosylations. Chrysomela populi SAO is blotted (lane 3), demonstrating a similar size of the SAO in both genera after expression.
(b). Salicyl alcohol oxidase expression and activity
Phratora vitellinae SAO-like full-length cDNA starts with ATG AAA ATG AAG. We used a six-nucleotide shorter transcript for characterization because the methionine-encoding third triplet turned out as the most likely start codon in comparison with Chrysomela SAOs after multiple alignment analysis (electronic supplementary material, figure S1). The SAO-like protein was expressed in Sf9 insect cells and a protein of the predicted size was detected by Western blot analysis in the Sf9 cultural medium (figure 3). This demonstrated the functional N-terminal signal peptide for the secretory pathway and similar post-translational modifications indicated by a comparable size of Sf9 cell and beetle-expressed SAO.
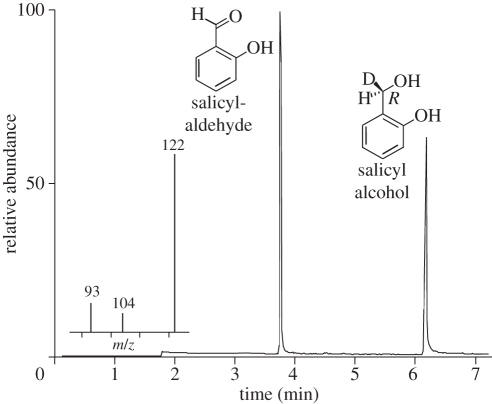
Enzyme assays of the heterologously expressed SAO-like protein revealed SAO activity (figure 4). The oxidation proceeded Re specifically and removed exclusively the deuterium atom and yielded [1-1H]-salicylaldehyde from the 1R-[1-2H1]-salicyl alcohol precursor. This stereochemical course is in agreement with previous studies using the glandular secretion of P. vitellinae [20] and confirmed that the oxidation is not an autoxidative artefact. By contrast, the previously described oxidation of 8-hydroxygeraniol, for which glandular secretion of P. vitellinae was used [20], was not detectable using the heterologously expressed enzyme. In addition, from MS/MS analyses, no peptides matching alternative GMC oxidoreductases, other than the SAO present in the glandular secretion of P. vitellinae, were identified.
Figure 4.
GC chromatogram of heterologously expressed P. vitellinae SAO activity assay after extraction. The formation of salicylaldehyde via a stereoselective deuterium removal of salicyl alcohol is indicated by the mass spectrum of the aldehyde.
(c). Expression pattern of the P. vitellinae salicyl alcohol oxidase
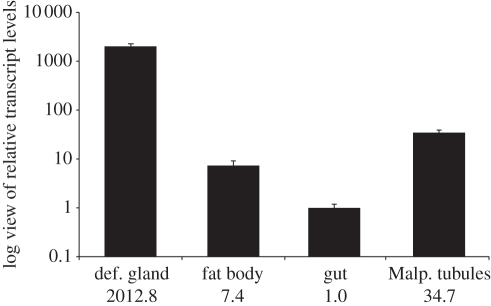
SAO gene expression levels were compared in different larval tissues. The SAO is specifically expressed in the glandular tissue (figure 5) with an approximately 2000-fold higher expression level compared with the fat body, gut and Malpighian tubules, where SAO gene expression levels are close to the background level. The large amount of protein present in the glandular secretion (§3a and figure 2) coincides with the high transcript level of SAO in the glandular tissue, verifying that the enzyme is expressed in a tissue-specific manner and most probably secreted into the glandular reservoir.
Figure 5.
Expression pattern of P. vitellinae SAO. Relative transcript levels in different larval tissues display glandular tissue specificity of the SAO with an approximately 2000-fold higher transcript abundance compared with fat body, gut and Malpighian tubules. Transcript levels are indicated logarithmic and average values are shown below each column. For normalization of transcript quantities, EF1α and eIF4A were used. Error bars indicate the SEM.
