Abstract
Repression of gene transcription is a fundamental property of nuclear hormone receptors. We report here that cell-specific repression by nuclear receptors correlates with levels of nuclear receptor corepressor (N-CoR) protein. N-CoR protein levels are regulated by mSiah2, a mammalian homolog of Drosophila Seven in absentia that targets N-CoR for proteasomal degradation. mSiah2 expression is cell-type specific and differentially regulates the repressive activities of nuclear receptors. These findings establish targeted proteolysis of transcriptional coregulators as a mechanism for cell-specific regulation of gene transcription.
Keywords: Nuclear receptor corepressor, proteasomal regulation, repression, RevErb, thyroid hormone receptor, cell specificity
Nuclear hormone receptors are among the best studied transcriptional repressors (Mangelsdorf et al. 1995; Shibata et al. 1997b). Thyroid hormone receptor (TR) and retinoic acid receptors (RAR) repress transcription in the absence of ligand, thus amplifying the magnitude of ligand-induced activation (Graupner et al. 1989; Baniahmad et al. 1992). Orphan members of the nuclear receptor superfamily, such as RevErb and COUP-TF, are ligand-independent repressors of transcription (Harding and Lazar 1995; Shibata et al. 1997a). RevErb does not possess the carboxy-terminal amphipathic helix characteristic of ligand-activated receptors, and thus its main function is to constitutively repress transcription. This is likely to serve major developmental roles in a number of organisms; RevErb antagonizes the effects of RORα, whose mutation leads to the cerebellar degeneration of the Staggerer mouse (Hamilton et al. 1996). In Drosophila, the RevErb homolog E75 regulates metamorphosis (Segraves and Hogness 1990; White et al. 1997).
Corepressors N-CoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoid and thyroid receptors) bind to nuclear receptor dimers and serve as a scaffold for additional proteins involved in repression, including Sin3 and histone deacetylases (Chen and Evans 1995; Horlein et al. 1995; Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997; Nagy et al. 1997). Receptor specificity of corepressor interaction on DNA is exemplified by the observation that RevErb specifically utilizes N-CoR, whereas thyroid hormone receptor TR may use both SMRT and N-CoR to mediate repression (Zamir et al. 1997b).
The amino terminus of N-CoR contains two repression domains that have not been identified in the reported sequence of SMRT (Chen and Evans 1995). However, the function of the amino terminus of N-CoR has not been well characterized. Here we show that the extreme amino terminus of N-CoR targets the protein for proteasomal degradation because of interaction with mSiah2, the mammalian homolog of Drosophila Seven in absentia (sina). This results in a receptor-specific antirepression function for mSiah2. Expression of mSiah2 is cell-type specific, explaining the cell specificity of repression by RevErb.
Results and Discussion
N-CoR interacts with mSiah2
To test the hypothesis that the unique amino terminus of N-CoR has a regulatory function, amino acids 1–160 were used as bait in a yeast dihybrid screen of a 17-day mouse embryo library. Eleven positive clones were identical to mSiah2, a homolog of sina (Carthew and Rubin 1990). The clones (mSiah2ΔN) began at amino acid 108 of the full-length mSiah2 protein (Fig. 1A). Polypeptides containing N-CoR amino acids 1–160 interacted with mSiah2 in yeast, whereas other regions of the amino terminus of N-CoR as well as that of SMRT did not (Fig. 1B). The in vivo interaction in yeast was recapitulated in vitro, as a GST fusion of N-CoR interacted with in vitro-translated mSiah2 (Fig. 1C). Although N-CoR amino acids 1–312 interacted with both mSiah2 and mSin3B, amino acids 1–160 of N-CoR interacted strongly with mSiah2 but not with mSin3B. N-CoR (1–312) also interacted strongly with Sina, whereas the amino terminal repression domain of SMRT did not (Fig. 1D). The ability of N-CoR to interact with mSiah2 was also observed in mammalian cells. Consistent with an in vivo interaction, transfected HA-tagged mSiah2ΔN coimmunoprecipitated with endogenous N-CoR in 293T cells (Fig. 1E). As this interaction might have occurred in vitro during processing of the cells, we also tested for in vivo interaction using a mammalian two-hybrid assay. Figure 1F shows that Gal4–N-CoR(1–160) but not Gal4–SMRT(1–483) was activated by VP16–mSiah2ΔN, consistent with in vivo interaction between N-CoR and mSiah2.
Figure 1.
