Abstract
Allelic exclusion in κ light-chain synthesis is thought to result from a feedback mechanism by which the expression of a functional κ light chain on the surface of the B cell leads to an intracellular signal that down-regulates the V(D)J recombinase, thus precluding rearrangement of the other allele. Whereas such a feedback mechanism clearly plays a role in the maintenance of allelic exclusion, here we provide evidence suggesting that the initial establishment of allelic exclusion involves differential availability of the two κ alleles for rearrangement. Analysis of κ+ B-cell populations and of individual κ+ B cells that have rearranged only one allele demonstrates that in these cells, critical sites on the rearranged allele are unmethylated, whereas the nonrearranged allele remains methylated. This pattern is apparently generated by demethylation that is initiated at the small pre-B cell stage, on a single allele, in a process that occurs prior to rearrangement and requires the presence in cis of both the intronic and 3′ κ enhancers. Taken together with data demonstrating that undermethylation is required for rearrangement, these results indicate that demethylation may actually underly the process of allelic exclusion by directing the initial choice of a single κ allele for rearrangement.
Keywords: Methylation, allelic exclusion, immunoglobulin, B lymphocytes
The generation of the vast repertoire of antibody molecules during B-cell differentiation is complex and is based on a program of gene-specific rearrangement events (for review, see Lewis 1994; Rajewsky 1996). Ultimately, this program leads to the expression of a single heavy chain (μ) and a single light chain (either κ or λ) in each mature B cell. All of these rearrangement events are mediated by conserved recombination signal sequences (RSSs) on the DNA and are carried out by a single set of enzymes (RAG-1 and RAG-2) (Schatz et al. 1989; Oettinger et al. 1990; for review, see Gellert 1996). In spite of the sharing of the enzymes and RSSs, recombination takes place in a programmed, sequential manner that involves the ordered selection of the proper gene loci as well as a mechanism for choosing only one allele at a time. This poses an interesting problem in the regulation of the rearrangement process.
Many studies of the regulation of rearrangement have focused on the mechanisms underlying the ordered selection of the proper gene loci. These studies have identified enhancer-mediated changes in chromatin structure as well as changes in the extent of DNA methylation. At various stages of B-cell development, different trans-acting proteins clearly are involved in rendering the μ, κ, and λ immunoglobulin loci available to the recombination apparatus. Responding to these trans-acting proteins, enhancers and other cis-regulatory sequences mediate local changes in chromatin structure that underlie increased accessibility to the recombinase (Stanhope-Baker et al. 1996). For example, experiments in transgenic mice have clearly shown that enhancer elements can indeed mediate changes in chromatin structure (Jenuwein et al. 1993; Forrester et al. 1994), and in vivo targeted deletions of these sequences strongly repress rearrangement (Chen et al. 1993; Serwe and Sablitzky 1993; Takeda et al. 1993; Xu et al. 1996; Gorman et al. 1996).
DNA methylation plays an important role in regulating transcription and chromatin structure in a number of genes (for review, see Mostoslavsky and Bergman 1997). Methylation has also been correlated with immunoglobulin gene expression; both the μ and κ chains have been shown to be heavily modified in most tissues, but unmethylated in mature B cells (Mather and Perry 1983; Storb and Arp 1983; Kelley et al. 1988; Goodhardt et al. 1993). Using a transfection assay, we previously demonstrated that this pattern is brought about by stage-specific demethylation events that are directed by local cis-acting elements. Demethylation of sequences adjacent to the Jκ region, for example, can be induced by the presence in cis of a complex DNA signal containing enhancer and matrix attachment region (MAR) sequence elements (Lichtenstein et al. 1994). Additionally, in vitro experiments have demonstrated that methylation inhibits rearrangement (Engler et al. 1991; Hsieh and Lieber 1992). These experiments suggest an important role for demethylation in the regulation of κ gene rearrangement. However, the in vivo role of demethylation in the process of B-cell development has not yet been clarified.
Until now, the random choice of one allele for rearrangement (at a given locus) has not been addressed in studies of chromatin structure or methylation, but rather in experiments that have demonstrated the existence of a feedback mechanism by which the expression of a functional κ light chain on the surface of the B cell leads to an intracellular signal that down-regulates the V(D)J recombinase, thus precluding rearrangement of the other allele. The widely accepted theory is that (for a given immunoglobulin gene locus: μ, κ, or λ) inefficiency in the rearrangement process allows one allele to rearrange prior to the other (Coleclough et al. 1981). Then, allelic exclusion results from the feedback down-regulation mechanism discussed above. Whereas such a feedback mechanism clearly plays a role in the maintenance of allelic exclusion, here we provide evidence suggesting that the initial establishment of allelic exclusion involves differential availability of the two κ alleles for rearrangement. We demonstrate that demethylation of the κ gene occurs initially on only one allele in each B cell. We also define two other aspects of the allele-specific demethylation that are critical to its role in allelic exclusion by demonstrating that, in vivo, demethylation takes place prior to and independently of rearrangement. We also demonstrate that the demethylation is driven, in vivo, by the intronic and 3′ enhancer sequences that were implicated previously (Lichtenstein et al. 1994; Kirillov et al. 1996) in driving the process in cell lines. These results, when taken together with direct experiments demonstrating that methylation inhibits rearrangement (Engler et al. 1991; Hsieh and Lieber 1992), suggest that demethylation itself may be the rate-limiting step in the rearrangement process. Thus, our results strengthen the model in which changes in DNA methylation play a major role in controlling the program of sequential rearrangement events. Additionally, our data indicating that the demethylation is monoallelic in individual B cells suggest a novel primary basis for allelic exclusion.
Results
Allelic differences in Igκ gene methylation status in normal B cells
To evaluate Igκ methylation patterns in vivo, κ-expressing B cells were isolated from adult mouse bone marrow or spleen and their DNA was subjected to methylation analysis (see map, Fig. 1). DNA was first digested with HindIII, which allows one to visualize both the germ-line (discrete 2.7-kb band) and rearranged alleles. Further digestion with the methyl-sensitive restriction enzyme HhaI revealed that (in a large fraction of DNA molecules) sites located in the junction (J) region upstream of the intronic enhancer are unmethylated, whereas the germ-line band is mostly methylated (Fig. 1a, lanes 1,2). Next, we examined a 1.0-kb BglII–HindIII restriction fragment at the 3′ end of the J cluster that is present in both rearranged and germ-line κ alleles. Analysis with HhaI showed clearly that ∼50% of the gene copies are indeed unmethylated (Fig. 1a, lanes 3,4).
Figure 1.