(d). Salicyl alcohol oxidase-like sequences in iridoid-producing species
Phratora vitellinae SAO is closely related to Chrysomela SAOs in sequence but isolated from these according to species phylogeny. To address the question of the origin of SAO, we searched for SAO-like sequences in iridoid producers first in a species that is, based on phylogenetic data, located between Phratora and Chrysomela (Phaedon cochleariae) and second within the genus Phratora (P. laticollis). The analysis of a BLAST search of chrysomeline SAOs against a P. cochleariae in-house EST library led to a single partial ORF encoding an SAO-like protein. Full-length sequencing after RACE PCR showed about 58 per cent sequence identity to known SAOs and related proteins. However, there is no indication for the presence of this protein in the glandular reservoir (see §4 for details). From genomic DNA of P. laticollis, we were able to amplify an 8 kb SAO-like gene fragment, using P. vitellinae SAO-derived primers. The predicted encoded protein possesses about 73 per cent sequence identity to Chrysomela and 90 per cent to P. vitellinae SAOs (electronic supplementary material, figure S1 and table S1).
(e). Comparative salicyl alcohol oxidase gene architecture
Based on the SAO cDNA sequence of P. vitellinae, we were able to amplify about 8 kb from genomic DNA, which covers most of the SAO-encoding gene. The alignment of cDNA and the corresponding genomic region showed that SAO possesses at least eight exons. In comparison with known Chrysomela SAO genes [18], both a highly similar gene length and number of exons could be identified. Most remarkably, an identical length of almost all exons in both genera was obvious. The P. laticollis SAO-like gene fragment (§3d) also possesses identical exon lengths of its predicted coding sequence. In summary, comparative SAO gene architecture indicates a common SAO origin.
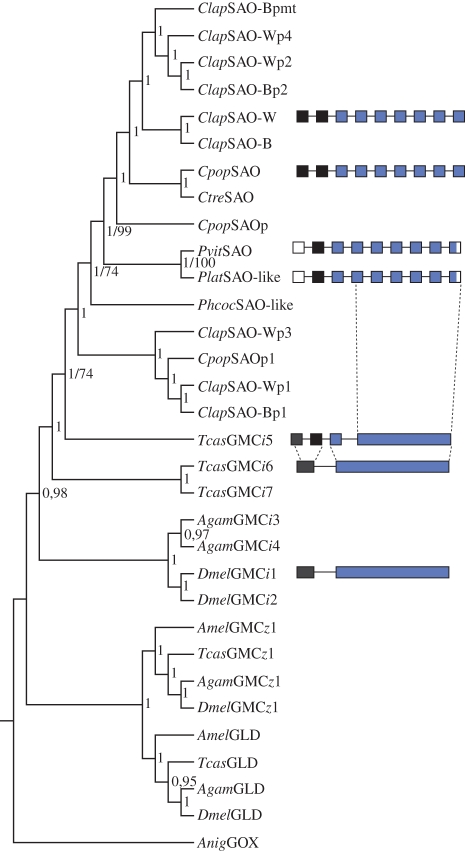
Further analysis of genomic DNA sequences of GMC oxidoreductases in Tribolium castaneum and Drosophila melanogaster demonstrated one to three encoding exons of variable length. Furthermore, whereas all the fruitfly and red flour beetle GMC genes belonging to the GMCi subfamily possess two exons, the closest relative to the chrysomeline SAOs (TcasGMCi5) possesses four exons (figure 6 and electronic supplementary material, figure S3). Those findings not only show an accumulation of introns common to chrysomeline GMCi genes, but also indicate that this increase in gene architecture complexity may have arisen in the most recent common ancestral gene.
Figure 6.
Phylogeny of chrysomeline SAOs and related GMC oxidoreductases, including other insects. The topology was generated using a Bayesian analysis. Posterior probability values are shown next to each node. Bootstrap values are exemplary stated as second numbers (for details of parameters and sequence information, see §2 and electronic supplementary material, table S3). Chrysomeline SAO and other insect GMCi gene architectures are depicted next to the phylogenetic tree. Exons are shown with boxes, the same colour coding indicates common origin among different species, empty boxes mark gaps in our dataset and the dashed lines mark intron accumulations. Clap (C. lapponica), Cpop (C. populi), Ctre (C. tremulae), Pvit (P. vitellinae), Plat (P. laticollis), Phcoc (Phaedon cochleariae), Tcas (T. castaneum), Anig (Aspergillus niger), SAO-W/B (salicyl alcohol oxidase of willow/birch-feeder), GMC (glucose–methanol–choline oxidoreductase), GLD (glucose dehydrogenase), GOX (glucose oxidase), p (SAO paralogues), pmt (SAO paralogues Malpighian tubule specific).