In vivo and in vitro interactions between N-CoR and mSiah2. (A) Comparison of library clone (mSiah2ΔN), mSiah2, and Sina. The location of conserved Ring finger in mSiah/Sina is shown. (B) Specificity of interactions between N-CoR and mSiah2 in vivo in yeast. SMRT amino acids 1–483 are homologous to N-CoR(1030–1445). (C,D) In vitro interaction studies using GST-pulldown assay. mSiah2(108–325), Sina, and mSin3B (provided by R. Eisenman, Fred Hutchinson Cancer Research Center, Seattle, WA) were 35S-labeled and translated in vitro. The input lane contained 10% of total protein. (E) Coimmunoprecipitation of transfected HA–mSiah2ΔN with N-CoR was performed with nonimmune or rabbit serum containing antibodies raised against N-CoR amino and carboxyl termini. The immunoprecipitated protein was visualized by Western analysis with monoclonal α-HA antibody 12CA5. (F) Mammalian two-hybrid studies. Interaction between cotransfected Gal4–N-CoR(1–160) and VP16–mSiah2ΔN as indicated by activation of a luciferase reporter containing a Gal4 binding site. Gal4 DBD–SMRT(1–483) interacted negligibly with VP16–mSiah2ΔN as shown. Similar results were obtained with Gal4 DBD alone (not shown).
mSiah2 targets N-CoR for proteolytic degradation
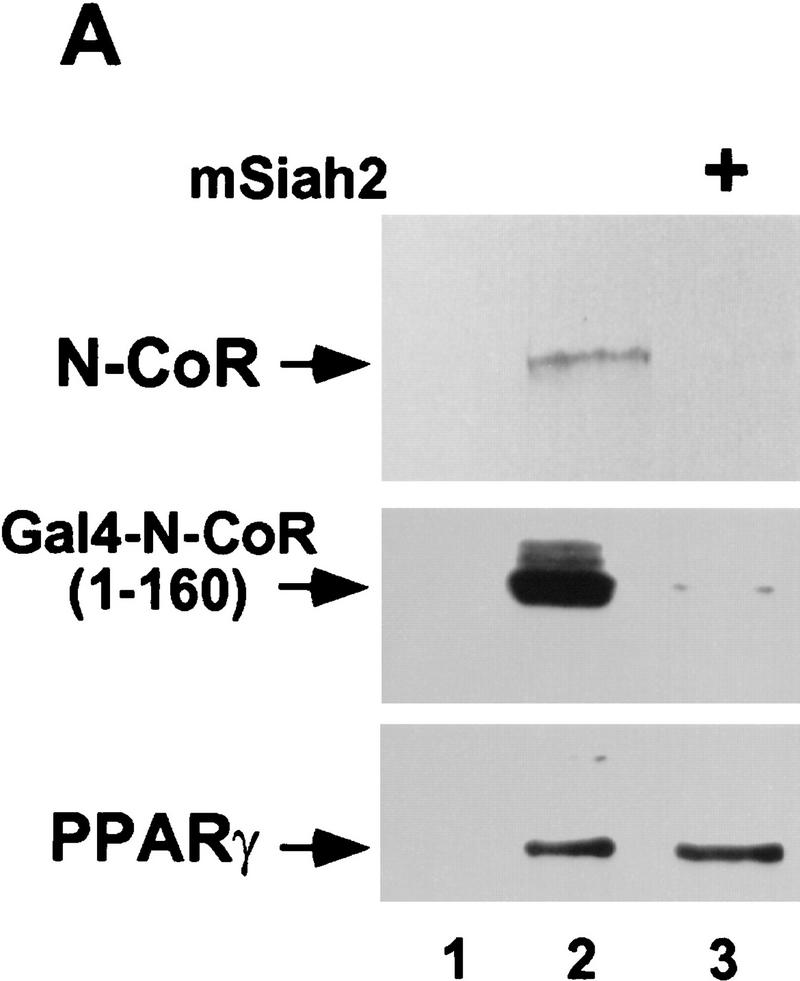
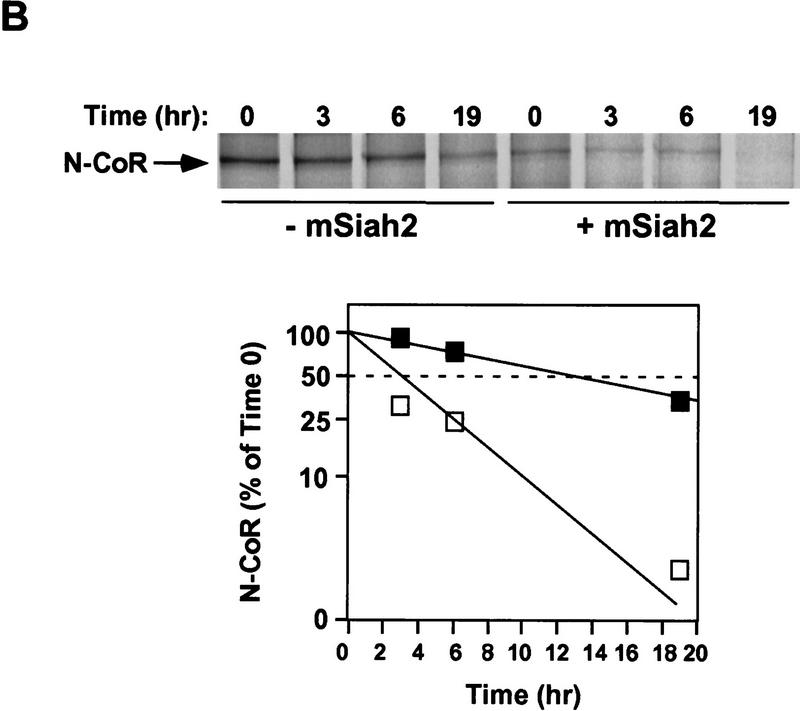
Both Sina and mSiah2 have been implicated in regulating proteasomal degradation of certain proteins to which they bind (Hu et al. 1997; Li et al. 1997; Tang et al. 1997). Transfection of mSiah2 with N-CoR in 293T cells led to near complete loss of N-CoR protein (Fig. 2A). mSiah2 also markedly reduced levels of N-CoR amino acids 1–160 fused to Gal4 DBD, indicating that this effect did not require amino acids 161–2453 of N-CoR. The specificity of mSiah2 for N-CoR was demonstrated by its lack of effect on PPARγ (Fig. 2A). Similar results were obtained with cotransfections of N-CoR and Sina (data not shown). mSiah2-induced reduction in N-CoR levels was caused by increased degradation of N-CoR (Fig. 2B). mSiah2 reduced the half-life of newly synthesized N-CoR protein from ∼14 hr to ∼3 hr.