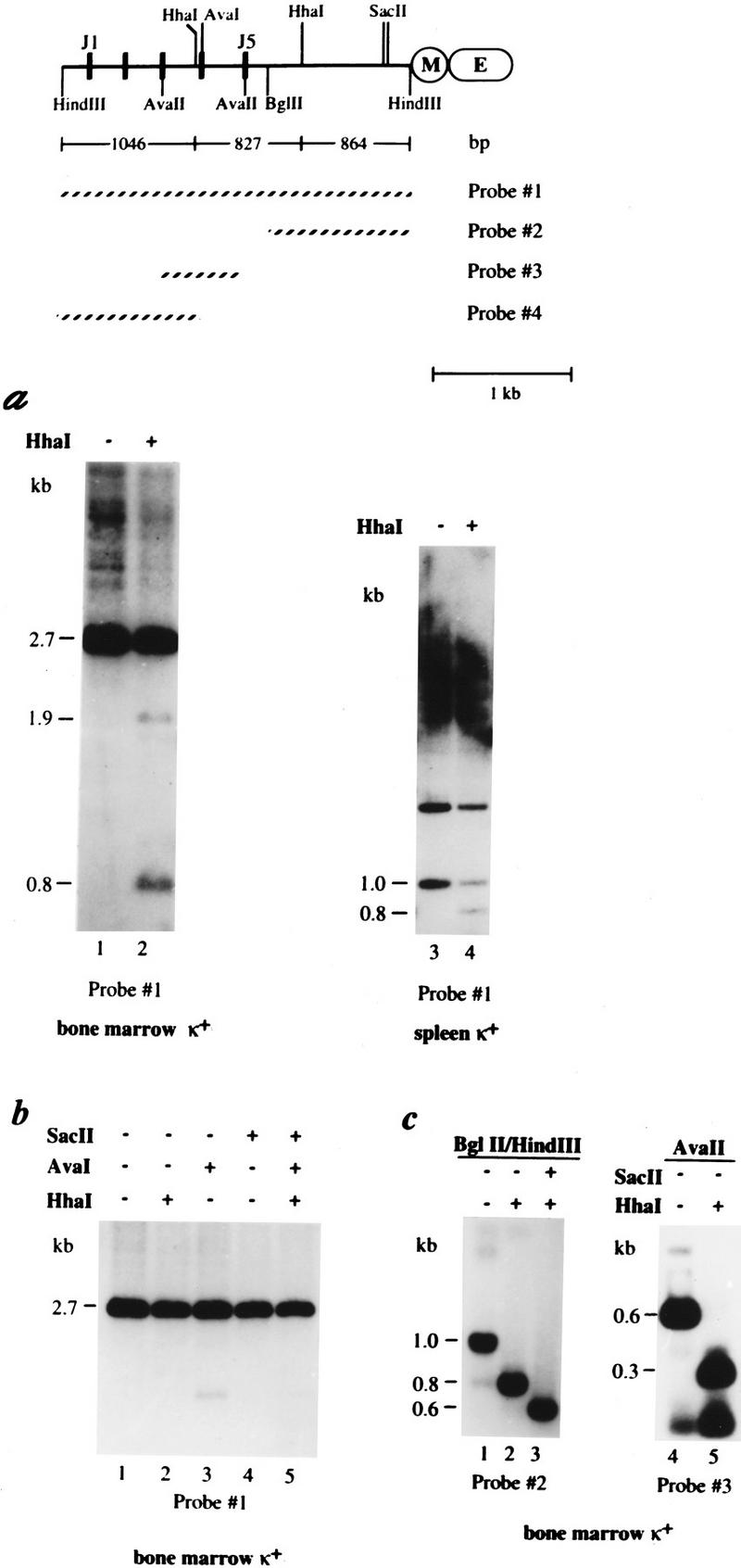
In vivo methylation pattern of the κ chain gene. A map of the κ chain gene locus shows the J segments, the intronic MAR (M) and enhancer (E) sequences, the relevant restriction sites, fragment sizes, and the probes used in these experiments. (a) DNA from bone marrow (lanes 1,2) or spleen (lanes 3,4) κ+ cells derived from wild-type mice was digested with HindIII (lanes 1–2) or HindIII–BglII (lanes 3–4) and then tested for methylation by additional restriction with HhaI (+). These samples were then subjected to gel electrophoresis and analyzed by blot hybridization using probe 1. It should be noted that equal amounts of DNA were loaded in both lanes 1 and 2 as indicated by ethidium bromide staining (data not shown). When cut with HindIII the bone marrow DNA shows a germ-line band (2.7 kb) as well as numerous rearranged bands (seen as a smear). Full undermethylation results in 1046-, 827-, and 864-bp fragments, but partial digests can also be seen on the gel (1.6 and 1.9 kb). It should be noted that the appearance of a 1.9-kb band indicates that a small fraction of the germ-line allele is also unmethylated at some sites. Similar results were seen when DNA from κ+ spleen cells was analyzed in the same manner (data not shown). Digestion with HindIII–BglII concentrates all κ gene copies into a single band (1.0 kb), which if unmethylated is reduced to 0.8 kb when cleaved with HhaI. This site is ∼50% methylated in normal κ+ cells (lane 4). In this gel, the 1.0-kb band has a lower intensity than the 1.7-kb band, mainly because it hybridizes to a smaller part of the probe and because gel transfer is not as efficient at this molecular weight (as shown by analysis of HindIII–BglII digests of non-B-cell DNA). The upper (1.7-kb) band seen in this gel represents the 5′ HindIII–BglII fragment derived from germ-line copies of the gene. (b) DNAs from κ+ bone marrow cells were treated with HindIII and further digested with the methyl-sensitive restriction enzymes, HhaI, AvaI, or SacII and analyzed as in a. (c) Because the degree of undermethylation on the rearranged κ alleles could not be determined quantitatively from the data shown in Figure 1a, DNA was extracted (Quiagen Kit) from the smear region (>2.7 kb; a, lane 1), further digested with either BglII (lanes 1–3) or AvaII (lanes 4,5) plus methyl-sensitive restriction enzymes SacII or HhaI (indicated by a + or − at the top), and analyzed using either probe 2 or probe 3, as indicated. The resulting BglII–HindIII fragment (region covered by probe 2) yielded a 0.8- or 0.6-kb band when the respective HhaI and SacII sites are unmethylated. The AvaII fragment (region covered by probe 3) was cleaved by HhaI to yield a 0.3-kb band if unmethylated.
To further assess the relationship between κ chain gene rearrangement and methylation, κ+ B-cell DNA was analyzed using several different methyl-sensitive restriction enzymes (see map). The unrearranged germ-line copies (2.7-kb Hind III band) are strikingly resistant to digestion at five individual CpG sites, indicating that most of these molecules are highly methylated (Fig. 1b). In contrast, the rearranged HindIII-treated κ alleles, extracted from the smear region (>2.7 kb) on the original agarose gel (Fig. 1a, lane 1) and further digested with methyl-sensitive restriction enzymes, proved to be completely unmethylated over the entire Jκ and intronic region (Fig. 1c). This is most clearly seen in the complete digestion by both SacII and HhaI of the 1.0-kb BglII–HindIII fragment (Fig. 1c, lanes 1–3). Methyl sensitivity is also observed in the HhaI site near J4 where most of the 0.6-kb AvaII band disappears upon HhaI digestion (Fig. 1c, lanes 4,5). These results demonstrate the presence of rearranged unmethylated alleles and germ-line methylated alleles in κ+ B cells. Since each cell expresses κ light-chain protein on its surface, each must have at least one rearranged κ allele. Thus, these experiments demonstrate clearly that in a majority of κ+ B cells, a fully methylated germ-line copy coexists with an undermethylated, productively rearranged κ allele.