(f). Salicyl alcohol oxidase evolution in chrysomelines
Phylogenetic analyses, including members of different GMC oxidoreductase subfamilies, a subset of SAO and related genes from previous work [18], showed a common origin of both P. vitellinae and Chrysomela ssp. SAOs within the GMCi subfamily (figure 6). This is indicated by their affiliation to TcasGMCi5 supported by a high posterior probability value (1). Moreover, within the leaf beetle GMCi members, a clear separation of P. vitellinae SAO from Chrysomela SAO-related genes (paralogues 1 and 3) and the clustering of the P. vitellinae SAO with Chrysomela SAOs (posterior probability value of 1 for each node) indicate a single ancestral gene in both genera. Phratora vitellinae SAO is not located inside the Chrysomela SAO clade but represents a sister-group and clusters together with the P. laticollis SAO-like protein, which is reflective of the overall species phylogeny.
In addition, we included the SAO-like protein of P. cochleariae (§3d) in our analysis, which also showed unambiguously affiliation to the chrysomeline SAO clade. This generally indicates the presence of SAO-like proteins in iridoid producers and probably reflects GMCi subfamily expansion by gene duplications early in chrysomeline evolution.
4. Discussion
Chrysomela species are known to produce salicylaldehyde for their chemical defence, a compound shown to be highly deterrent against generalist predators and microbial infestations. As the precursor salicin acts as a general feeding repellent of their salicaceous host plants (reviewed in [24]), the sequestration of this secondary compound is a remarkable example of an engaging detoxification and economical defence in leaf beetle larvae. Interestingly, beside the genus Chrysomela, this remarkable derived state of chemical defence strategy is only present in P. vitellinae [15]. This is noteworthy, because the two leaf beetle genera are not close relatives, and therefore a convergent evolution of host-derived chemical defence based on salicin sequestration has been proposed [12]. However, despite the evolutionary distance between these species, the mode of sequestration as well as salicylaldehyde biosynthesis and SAO activity in particular are the same in both genera [6,10,17,20].
To shed light onto salicin-dependent chemical defence evolution, we first identified and functionally expressed the SAO of P. vitellinae. The SAO is the most abundant protein in larval glandular secretions, consists of 626 amino acids including the N-terminal signal peptide addressing the secretory pathway and shows a complex N-glycosylation pattern of about 7 kDa. The highly abundant transcript specific for the corresponding glandular tissue is consistent with both the proportion of oxidative capacity inside the glandular secretion and the prominent band in the one-dimensional protein gel. Sequence comparisons verified that the SAO belongs to the GMC oxidoreductase family and possess a 72 per cent amino acid identity compared with Chrysomela SAOs. The Re specificity of the heterologously expressed protein was verified for the first time, which fits to previous findings with in vitro assays of the glandular secretion [20]. We showed that P. vitellinae and Chrysomela SAOs not only share a highly tissue-specific expression and high amounts of protein in the glandular system, the same protein size, similar post-translational modifications and the affiliation to GMC oxidoreductases, but also a well conserved gene architecture (number and lengths of exons). These findings provide additional support for the results of our phylogenetic analyses demonstrating a single origin of P. vitellinae and Chrysomela ssp. SAO genes in the GMCi subfamily.