Figure 2.
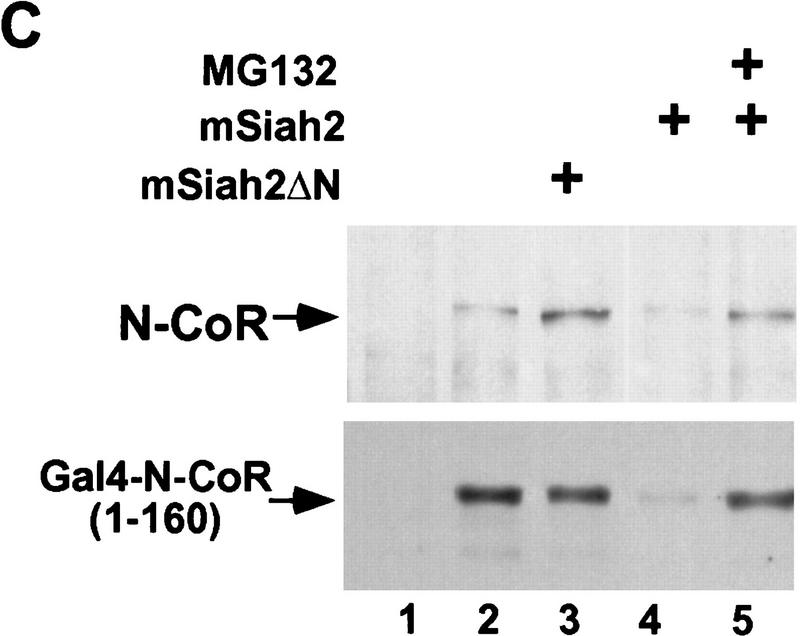
mSiah2 targets N-CoR for proteasomal degradation. (A) mSiah2 blocks expression of N-CoR in 293T cells. Vectors expressing Flag–N-CoR (20 μg, lanes 2,3), Gal4–N-CoR(1–160) (20 μg, lanes 2,3), and PPARγ2 (10 μg, lanes 2,3) or control (lane 1) were transfected into 293T cells with mSiah2 expression vector (lane 3) or control. Gal4–N-CoR and PPARγ were cotransfected. For Flag–N-CoR detection, protein extracts (200 μg) were immunoblotted with monoclonal α-Flag antibody M2. For Gal4–N-CoR and PPARγ detection, 8 μg protein extracts were immunoblotted with either antibody to Gal4 or to PPARγ. (B) Half-life of N-CoR determined by pulse chase experiment. Results were quantitated in a PhosphorImager (Molecular Dynamics), normalized to t = 0 levels, and plotted in semilog format. The level of labeled N-CoR at t = 0 was reduced in mSiah2-transfected cells, attributable to increased degradation of protein synthesized during the time of the pulse. (C) 293T cells were transfected with expression vectors for indicated proteins and either no N-CoR/Gal4–N-CoR(1–160) (lane 1; top, 300 μg protein extract; bottom 29 μg extract), or Flag–N-CoR (300 μg extract, lanes 2–5, top) or Gal4–N-CoR(1–160) (29 μg extract, lanes 2–5, bottom), and treated with or without MG132 (20 μm). Cell extracts were analyzed by immunoblot analysis.