Single-cell analysis of allelic differences in Igκ gene methylation
We have developed an assay to directly determine the methylation status of the two κ alleles present in individual κ+ B cells. In order to accomplish this we first isolated the Mus spretus κ gene and identified polymorphisms with respect to the Mus musculus allele (C57BL/6). Thus, in F1 progeny from a cross between M. spretus and M. musculus, it is possible to distinguish the two alleles. Spleen cells were harvested from F1 progeny and κ+ B lymphocytes were sorted as single cells directly into PCR tubes using a fluorescence-activated cell sorter (FACS). Genomic DNA from these single cells was subjected to digestion by the methyl-sensitive restriction enzyme AvaI to analyze the methylation status of the AvaI site in J4 (Fig. 2a). After AvaI digestion, oligonucleotide primers flanking the AvaI site were used for nested PCR (primers 1–4; see Fig. 2a). Because these primers will not amplify DNA that has been AvaI digested, only DNA molecules that are methylated at the AvaI site will amplify. We will refer to this methyl-sensitive restriction enzyme-dependent PCR assay as MSRE–PCR.
Figure 2.
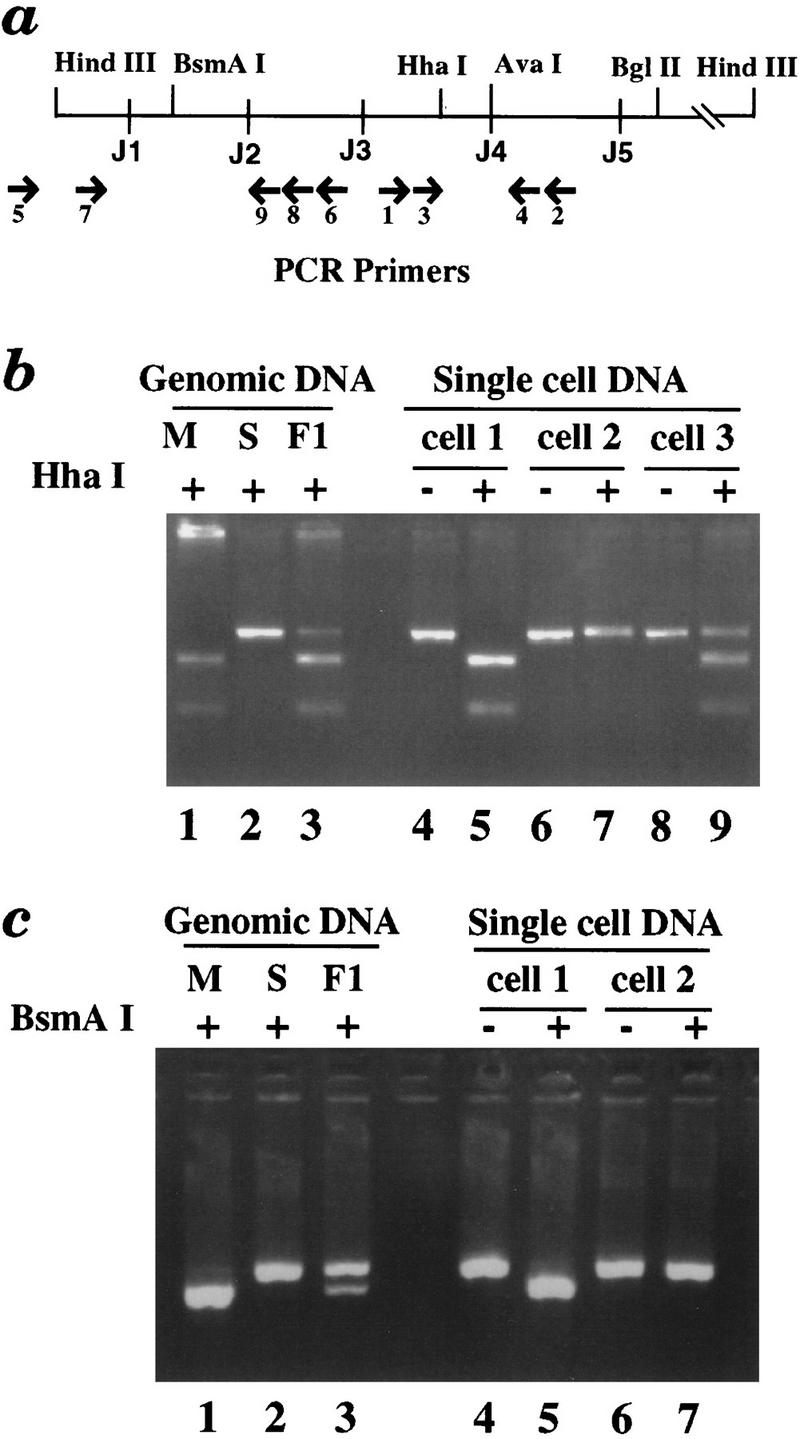
MSRE–PCR analysis of the κ gene in single cells. (a) A map of the Jκ region showing the location of primers used for MSRE–PCR analysis and relevant restriction enzyme sites. AvaI was used to detect DNA methylation, whereas the polymorphic HhaI and BsmAI were employed to distinguish between the two κ alleles. (b) (Lanes 1–3) As a control, genomic DNAs from M. musculus, M. spretus, or F1 mice were PCR amplified with primers P3 and P4, digested with HhaI (used here because of a polymorphism, not because of its methyl-sensitivity), and analyzed on a 4% agarose gel. The M. musculus gene is digested, the M. spretus gene does not cut, and the F1 DNA yields a product that shows 50% digestion. (Lanes 4–7) DNA from two single κ+ B cells was digested with AvaI and PCR amplified. (The secondary PCR primers were P1 and P2.) Uncut PCR products are in lanes 4 and 6; HhaI digested PCR products are in lanes 5 and 7. Cells 1 and 2 are monoallelic (M. musculus and M. spretus allele, respectively). Most κ+ B cells analyzed were monoallelic. (Lanes 8, 9) The same analysis of a single midbrain cell (cell 3). Both alleles were amplified from this cell and in most of the control cells analyzed. (c) (Lanes 1–3) Control M. musculus, M. spretus, or F1 genomic DNAs were PCR amplified by primers P7 and P9, digested with BsmAI, and analyzed on a 4% agarose gel. (Lanes 4–7) Tertiary PCR products derived from cells 1 and 2 (from b) using primers P7 and P9. The tertiary amplification signal is dependent on the germ-line-specific primary PCR (using primers P5 and P6). Uncut PCR products are in lanes 4 and 6, and corresponding BsmAI-digested PCR products are in lanes 5 and 7. Cell 1 (shown previously to have a methylated M. musculus allele) has an unrearranged M. musculus allele; cell 2 (shown previously to have a methylated M. spretus allele) has an unrearranged M. spretus allele. All together, 15 individual cells with PCR products from both regions were analyzed.