Taking leaf beetle species phylogenies into account, which all support the notion that Chrysomela and Phratora are not sister genera [12,25,26], the following evolutionary scenarios leading to SAO activity are conceivable. A single gene duplication event led to (gave birth to) the evolution of the SAO gene and activity in Phratora and Chrysomela. Whether this proceeded via one sub-/neofunctionalization in their most recent common ancestor or convergent SAO acquisitions (and additional genera-specific gene duplications of a ‘precursor’ gene) cannot be resolved. Therefore, we cannot exclude the possibility that Chrysomela and Phratora SAO are not true orthologues in a strict sense. However, for both scenarios, the persistence of the SAO or SAO ‘precursor’ gene (e.g. through retaining the original function of the SAO ‘precursor’ gene) in the iridoid-producing genera between Chrysomela and Phratora (e.g. Phaedon, Gastrophysa) is likely a prerequisite.
Because of a common glucosidase–oxidase pathway leading to salicylaldehyde and iridoids, shifts from iridoid to salicylaldehyde biosynthesis in leaf beetle chemical defence evolution has been proposed to take place via changing substrate specificity of the oxidase [10,12,20]. This change of substrate specificity is supported by in vitro oxidation of 8-hydroxygeraniol by the secretion of salicylaldehyde producing P. vitellinae [20], which is seen as an argument for the evolution of SAOs from oxidases of iridoid-producing ancestors.
In contrast to these findings, we were not able to verify 8-hydroxygeraniol oxidation with enzyme assays of the heterologously expressed P. vitellinae SAO. Furthermore, we found neither an oxidase with affiliation to the GMCi subfamily (e.g. P. cochleariae SAO-like protein: §3d) nor a GMC oxidoreductase of the conserved insect gene cluster at all, present in the secretion of iridoid-producing P. cochleariae, Gastrophysa viridula or Gastrophysa cyanea larvae (R. Kirsch 2010, unpublished data). Several chemical and biochemical properties identified (oxygen dependence and Re specificity) are consistent between SAO and 8-hydroxygeraniol oxidase [27]. However, our findings strongly argue for distinct, non-SAO related oxidases converting 8-hydroxygeraniol to 8-oxocitral in iridoid-producing species and, moreover, an independent evolution of the oxidative step in salicylaldehyde and iridoid biosynthesis. In this context, the elucidation of the SAO-like EST in P. cochleariae and the SAO-like gene in P. laticollis, both iridoid-producing species, is important. The presence of SAO-like sequences in these species indicate that gene duplications in the GMCi subfamily started early in chrysomeline speciation followed by species specific gene duplications (shown for C. lapponica in [18]). Furthermore, the persistence of SAO-like genes in the iridoid producers is probably due to the acquisition of functions different from SAO activities (i.e. functions not related to chemical defence).
We clearly showed a common origin of SAOs in Chrysomela and P. vitellinae and their most likely independent evolution from iridoid biosynthesis. However, characterizations of SAO-like proteins in iridoid producers and the GMCi5 in T. castaneum are needed to resolve molecular functional origins and gene-family dynamics of SAO and related genes.
Acknowledgements
We thank Domenica Schnabelrauch for shotgun sequencing and Antje Loele for protein sample preparation for MS analyses. We are grateful to Dr Nicole Joussen for preparing the Sf9 insect cells for transfections. This work was supported by the Max Planck Society and the Hessian Ministry for Science and Arts funding the LOEWE research focus Insect Biotechnology.
Author contributions: R.K., H.V. and W.B designed research; R.K., H.V., A.M., J.M.P. and W.B wrote the paper; R.K., H.V., A.M. and A.V. performed research and analysed data.