The amino terminus of mSiah2 was required for its ability to target N-CoR protein for degradation, as mSiah2ΔN did not lead to inhibition of N-CoR expression (Fig. 2C, lane 3). This indicates an active role for a region of mSiah2 that is required for interacting with E2 ubiquitin conjugating enzymes (Tang et al. 1997). The hypothesis that mSiah2 was targeting N-CoR for proteolytic degradation via a proteasomal pathway was supported by the observation that the proteasome inhibitor MG132 (Rock et al. 1994) abolished the ability of mSiah2 to reduce levels of N-CoR and Gal4–N-CoR(1–160) proteins (Fig. 2C). This indicated that interaction with mSiah2 targeted N-CoR for degradation via a proteasomal pathway.
mSiah2 functions as an antirepressor for nuclear receptors
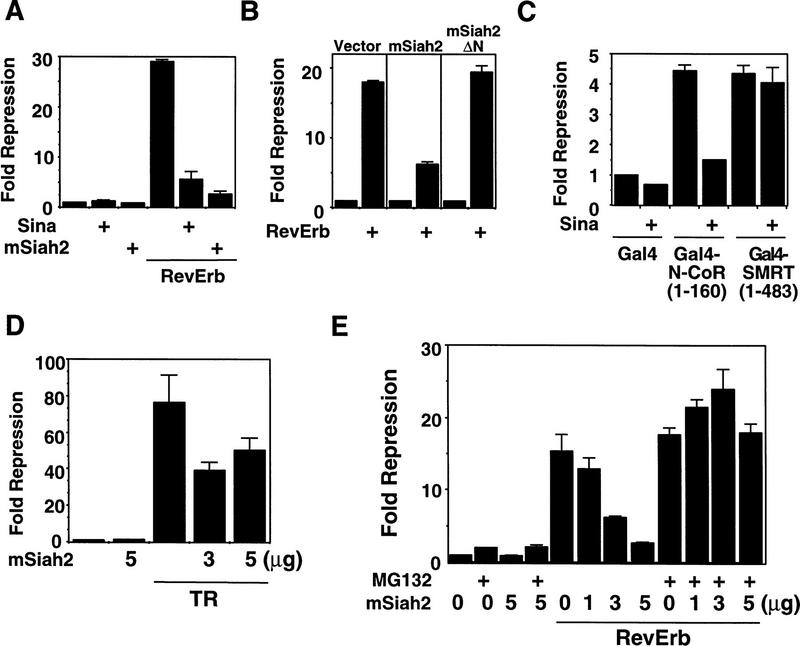
The ability of mSiah2 to reduce N-CoR protein levels by targeting N-CoR for degradation suggested that mSiah2 should block functional repression caused by N-CoR. RevErb provides an excellent means to test this hypothesis, as N-CoR is required for mediating RevErb repression in 293T cells (Zamir et al. 1997b). Both mSiah2 and Sina nearly completely abolished the repression activity of RevErb in 293T cells (Fig. 3A). In contrast, the inability of mSiah2ΔN to block RevErb activity was consistent with its inability to target N-CoR for degradation (Fig. 3B). Both Sina (Fig. 3C) and mSiah2 (not shown) also blocked repression by Gal4–N-CoR(1–160). Sina had no significant effect on repression mediated by the amino terminal repression domain of SMRT (Fig. 3C). This was expected, as the region of N-CoR that interacts with mSiah2/Sina is absent in full-length, recombinant SMRT and neither mSiah2 nor Sina interacted with this protein (data not shown). In contrast to its ability to nearly completely abolish repression by RevErb, mSiah2 blocked TR repression by ∼50% (Fig. 3C). This result is consistent with the N-CoR selectivity of RevErb, whereas TR may utilize other corepressor(s) as well.
Figure 3.
mSiah2 is a nuclear receptor antirepressor. (A) Transfection of mSiah2 and Sina inhibits RevErb repression in 293T cells. Cells were transfected with RevErb expression vector and RevDR2–SV40–luciferase reporter as described previously (Harding and Lazar 1995). (B) mSiah2ΔN does not inhibit RevErb repression in 293T cells. Five micrograms of mSiah2 or mSiah2ΔN expression plasmid was cotransfected where indicated in A and B. (C) Sina also functions as antirepressor for Gal4–N-CoR (1–160) but not Gal4–SMRT(1–483). (D) mSiah2 partially blocks TR repression in 293T cells. Cells were transfected with (Gal4 × 5)–SV40–luciferase and Gal4–TR as described previously (Zhang et al. 1997). (E) MG132 blocks mSiah2 effects on RevErb repression in 293T cells. mSiah2 inhibits RevErb repression in a concentration-dependent manner, and this is prevented by treatment with MG132.