Using MSRE–PCR, we obtained PCR products from 80% of sorted κ+ B cells. To determine which alleles have been PCR amplified, we took advantage of a polymorphism between the M. spretus and M. musculus κ genes that leads to the presence of an HhaI site in the M. musculus allele and absence of this site in the M. spretus allele. Whereas HhaI is itself a methyl-sensitive restriction enzyme, here it is used after PCR amplification to distinguish the two alleles. PCR products from 86% (89/103) of the κ+ B cells revealed only one allele in this assay (Table 1). Representative analyzed PCR products are shown in Figure 2b. Half the alleles are maternally (M. musculus) derived and half the alleles are paternally (M. spretus) derived. The small number of biallelic PCR products likely derive from instances in which the AvaI enzyme has failed to cut an unmethylated allele.
Table 1.
Allelic methylation of the κ gene in single cells
| Cells with monoallelic PCR product | ||
|---|---|---|
| κ+ B cells | Nonlymphoid cells | |
| (+AvaI) | (−AvaI) | (+AvaI) |
| 89/103 (86%) | 4/12 (33%) | 6/39 (15%) |
Single-cell PCR using primer sets P1–P4 was carried out as described in Materials and Methods with (+) or without (−) pretreatment with AvaI, and the products were digested with HhaI to determine the contribution of the M. spretus and M. musculus alleles (examples shown in Fig. 2b). It should be noted that the monoallelic products (15%) seen in the nonlymphoid control cells are probably due to inefficient PCR reactions. In the absence of AvaI, B-cell DNA yields a high background level (33%) of monoallelic products primarily because of rearrangements to J4 or J5, which remove the primer sequences.
We next performed a variety of control experiments to validate our results and show that this assay is efficient enough to detect both alleles in each cell. When AvaI digestion is omitted, for example, only 33% (4/12) of κ+ cells yield a monoallelic PCR product (Table 1). These monoallelic products reflect either inefficiency in the PCR or the small fraction of κ+ B cells that have used J4 or J5 for rearrangement. An additional control involved analyzing nonlymphoid cells. When analyzed in a similar manner with AvaI digestion preceding PCR, 85% (33/39) of these cells yielded biallelic PCR products; only 15% were monoallelic (Table 1). The 15% of products that are monoallelic probably reflect inefficiency in the PCR, rather than undermethylation in these nonlymphoid cells. These control experiments strengthen the conclusion that one allele is methylated and one allele is unmethylated in many individual κ+ B cells.
In a second series of experiments, we expanded the MSRE–PCR single-cell assay to include a determination of which allele is rearranged. After AvaI digestion, PCR primers designed to amplify only germ-line (unrearranged) DNA upstream of J3 (primers 5 and 6) were used in addition to the primers flanking the AvaI site (primers 1 and 2) that we used in the previous experiments. In subsequent nested PCR reactions, the two regions were amplified separately. We then determined the parental origin of the resultant PCR products by digesting them with restriction enzymes whose sites are polymorphic between M. spretus and M. musculus. [There is a polymorphic BsmAI site in the region amplified by the primers (5 and 6) that only detect the germ-line configuration]. Figure 2c shows representative BsmAI digested PCR products. All 15 times we observed a PCR product for both regions, the monoallelic product reflecting methylation of the AvaI site was associated with a monoallelic germ-line product from the same allele. Thus, these single-cell experiments demonstrate that the methylated allele is the germ-line allele.
The kinetics of κ chain demethylation in vivo
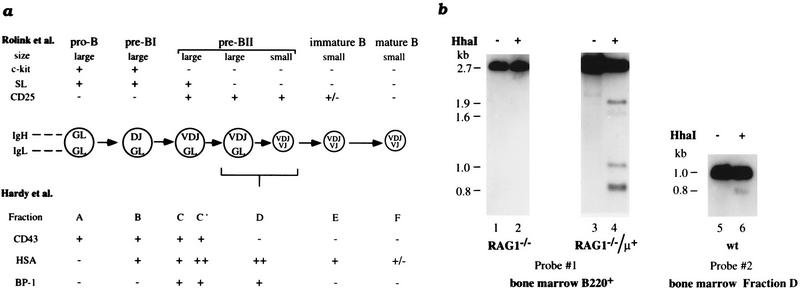
We have analyzed populations of B cells at various stages of B-cell development to determine the timing of κ demethylation relative to rearrangement. The κ locus is methylated in RAG-1−/− mouse B220+ bone marrow cells (Fig. 3b, lanes 1,2), which are known to be arrested at the pro-B cell stage of differentiation (fraction C′, Fig. 3a). We also examined IL-7 dependent cultured bone-marrow-derived cells (Rolink et al. 1991), which represent one of the earliest stages in B-cell development (fraction B, Fig. 3a), and observed full methylation of the κ locus (data not shown). In contrast, analysis of bone marrow cells derived from Rag-1−/− mice carrying a prerearranged μ transgene (Rag-1−/−/μ+; Spanopoulou et al. 1994), demonstrated clear-cut κ gene undermethylation (Fig. 3b, lanes 3,4), suggesting that demethylation occurs for the first time in these pre-B cells (fraction D, Fig. 3a). This was, in fact, confirmed by the observation of Jκ undermethylation in normal pre-B cells isolated from bone marrow (Fig. 3b, lanes 5,6). The low level of demethylation observed is expected as fraction D is made up of a mixed population containing both large and small pre-B cells and κ rearrangement is restricted primarily to the smaller cells (ten Boekel et al. 1995). These kinetic studies demonstrate that demethylation of the Jκ region occurs at about the same developmental stage in which the first κ rearrangement events take place (Hardy et al. 1991; Ehlich et al. 1993; ten Boekel et al. 1995).
Figure 3.
κ chain demethylation in B-cell development. (a) Schematic diagram of murine B-cell development (adapted from Nutt et al. 1997). The different developmental stages of B lymphopoiesis are shown together with their characteristic cell surface markers, which are used for classification according to Rolink et al. (1994) (top) or Hardy et al. (1991) (bottom). As the correlation between the two classification systems is not straightforward in all aspects, the reader is referred to the original literature for details. (b) DNAs from bone marrow B220+ cells at different stages of B-cell development (see the diagram) were tested for methylation by digestion with HhaI (+). B-cell DNA from Rag1−/− mice (arrested at the pro-B stage) (Mombaerts et al. 1992) and from Rag1−/− mice carrying a μ transgene (arrested at the small pre-B stage) (Spanopoulou et al. 1994) was digested by HindIII with (+) or without (−) HhaI. DNA from wild-type (wt) fraction D cells (pre-B stage, Hardy et al. 1991; Ehlich et al. 1993) was analyzed by digestion using HindIII–BglII with (+) or without (−) HhaI.