References
- 1.Blum M. S., Brand J. M., Wallace J. B., Fales H. M. 1972. Chemical characterization of the defensive secretion of a chrysomelid larva. Life Sci. 11, 525–531 10.1016/0024-3205(72)90286-X (doi:10.1016/0024-3205(72)90286-X) [DOI] [Google Scholar]
- 2.Denno R. F., Larsson S., Olmstead K. L. 1990. Role of enemy-free space and plant quality in host-plant selection by willow beetles. Ecology 71, 124–137 10.2307/1940253 (doi:10.2307/1940253) [DOI] [Google Scholar]
- 3.Gross J., Podsiadlowski L., Hilker M. 2002. Antimicrobial activity of exocrine glandular secretion of Chrysomela larvae. J. Chem. Ecol. 28, 317–331 10.1023/A:1017934124650 (doi:10.1023/A:1017934124650) [DOI] [PubMed] [Google Scholar]
- 4.Hilker M., Schulz S. 1994. Composition of larval secretion of Chrysomela lapponica (Coleoptera, Chrysomelidae) and its dependence on host plant. J. Chem. Ecol. 20, 1075–1093 10.1007/BF02059744 (doi:10.1007/BF02059744) [DOI] [PubMed] [Google Scholar]
- 5.Palokangas P., Neuvonen S. 1992. Differences between species and instars of Phratora leaf beetles (Coleoptera, Chrysomelidae) in the probability of being preyed on. Ann. Zool. Fennici 29, 273–278 [Google Scholar]
- 6.Pasteels J. M., Rowell-Rahier M., Braekman J. C., Dupont A. 1983. Salicin from host plant as precursor of salicylaldehyde in defensive secretion of Chrysomeline larvae. Physiol. Entomol. 8, 307–314 10.1111/j.1365-3032.1983.tb00362.x (doi:10.1111/j.1365-3032.1983.tb00362.x) [DOI] [Google Scholar]
- 7.Smiley J. T., Horn J. M., Rank N. E. 1985. Ecological effects of salicin at three trophic levels: new problems from old adaptations. Science 229, 649–651 10.1126/science.229.4714.649 (doi:10.1126/science.229.4714.649) [DOI] [PubMed] [Google Scholar]
- 8.Wallace J. B., Blum M. S. 1969. Refined defensive mechanisms in Chrysomela scripta. Ann. Entomol. Soc. Am. 62, 503–506 [Google Scholar]
- 9.Gross J., Müller C., Vilcinskas A., Hilker M. 1998. Antimicrobial activity of exocrine glandular secretions, hemolymph, and larval regurgitate of the mustard leaf beetle Phaedon cochleariae. J. Invertebr. Pathol. 72, 296–303 10.1006/jipa.1998.4781 (doi:10.1006/jipa.1998.4781) [DOI] [PubMed] [Google Scholar]
- 10.Pasteels J. M., Duffey S., Rowell-Rahier M. 1990. Toxins in Chrysomelid beetles: possible evolutionary sequence from de novo synthesis to derivation from food-plant chemicals. J. Chem. Ecol. 16, 211–222 10.1007/BF01021280 (doi:10.1007/BF01021280) [DOI] [PubMed] [Google Scholar]
- 11.Köpf A., Rank N. E., Roininen H., Julkunen-Tiitto R., Pasteels J. M., Tahvanainen J. 1998. The evolution of host-plant use and sequestration in the leaf beetle genus Phratora (Coleoptera: Chrysomelidae). Evolution 52, 517–528 10.2307/2411087 (doi:10.2307/2411087) [DOI] [PubMed] [Google Scholar]
- 12.Termonia A., Hsiao T. H., Pasteels J. M., Milinkovitch M. 2001. Feeding specialization and host-derived chemical defense in Chrysomeline leaf beetles did not lead to an evolutionary dead end. Proc. Natl Acad. Sci. USA 98, 3909–3914 10.1073/pnas.061034598 (doi:10.1073/pnas.061034598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasteels J. M., Braekman J. C., Daloze D. 1988. Chemical defense in the Chrysomelidae. In Biology of Chrysomelidae (eds Jolivet P., Petitpierre E., Hsiao T.), pp. 233–252 Dordrecht, The Netherlands: Kluwer [Google Scholar]
- 14.Pasteels J. M., Rowell-Rahier M., Raupp M. J. 1988. Plant-derived defense in chrysomelid beetles. In Novel aspects of insect-plant interactions (eds Barbosa P., Letorneau D.), pp. 235–272 New York, NY: John Wiley & Sons [Google Scholar]