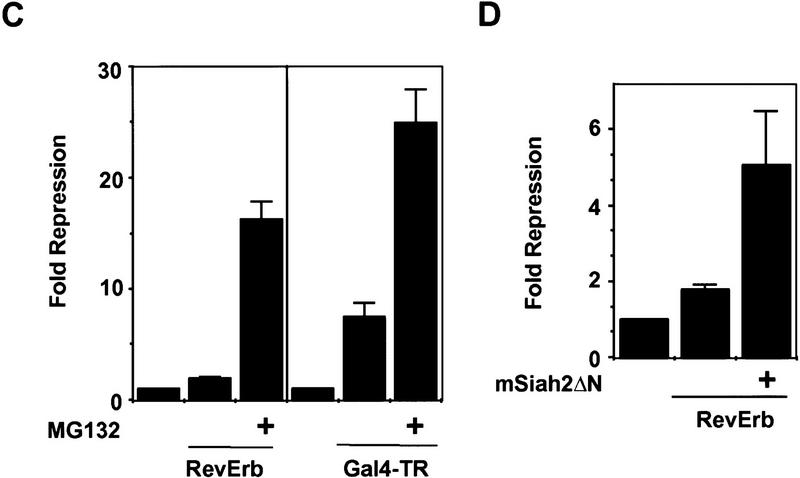
If N-CoR degradation was the mechanism by which mSiah2 blocked repression, then MG132, which prevented the mSiah2-targeted proteasomal degradation of N-CoR, should also prevent mSiah2 from functioning as an antirepressor. The ability of mSiah2 to block the repressive activity of RevErb was prevented by treatment with MG132 (Fig. 3D). The protease inhibitor had little or no effect on RevErb repression activity in the absence of mSiah2 transfection (Fig. 3D), suggesting that in 293T cells there was little or no endogenous mSiah2-mediated proteasomal degradation of N-CoR in the absence of exogenous mSiah2.
Proteolysis of N-CoR underlies cell specificity of repression
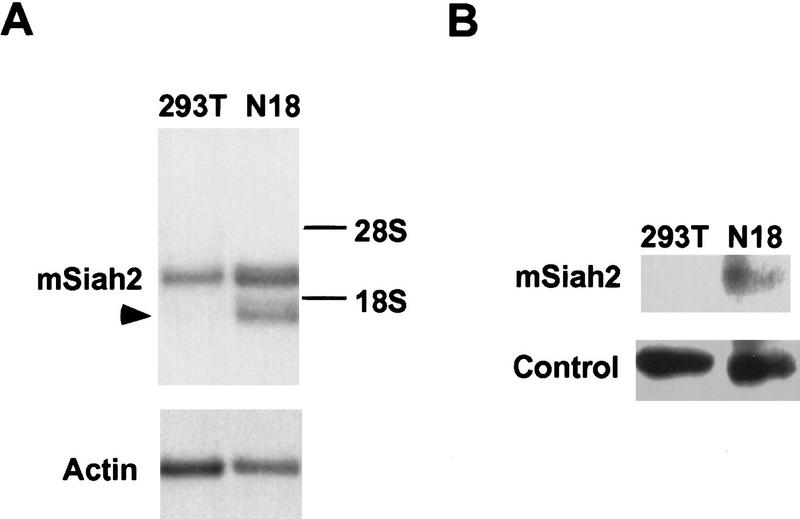
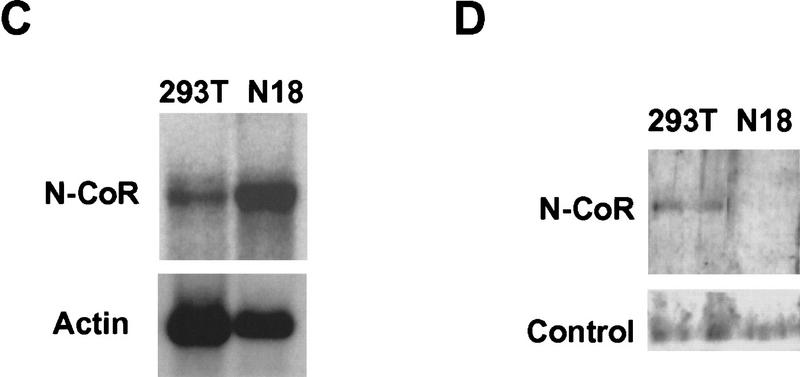
Previously, we observed that RevErb repression varies widely as a function of cell type (Harding and Lazar 1995). Because RevErb utilizes N-CoR as corepressor, we hypothesized that mSiah2 expression could play a role in this cell-specific repression. mSiah2 mRNA is most abundant in cells of the nervous system that typically express two distinct mSiah2-related mRNAs (Della et al. 1993). We found that 293T cells contained a single mSiah2 mRNA species, corresponding to the larger of the two reported species. In contrast, N18 cells contained more of this mSiah2 mRNA and, more strikingly, also contained the second mSiah2 mRNA (Fig. 4A). Moreover, only the N18 neuroblastoma cells contained detectable levels of mSiah2 protein (Fig. 4B). The N18 cells contained N-CoR mRNA at levels comparable to those in 293T cells (Fig. 4C). However, consistent with the expression pattern of mSiah2, immunoblot analysis indicated that N-CoR protein was detectable in 293T cells but undetectable in N18 cells (Fig. 4D).
Figure 4.