κ chain gene allelic demethylation is independent of rearrangement
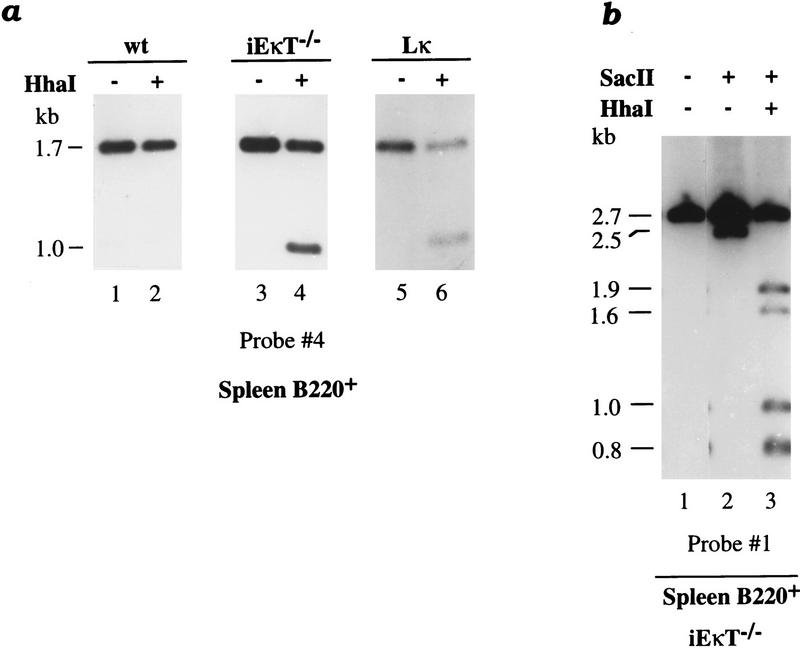
To determine whether demethylation takes place before or after rearrangement, we studied κ gene methylation patterns in cells derived from mice in which rearrangement of the endogenous locus is prevented. In iEκT−/− mice, where both copies of the κ intronic enhancer are replaced by the neomycin resistance (neo) gene, rearrangement of the κ locus is strongly inhibited and, as a result, immunoglobulin-producing cells express the λ light chain exclusively (Takeda et al. 1993). It is likely that this dominant effect is mainly caused by the presence of the foreign neo gene, since this bacterial sequence has been shown to inhibit rearrangement even when placed at other positions in this gene (Takeda et al. 1993). We have isolated B220+ splenic B cells from these animals and analyzed their methylation pattern at the endogenous κ locus. Despite the fact that both alleles remain in the germ-line configuration in these cells, ∼50% of the κ gene copies undergo demethylation at HhaI sites in the J region (Fig. 4a, lanes 3,4). In addition, alleles that are unmethylated at the SacII site are also completely unmethylated at both HhaI sites (Fig. 4b, lane 3). These multiple demethylation events take place although ∼50% of the κ molecules remain highly modified, indicating that in vivo demethylation mainly behaves cooperatively and occurs on one allele at a time.
Figure 4.
κ chain demethylation and rearrangement. (a) B220+ spleen cell DNA from wild-type (wt) mice (lanes 1,2), from mice carrying a neomycin resistance gene replacing the κ intronic enhancer [iEκT−/−] (Takeda et al. 1993) (lanes 3,4), and from mice carrying a prerearranged κ-transgene [Lκ] (Sharpe et al. 1991; Betz et al. 1994) (lanes 5,6) was digested by HindIII–BglII with (+) or without (−) HhaI. Blot hybridization was performed with probe 4 (see legend to Fig. 1) to visualize the endogenous κ genes exclusively. Both the endogenous κ alleles in iEκT−/− and Lκ mice were found to be fully methylated in non-B cells (data not shown). (b) B220+ spleen cell DNA taken from iEκT−/− mice was digested by HindIII (lane 1) with (+) or without (−) SacII (lane 2) or SacII–HhaI (lane 3) and analyzed by blot hybridization using probe 1. Note that although <50% of the κ alleles have the SacII site unmethylated (2.5 kb), all of these molecules are also unmethylated at the HhaI sites, suggesting that the demethylation is of an allelic nature.
Endogenous κ gene rearrangement is also prevented in mice carrying a prerearranged κ transgene (Betz et al. 1994). This effect presumably occurs because the readily synthesized transgene product constitutively turns off RAG-1 and RAG-2 expression in the B-cell lineage. We analyzed the methylation patterns of the endogenous alleles present in these mice and found that ∼50% of these κ alleles underwent demethylation (Fig. 4a, lanes 5,6).
Whereas these results indicate that demethylation can take place prior to rearrangement, we still wanted to rule out the possibility that the rearranged state itself can trigger demethylation. To this end, we examined heterozygous mice carrying a single VκJκ prerearranged gene (3–83κi) permanently targeted (via homologous recombination) into the normal endogenous κ locus (Fig. 5a) (Pelanda et al. 1996). It should be noted that in these animals, the rearranged construct is actually under the control of the same endogenous cis-acting DNA elements that normally regulate κ chain expression. As shown in figure 5b, the allele that