- 15.Pasteels J. M., Rowell-Rahier M., Braekman J. C., Daloze D. 1984. Chemical defences in leaf beetles and their larvae: the ecological, evolutionary and taxonomic significance. Biochem. Syst. Ecol. 12, 395–406 10.1016/0305-1978(84)90071-1 (doi:10.1016/0305-1978(84)90071-1) [DOI] [Google Scholar]
- 16.Wain R. L. 1943. The secretion of salicylaldehyde by the larvae of the brassy willow beetle (Phyllodecta vitellinae L.). Annu. Rep. Agric. Hortic. Res. Stn, 108–110 [Google Scholar]
- 17.Brückmann M., Termonia A., Pasteels J. M., Hartmann T. 2002. Characterization of an extracellular salicyl alcohol oxidase from larval defensive secretions of Chrysomela populi and Phratora vitellinae (Chrysomelina). Insect Biochem. Mol. Biol. 32, 1517–1523 10.1016/S0965-1748(02)00072-3 (doi:10.1016/S0965-1748(02)00072-3) [DOI] [PubMed] [Google Scholar]
- 18.Kirsch R., Vogel H., Muck A., Reichwald K., Pasteels J. M., Boland W. In press Host plant shifts affect a major defense enzyme in Chrysomela lapponica. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1013846108 (doi:10.1073/pnas.1013846108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalski C., Mohagheghi H., Nimtz M., Pasteels J. M., Ober D. 2008. Salicyl alcohol oxidase of the chemical defense secretion of two chrysomelid leaf beetles. J. Biol. Chem. 283, 19 219–19 228 10.1074/jbc.M802236200 (doi:10.1074/jbc.M802236200) [DOI] [PubMed] [Google Scholar]
- 20.Veith M., Oldham N. J., Dettner K., Pasteels J. M., Boland W. 1997. Biosynthesis of defensive allomones in leaf beetle larvae: stereochemistry of salicylalcohol oxidation in Phratora vitellinae and comparison of enzyme substrate and stereospecificity with alcohol oxidases from several iridoid producing leaf beetles. J. Chem. Ecol. 23, 429–443 10.1023/B:JOEC.0000006369.26490.c6 (doi:10.1023/B:JOEC.0000006369.26490.c6) [DOI] [Google Scholar]
- 21.Feld B., Pasteels J. M., Boland W. 2001. Phaedon cochleariae and Gastrophysa viridula (Coleoptera: Chrysomelidae) produce defensive iridoid monoterpenes de novo and are able to sequester glycosidically bound terpenoid precursors. Chemoecology 11, 191–198 10.1007/PL00001851 (doi:10.1007/PL00001851) [DOI] [Google Scholar]
- 22.Thompson J. D., Higgins D. G., Gibson T. J. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 24.Ruuhola T. M., Sipura M., Nousiainen O., Tahvanainen J. 2001. Systemic induction of salicylates in Salix myrsinifolia (Salisb.). Ann. Bot. 88, 483–497 10.1006/anbo.2001.1491 (doi:10.1006/anbo.2001.1491) [DOI] [Google Scholar]
- 25.Gómez-Zurita J., Hunt T., Kopliku F., Vogler A. P. 2007. Recalibrated tree of leaf beetles (Chrysomelidae) indicates independent diversification of angiosperms and their insect herbivores. PLoS ONE 2, e360. (doi:10.1071/journal.pone.0000360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gómez-Zurita J., Hunt T., Vogler A. P. 2008. Multilocus ribosomal RNA phylogeny of the leaf beetles (Chrysomelidae). Cladistics 24, 34–50 10.1111/j.1096-0031.2007.00167.x (doi:10.1111/j.1096-0031.2007.00167.x) [DOI] [Google Scholar]
- 27.Veith M., Dettner K., Boland W. 1996. Stereochemistry of an alcohol oxidase from the defensive secretion of larvae of the leaf beetle Phaedon armoraciae (Coleoptera: Chrysomelidae). Tetrahedron 52, 6601–6612 10.1016/0040-4020(96)00298-0 (doi:10.1016/0040-4020(96)00298-0) [DOI] [Google Scholar]
- 28.Iida K., Cox-Forster D. L., Yang X., Ko W.-Y., Cavener D. R. 2007. Expansion and evolution of insect GMC oxidoreductases. BMC Evol. Biol. 7, 75. 10.1186/1471-2148-7-75 (doi:10.1186/1471-2148-7-75) [DOI] [PMC free article] [PubMed] [Google Scholar]