Cell specificity of mSiah2 and N-CoR expression. Northern analysis of mSiah2 (A) and N-CoR (C) mRNA expression. Actin hybridization is shown as loading control. (Arrowhead in A) mSiah2 mRNA species present in N18 cells and absent from 293T cells. Immunoblot analysis of mSiah2 (B) and N-CoR (D) proteins. Total protein (400 μg in B; 200 μg in D) was anlayzed, and equal loading and transfer to nitrocellulose was determined by Ponceau S stain (not shown). Rabbit antiserum used was raised against Sina, but similar results were obtained with antisera of lower titer raised against mSiah2. (B, control) Unidentified protein band (∼42 kD) detected by the Sina antiserum in both N18 and 293T cells. The mSiah2 band comigrated with the in vitro-translated standard (not shown) at ∼36 kD. (D, control) Unidentified protein band (∼215 kD) detected by the N-CoR antiserum in both N18 and 293T cells. The N-CoR band comigrated with in vitro-translated standard (not shown) at ∼270 kD.
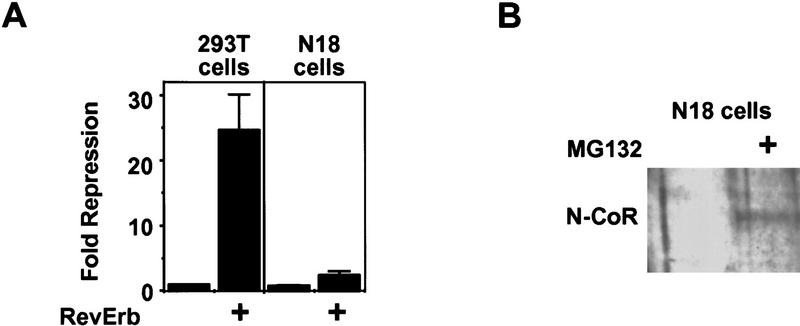
The role of regulated N-CoR proteolysis in cell specificity of repression was examined by studying RevErb and TR repression activities in the N18 cells. Unlike non-neuronal 293T cells, and correlating with the expression of mSiah2, RevErb was minimally active in N18 cells (Fig. 5A). Treatment with MG132 dramatically increased endogenous N-CoR protein levels in N18 cells (Fig. 5B), consistent with the hypothesis that endogenous mSiah2 was targeting endogenous N-CoR for proteolytic degradation. Moreover, treatment with the proteasome inhibitor increased RevErb repression activity in N18 cells by >8-fold (from 1.9- to 16.2-fold, Fig. 5C). Thus, the inability of RevErb to repress gene transcription in N18 cells was consistent with the expression of mSiah2 and the proteasome-dependent instability of N-CoR protein in these cells. In contrast to RevErb, Figure 5C shows that TR was better able to repress transcription in N18 cells (mean of 5.9-fold in two experiments). This is consistent with the ability of TR to utilize corepressors other than N-CoR, such as SMRT. Nevertheless, proteolytic regulation played a modest role in TR repression, such that MG132 increased TR repression by ∼3- to 4-fold (mean of 3.4-fold in two experiments) in N18 cells.
Figure 5.
Role of mSiah2-directed proteasomal degradation of N-CoR in nuclear receptor repression in N18 cells. (A) RevErb is a potent repressor in 293T but not in N18 cells. (B) MG132 increases N-CoR protein expression in N18 cells. Western blot performed as in Fig. 4. (C) Effects of MG132 on repression. (Left) MG132 increases RevErb repression in N18 cells; (right) TR represses in N18 cells, and MG132 potentiates TR repression. (D) mSiah2ΔN cotransfection increases RevErb repression in N18 cells. Three micrograms of mSiah2ΔN expression plasmid was transfected.
The putative role of endogenous mSiah2 in this process suggested that expression of mSiah2ΔN, which interacts with N-CoR but lacks the ability to target it for degradation, might interfere with interaction between N-CoR and endogenous mSiah2, and thus allow RevErb to function as a repressor in N18 cells. Consistent with this hypothesis, mSiah2ΔN expression in N18 cells increased RevErb repression to about five fold over basal (Fig. 5D). We note that this is not as dramatic as the effect of proteasomal inhibition. This could be because of insufficient expression of the mSiah2ΔN, but a more likely explanation is that although interference by mSiah2ΔN is competitive, interaction between endogenous mSiah2 and N-CoR is likely to be noncompetitive, because the N-CoR is degraded in a short time frame.
The proteolytic mechanism underlying the function of mSiah2 as an antagonist of N-CoR provides a novel mode of regulating transcriptional repression in mammalian cells. Sina was shown recently to bind to a sequence-specific DNA-binding protein, Tramtrack, and target it for degradation through what appears to be a similar pathway (Li et al. 1997; Tang et al. 1997). Sina function in Drosophila involves an additional protein, Phyllopod, which has no known mammalian homolog (Chang et al. 1995; Dickson et al. 1995). Our results indicate that targeting of N-CoR degradation in mammalian cells by mSiah2 is either independent of such an activity or due to an endogenous Phyllopod-like activity.