underwent homologous recombination undergoes ∼40%–60% demethylation in both bone marrow and spleen B220+ cells, and thus appears to behave in the same manner as κ gene copies in normal B cells (Fig. 1). Furthermore, in light of the fact that many targeted molecules remain modified, it is clear that a rearranged configuration itself is not sufficient to bring about demethylation. Similar results were also obtained in homozygous animals and in a mouse line containing a different prerearranged κ allele, Vκ8R (Luning-Prak and Weigert 1995; data not shown).
Figure 5.
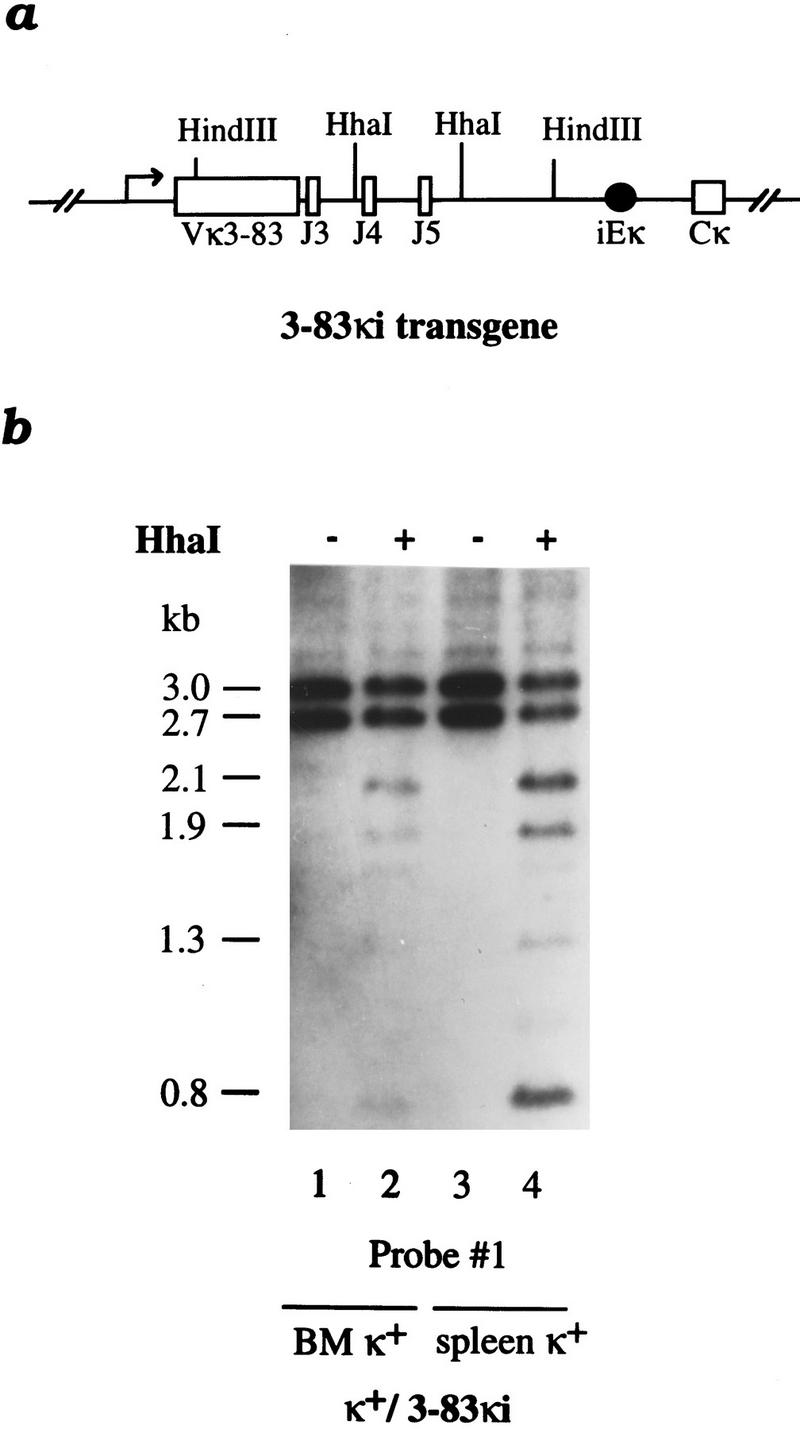
Demethylation in a targeted prerearranged κ chain gene. (a) A map shows the Vκ3–83 targeted gene including the intronic enhancer (iEκ) and the constant region (Cκ) (Pelanda et al. 1996). Note that the rearranged construct is under the control of the same endogenous cis-acting DNA elements that normally regulate κ chain gene expression. (b) DNA from κ+ bone marrow (BM) (lanes 1,2) or spleen (lanes 3,4) cells of mice carrying a rearranged 3–83κ immunoglobulin light gene replacing one endogenous Igκ allele (κ+/3–83κi) (Pelanda et al. 1996) was digested by HindIII with (+) or without (−) HhaI and analyzed by blot hybridization using probe 1. Both the wild-type germ-line copy (2.7 kb) and the rearranged allele (3.0 kb) can be seen on the gel. The HhaI digestion products include the 2.1-kb band that is derived exclusively from the rearranged allele and the 0.8-kb band that comes from both alleles. Similar results were obtained with DNA from homozygous 3–83κi mice (data not shown).
Both κ enhancers are needed for demethylation in vivo
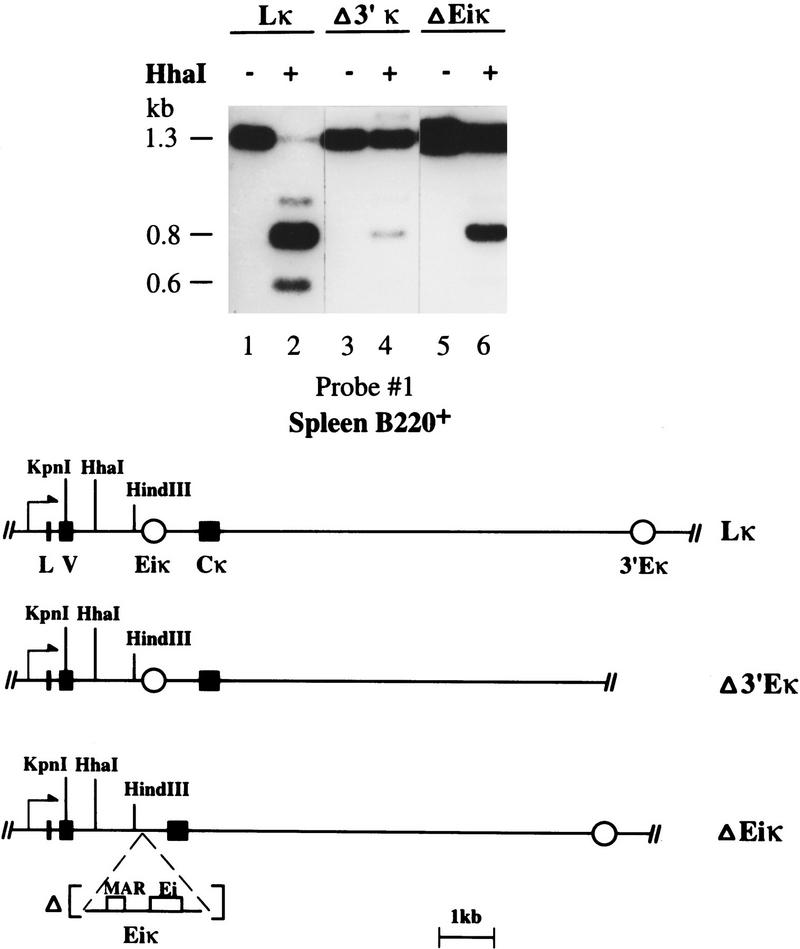
A recent series of mouse knockout experiments has demonstrated that both the intronic and 3′ enhancers play a role in κ rearrangement, since a single deletion of either element (Gorman et al. 1996; Xu et al. 1996) partially inhibits this process. In parallel, we have shown by transfection experiments that both the intronic and 3′ enhancer sequences can individually induce κ demethylation in B-cell lines (Lichtenstein et al. 1994; Kirillov et al. 1996). To assess the precise role of these enhancers in the demethylation process in vivo, we analyzed a series of transgenic mice carrying constructs with a prerearranged κ light-chain gene (Sharpe et al. 1991; Betz et al. 1994). In all of these transgenes, the VκOx1 sequence is present in the rearranged configuration adjacent to Jκ5 which is linked to a 3′ region, which is either wild-type (Lκ), lacking the 3′ enhancer (Δ3′Eκ), or lacking the intronic enhancer (ΔEiκ) (Fig. 6). To study tissue-specific demethylation, B220+ spleen cells were isolated from these three mouse lines and the exogenous sequences then analyzed for their methylation patterns. It should be noted that transgenes in general undergo de novo methylation at the stage of implantation in the same manner as their endogenous counterparts (Frank et al. 1991). Although some B-cell-specific demethylation is observed when either the intronic or 3′ κ enhancer is present alone (Fig. 6, lanes 3–6), full demethylation was only obtained in the wild-type construct, where both enhancers are present on the same molecule (Fig. 6, lanes 1,2). Thus, although either the intronic or 3′ κ enhancer is sufficient to induce demethylation in B-cell cultures (Kirillov et al. 1996), both enhancers are required for optimal demethylation in vivo. These experiments demonstrate that both demethylation and rearrangement are controlled by the same combination of genetic elements.
Figure 6.