N-CoR and SMRT differ in their abilities to function with different nuclear receptors (Zamir et al. 1997b). mSiah2-directed degradation of N-CoR provides a strategic mechanism for regulating repression in a receptor-specific manner. By preventing N-CoR accumulation, expression of mSiah2 may allow the specificity of SMRT or other putative nuclear receptor corepressors to dominate in a given cell type, such as the neuroblastoma cells. This regulatory mechanism may be even more general because N-CoR also functions as a corepressor for the transcription factor Mad (Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997) and for the PLZF-RARα oncoprotein (Grignani et al. 1998; Lin et al. 1998). RevErb is a model for an N-CoR-selective nuclear receptor, and mSiah2 functions as a potent antirepressor in this context. In contrast, repression by TR is only modestly affected by the presence of ectopic or endogenous mSiah2 (in 293T and N18 cells, respectively). This is consistent with the ability of TR to utilize other corepressors, most notably SMRT. Although recombinant SMRT was not degraded by mSiah2 (data not shown), we cannot rule out the possibility that endogenous SMRT is a target of mSiah2 and that some other molecule mediates the mSiah2-insensitive repression by TR. Importantly, whereas mSiah2-mediated corepressor proteolysis prevents a subset of repression complexes from forming, this does not prevent nuclear receptor heterodimers from binding to DNA, and the presence of mSiah2 has no effect upon the maximum transcriptional rate of TR in the presence of thyroid hormone (although the fold induction caused by hormone is reduced, as the baseline is not repressed; data not shown).
Cell specificity of transcriptional regulation has been demonstrated for a number of nuclear hormone receptors, as well as other transcription factors. In most cases, the mechanism of cell specificity is unknown. The present work strongly suggests that ubiquitin-mediated proteolysis of N-CoR is an important regulator of its function. It is tempting to speculate that similar regulation of coactivator protein levels might occur. Unlike N-CoR, mSiah2 is not ubiquitously expressed in differentiated mammalian cells, but rather is restricted primarily to cells of the nervous system. Multiple nuclear receptors that function as repressors are required for normal brain function and development (Forrest et al. 1996; Hamilton et al. 1996; Harding et al. 1997; Qiu et al. 1997). Thus, regulation of N-CoR activity by targeted proteolysis is likely to modulate repression in important developmental and functional contexts.
Materials and methods
Two-hybrid screen
A yeast dihybrid screen of a 17-day mouse embryo library using N-CoR(1–160) as bait was performed as described previously (Zamir et al. 1996). The mSiah2 clone obtained multiple times in this screen was not obtained from the same library using at least 10 other baits (Zamir et al. 1996; Cheng et al. 1997).
GST pulldown assays
Input proteins were translated in vitro and labeled with TNT kit (Promega). Preparation of GST fusion proteins and the protein interaction assays were performed as described previously (Zamir et al. 1997b). Bound proteins were eluted by boiling in 30 μl of SDS–PAGE loading buffer, resolved by electrophoresis, and visualized by autoradiography. GST fusion proteins were stained with Coomassie blue to ensure equal loading.
Cell culture and transfection
Expression vectors for RevErb (Harding and Lazar 1993), TR (Zhang et al. 1997), and N-CoR (Horlein et al. 1995) have been described previously. The coding regions of Sina and mSiah2 cDNAs (Li et al. 1997) were amplified by PCR and subcloned into pCMX expression vector. Gal4 and GST fusion proteins were prepared by standard methods and sequenced. Experiments utilizing Gal4 fusion proteins used (Gal4 × 5)–SV40–luciferase reporter (Harding and Lazar 1995) except for the mammalian two-hybrid experiment in which (Gal4 × 5)–E1B–luciferase was used. 293T and N18 cells were maintained in DMEM high glucose with 10% FCS and transfected with calcium phosphate as described previously (Zhang et al. 1997) or in some cases using Lipofectamine reagent (GIBCO) according to manufacturer’s instructions. Luciferase activity was normalized to β-galactosidase activity, which served as an internal control for transfection efficiency. Figures show the results of representative experiments in which individual data points were assayed in duplicate and the mean and range of the results is shown. Each experiment was repeated two to five times.
Immunoprecipitation and pulse chase
293T cells were transfected with pCMX–Flag–N-CoR and pCMX–mSiah2, and labeled for 3.5 hr with 35S-Translabel (NEN) 16 hr later after which medium was changed and protein extracts were made at the indicated times and immunoprecipitated as described previously (Zamir et al. 1996) with anti-Flag antibody (Research Diagnostics).
Immunoblotting
Immunoblotting was performed as described previously (Xue et al. 1996) on 8–400 μg of total protein from 293T or N18 cells treated with or without 20 μm MG132, using antibodies to Gal4 DBD (Santa Cruz), Flag (Research Diagnostics), PPARγ (Xue et al. 1996), and Sina (Li et al. 1997), as described previously (Harding and Lazar 1995). N-CoR polyclonal rabbit antiserum used for Western and coimmunoprecipitation (“α-amino-terminal”) was raised against N-CoR amino acids 150–425 fused to GST. Carboxy-terminal N-CoR antibody used in coimmuniprecipitation was raised against amino acids 1944–2453 fused to GST. Neither the amino- nor carboxy-terminal N-CoR antibodies immunoprecipitated in vitro-translated SMRT (not shown). Rabbit antiserum was also raised against GST–mSiah2. For coimmunoprecipitation, subconfluent 293T cells (100-mm dish) were transfected with HA–mSiah2ΔN (20 μg). Coimmunoprecipitation was performed as described previously (Zamir et al. 1997a).