Enhancers required for κ gene demethylation in vivo. B220+ spleen cell (>95% purity by FACS analysis) DNA from mice carrying a complete VκJκ rearranged transgene [Lκ] (lanes 1,2) or constructs individually lacking either the 3′ enhancer [Δ3′Eκ] (lanes 3,4) or the intronic enhancer [ΔEiκ] (lanes 5,6) (Betz et al. 1994) was digested by HindIII–KpnI with (+) or without (−) HhaI. The 1.3-kb HindIII–KpnI fragment and its 0.6-kb unmethylated digestion product are derived exclusively from the exogenous transgene; the 0.8-kb band is a common product of both the endogenous and transgenic copies. The accompanying map shows the various transgenes used in this experiment (Betz et al. 1994).
Discussion
In B cells functional rearrangement at a given immunoglobulin locus occurs on only one of two identical alleles, a phenomenon termed allelic exclusion. Inefficiency in the rearrangement process itself has been suggested as a potential mechanism underlying this selection (Coleclough et al. 1981). Inefficiency would then require a feedback mechanism to shut off further rearrangement when a functional receptor has been made. Otherwise, even an inefficient process would eventually result in rearrangement on the second allele. The well-documented existence of feedback down-regulation of the V(D)J recombinase maintains the viability of the inefficiency model explaining the establishment of allelic exclusion (Rajewsky 1996). However, another possibility is that when the trans-acting factors establish a cellular environment conducive to rearrangement of a given family, only one of the two alleles is available initially for rearrangement. Indeed, the experiments we present here lead us to suggest that the establishment of κ gene allelic exclusion involves a demethylation event that marks one of the two κ alleles for subsequent rearrangement.
κ demethylation is monoallelic
We have performed two distinct experiments, each of which on its own demonstrates that in normal κ+ B cells, κ demethylation is monoallelic. One analysis involved examining DNA prepared from populations of purified κ+ B cells using methyl-sensitive restriction enzymes and Southern blotting. We also developed a novel assay that allowed us to analyze the methylation status of the two κ alleles in individual κ+ B cells. These experiments demonstrate that κ+ B cells contain a rearranged, unmethylated allele coexisting with a germ-line allele that is methylated, and, to our knowledge, are the first to analyze the methylation status of the two κ alleles in individual cells in vivo. Previous studies using B lineage cell lines had suggested that germ-line alleles are undermethylated to the same extent as rearranged copies (Mather and Perry 1983; Storb and Arp 1983). However, in light of our in vivo results, it is likely that the generally unmethylated pattern they observed is caused by a cell culture or transformation artifact.
Demethylation precedes rearrangement
By analyzing B cells isolated from bone marrow of both normal and mutant mice, we determined that κ demethylation in vivo takes place at the pre-B cell transition stage (see Fig. 3a,b). It was not clear from these studies, however, whether the κ gene undergoes demethylation before or after the rearrangement event. We have used a genetic approach to distinguish between these two possibilities. We examined mice in which the rearrangement of the κ alleles is inhibited in cis (Takeda et al. 1993), and mice in which κ rearrangement is prevented by a trans-acting mechanism (Betz et al. 1994). In both cases, roughly half of the endogenous (germ-line) alleles undergo demethylation in κ+ lymphocytes, a pattern clearly reminiscent of the allele-specific demethylation observed in normal B cells. These results thus indicate that rearrangement is not necessary for the κ allele to undergo demethylation. We have also been able to prove that rearrangement is not sufficient to bring about demethylation. Indeed, our studies show that a prerearranged κ gene inserted into the normal κ locus (Pelanda et al. 1996) only undergoes partial demethylation in B cells. Taken together, these results strongly suggest that during normal B-cell development demethylation immediately precedes rearrangement.
Demethylation is necessary for rearrangement
The strong correlation between undermethylation and rearrangement raises the possibility that DNA methylation itself may inhibit the rearrangement process; several published experiments support this model. Using a simple transfection system, for example, it has been shown that plasmids containing the minimal heptamer/nonamer signal sequences readily undergo rearrangement in B cells. However, when these templates are first methylated in vitro and then allowed to undergo replication in the transfected cells, rearrangement is severely inhibited (Hsieh and Lieber 1992). More convincing evidence that methylation represses rearrangement was obtained by examining mice carrying a transgene construct containing basic rearrangement signals. In a C57BL/6 background, this transgene is apparently methylated in every tissue. However, when bred into the DBA/2 strain, this same transgene remains unmethylated throughout the organism and only in this unmethylated state does B-cell-specific rearrangement take place (Engler et al. 1991). It should be noted that in this experiment, the methylation pattern is controlled by modifier genes that have nothing to do with the signals normally used in B cells, and for this reason it provides convincing evidence that local DNA methylation itself plays a critical role in inhibiting gene rearrangement.
By employing transgenes containing a prerearranged κ gene configuration (Betz et al. 1994), it was possible to evaluate the effect of the intronic and 3′ κ enhancers on demethylation in vivo independent of the rearrangement process. Our results indicate that although each enhancer alone can induce a small amount of B-cell-specific demethylation, full demethylation of the κ locus is only seen when both are present together in cis. Recent mouse knockout experiments have demonstrated that single deletions of either the intronic or 3′ enhancer are sufficient to partially inhibit κ gene rearrangement (Takeda et al. 1993; Gorman et al. 1996; Xu et al. 1996). It thus appears that in vivo the same genetic elements are involved in both demethylation and rearrangement. One attractive explanation for these results is that the enhancers work by inducing demethylation which, in turn, allows rearrangement at the same locus.
It is interesting to consider data suggesting a role for methylation in regulating rearrangement at the heavy-chain gene locus in the mouse. The μ enhancer together with its accompanying MAR sequences have been shown to induce demethylation on transfected constructs in B-cell cultures and in vivo (Fernex et al. 1995; Kirillov et al. 1996). Furthermore, when this element is deleted from its normal locus in vivo, heavy-chain rearrangement is inhibited (Chen et al. 1993; Serwe and Sablitzky 1993). Because unmethylated constructs lacking either the heavy- or light-chain enhancer elements readily undergo rearrangement following transfection into pre-B cells in culture (Hesse et al. 1987; Lieber et al. 1987), it is clear that these cis-acting sequences are not required for carrying out the mechanical aspects of recombination. It is likely that in vivo the absence of the μ enhancer prevents this site from undergoing stage-specific demethylation, and it is for this reason that rearrangement is repressed. In a similar manner, DNA methylation may also play a role in the control of rearrangement at the T-cell-receptor locus (Oyashiki et al. 1992; Capone et al. 1993; Okada et al. 1994; Bories et al. 1996).
Role of demethylation in rearrangement and allelic exclusion
On the basis of our results it is now possible to understand how DNA methylation may play a role in regulating κ gene rearrangement. In pro-B cells, at a stage when the heavy-chain locus undergoes multiple rearrangement events, the κ alleles are still fully methylated, and it is probably this methylation, in the context of chromatin structure, which prevents their rearrangement despite the presence of RAG-1 and RAG-2 activity in these cells (Grawunder et al. 1995). The expression of a productive μ chain appears to bring about a number of dramatic changes in the κ locus, including the initiation of κ germ-line transcription (Spanopoulou et al. 1994), an increase in chromatin accessibility (Stanhope-Baker et al. 1996), and monoallelic demethylation of the sequences adjacent to the Jκ region (Fig. 3b). All of these events probably occur prior to rearrangement, and it is likely that they are mediated through trans-acting factors that are triggered by the presence of a functional μ chain. NF-κB, for example, is already bound to the κ intronic enhancer at this stage (Shaffer et al. 1997), and has been demonstrated to be required for κ demethylation in a transfection system (Demengeot et al. 1995; Kirillov et al. 1996).