Northern analysis
Poly(A)+ RNA was purified using the mini-oligo(dT) cellulose spin column (5 Prime-3 Prime, Boulder, CO). Northern analysis was performed as described previously (Zamir et al. 1997a), with 2 μg of RNA for mSiah2 and 3.2 μg of RNA for N-CoR detection.
Acknowledgments
This work was supported by National Institutes of Health grants DK43806 and DK45586 to M.A.L. We thank Iris Zamir for help in antibody production and for helpful discussions, and Diane Merry for providing N18 cells and advice. We also thank Myles Brown and Tom Kadesch for critically reading the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lazar@mail.med.upenn.edu; FAX (215) 898-5408.
References
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Kohne AC, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Rubin GM. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- Chang HC, Solomon NM, Wassarman DA, Karim FD, Therrien M, Rubin GM, Wolff T. phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell. 1995;80:463–472. doi: 10.1016/0092-8674(95)90497-2. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Cheng X, Reginato MJ, Andrews NC, Lazar MA. The transcriptional integrator CREB-binding protein mediates positive crosstalk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della NG, Senior PV, Botwell DD. Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina) Development. 1993;117:1333–1343. doi: 10.1242/dev.117.4.1333. [DOI] [PubMed] [Google Scholar]
- Dickson BJ, Dominguez M, van der Straten A, Hafen E. Control of Drosophila photoreceptor cell fates by phyllopod, a novel nuclear protein acting downstream of the Raf kinase. Cell. 1995;80:453–462. doi: 10.1016/0092-8674(95)90496-4. [DOI] [PubMed] [Google Scholar]
- Forrest D, Hanebuth E, Smeyne RJ, Everds N, Stewart CL, Wehner JM, Curran T. Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor beta; evidence for tissue-specific modulation of receptor function. EMBO J. 1996;12:3006–3015. [PMC free article] [PubMed] [Google Scholar]
- Graupner G, Wills KN, Tzukerman M, Zhang X-k, Pfahl M. Dual regulatory role for thyroid-hormone receptors allows control of retinoic-acid receptor activity. Nature. 1989;340:653–656. doi: 10.1038/340653a0. [DOI] [PubMed] [Google Scholar]
- Grignani F, DeMatteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Grignani F, Lazar MA, Minucci S, Pelicci PG. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WY, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, vanBerkel V, Birren BW, Kruglyak L, Lander ES. Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Harding HR, Lazar MA. The orphan receptor Rev-ErbAα activates transcription via a novel response element. Mol Cell Biol. 1993;13:3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Atkins GB, Jaffe AG, Seo WJ, Lazar MA. Transcriptional activation and repression by RORα, an orphan nuclear receptor required for cerebellar development. Mol Endocrinol. 1997;11:1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–348. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen T-M, Soderstrom M, Laherty CD, Torchia J, Yuang W-M, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu G, Zhang S, Vidal M, Baer JL, Xu T, Fearon ER. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin–proteasome pathway. Genes & Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li Y, Carthew RW, Lai ZC. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao H-Y, Chakvarkti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Pereira FA, DeMayo FJ, Lydon JP, Tsai SY, Tsai M-J. Null mutation of mCOUP–TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes & Dev. 1997;11:1925–1937. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–775. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes & Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Shibata H, Nawaz Z, Tsai SY, O’Malley BW, Tsai M-J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997a;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- Shibata H, Spencer TE, Onate SA, Jenster G, Tsai SY, Tsai MJ, O’Malley BW. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Rec Prog Horm Res. 1997b;52:141–164. [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Xue J-C, Schwarz EJ, Chawla A, Lazar MA. Distinct stages in adipogenesis revealed by retinoid inhibition of differentiation after induction of PPARγ. Mol Cell Biol. 1996;16:1567–1575. doi: 10.1128/mcb.16.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I, Harding HP, Atkins GB, Horlein A, Glass CK, Rosenfeld MG, Lazar MA. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with different repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I, Dawson J, Lavinsky RM, Glass CK, Rosenfeld MG, Lazar MA. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci. 1997a;94:14400–14495. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I, Zhang J, Lazar MA. Stoichiometric and steric principles govering repression by nuclear hormone receptors. Genes & Dev. 1997b;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zamir I, Lazar MA. Differential recognition of liganded and unliganded thyroid hormone receptor by retinoid X receptor regulates transcriptional repression. Mol Cell Biol. 1997;17:6887–6897. doi: 10.1128/mcb.17.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]