Our results clearly suggest that κ gene demethylation takes place preferentially on only one allele in each cell, and it is probably this demethylation that selects the gene copy that will ultimately undergo rearrangement. The synthesis of a functional κ chain completes the generation of the B-cell receptor and thus down-regulates the expression of RAG-1 and RAG-2. This down-regulation probably serves as a backup mechanism for preventing further rearrangement. In the absence of a productive κ chain it is possible that, given enough time, the second allele in the same cell will eventually undergo demethylation and subsequent rearrangement. This is consistent with our finding (Fig. 1) that all rearranged alleles in B cells are unmethylated.
A number of previous observations also support the idea that differences between the two alleles render one available for rearrangement prior to the other. For example, in IgM+ immature B cells, double-stranded DNA breaks were detected at the Jκ2 and Jκ5 signal sequences, but not at the initially preferred Jκ1 site, strongly suggesting that one allele can undergo repeated rearrangement events, whereas the other κ copy in the same cell is maintained in the germ-line configuration (Constantinescu and Schlissel 1997). These results are consistent with the observation that in cells carrying a prerearranged autoreactive κ gene, secondary rearrangements that introduce alternate variable regions take place preferentially on this same allele (Chen et al. 1997). This editing procedure presumably serves as a molecular mechanism for rescuing cells from deletion or anergy. It thus appears that complex regulation of κ gene rearrangement timing and allele selection is most likely mediated through changes in accessibility, and our data clearly indicate that demethylation plays an important role in this process. Yet another line of support comes from single-cell analysis of B cells carrying a prerearranged knock-in (Vκ8R or Vκ4R) on one allele (Luning-Prak and Weigert 1995). Our calculations show that in >90% of these cells, either the targeted or the natural allele, but not both, undergo rearrangement events. This suggests that there is indeed a mechanism for selecting one allele at a time which is independent of whether or not a productive κ gene can be synthesized. If such a primary mechanism did not exist, the rearranged construct should have brought about complete closure of the second allele in every cell.
There are several ways to explain how demethylation might initially occur on a single allele in each developing B cell. One possibility is that the demethylation enzyme machinery is present in limiting amounts in the cell and only suffices to remove methyl moieties from one allele at a time. In this model, demethylation itself would represent the rate-limiting step for allele selection. Alternatively, one κ allele in each B cell may already carry a preset cis-acting molecular marker that serves to direct the demethylation process. Although there is no direct evidence for either mechanism, our data show that a multi copy prerearranged κ transgene, integrated at an ectopic site in the genome, can undergo full demethylation in B cells, clearly suggesting that the demethylation potential is not rate limiting. In contrast to the κ transgene, a prerearranged construct knocked in to the endogenous κ locus itself became only partially unmethylated, probably because it is subject to cis-acting restrictions normally dictated by this gene region. In light of these data, we favor the hypothesis that the two alleles are differentially marked well before the initiation of κ gene rearrangement.
It thus appears that the process of allele selection at the κ locus may actually be analogous to the mechanism that controls olfactory receptor gene expression. In this case, as well, although the choice of receptor gene activation in any given olfactory neuron is carried out stochastically, chromosome structure limits this process to only a single allele (Chess et al. 1994). Thus, for both the immunoglobulin and olfactory receptor loci, structural allelic exclusion appears to be a primary mechanism for insuring the production of a single gene product from a large array.
Materials and methods
Animals
Wild-type C57BL/6 × BALB/c F1 mice were obtained from our own colony. iEκT−/− and 3–83κi mice were obtained from S. Takeda (Kyoto University, Japan) and K. Rajewsky (University of Cologne, Germany). Vκ8R mice were obtained from M. Weigert, and Lκ, Δ3′Eκ and ΔEiκ mice from M. Neuberger (MRC, Cambridge, UK). All mice were analyzed at the age of 8–12 weeks.
Immunofluorescence staining, cell sorting, purification, and analysis
Cell suspensions from bone marrow were prepared by flushing six to eight femura with PBS and then washing by centrifugation through PBS. Spleen cells were prepared by disruption in PBS, followed by gentle pipetting, and centrifuging through PBS.
Mouse spleen or bone marrow B cells were enriched by means of staining with either a biotinylated anti-κ or anti-B220 antibody (PharMingen), followed by incubation with streptavidin-coated magnetic beads (Miltenyi Biotec). Cells were sorted by magnetic cell sorting using the MACS system (Miltenyi et al. 1990). Cells bound to the column in the magnetic field were eluted and recovered for subsequent DNA analysis. Purity (> 90%) was assessed by flow cytometry (FACS) analysis after restaining with streptavidin-conjugated phycoerythrin (Jackson Laboratory). Wild-type fraction D cells were prepared by a dual laser-dye flow cytometer (FACStar, Becton-Dickinson). Cells were stained with anti-B220-PE, anti-CD43-FITC, and anti-IgM-FITC (provided by K. Rajewsky) and sorted for the B220+, CD43−, IgM− fraction (97% purity). Sorted cells were washed once with PBS and stored for subsequent DNA analysis. Cellular DNA (4–8 μg) was digested, electrophoresed in native (Tris-acetate) agarose gels, and transferred to nitrocellulose. DNA was then hybridized with the specific radioactive probes (see Fig. 1) and analyzed by autoradiography (Southern 1975). Hybridization was carried out at 65°C for 16 hr. In some cases, the degree of undermethylation was measured semiquantitavely using a PhosphorImager.
MSRE–PCR analysis on single cells
We modified the single-cell PCR method of Bertram et al. (1995) to include a MSRE analysis. Briefly, spleen cells from an F1 progeny of a cross between M. spretus and M. musculus were stained with a rat anti-mouse κ light chain monoclonal antibody (R5-240, PharMingen, FITC conjugated). Individual κ+ cells were FACS sorted directly into 25 μl of lysis buffer (50 mm KCl, 2.5 mm MgCl2, 0.5% Tween 20, 0.5% NP-40, 10 mm Tris-HCl at pH 8.3) in individual PCR tubes. The genomic DNA was liberated using proteinase K (0.001 mg/ml at 60°C for 2 hr), that was then inactivated with PMSF (0.002 mg/ml, 60°C, for 1 hr). The genomic DNA was subjected to digestion overnight at 37°C with 20 units of AvaI.
PCR was performed with Taq DNA polymerase (Promega) under standard conditions (50 mm KCl, 0.1% Triton X-100, 1.5 mm MgCl2, 10 mm Tris-HCl at pH 8.3, 200 μm each of all four dNTPs, 50 ng each of primer). The first-round PCR reaction was carried out in 50 μl using primers P1 (5′-TGAAGTTTTGGTCCCATTGTGTCC-3′) and P2 (5′-CCTAAGACGAGTTTTTCGCTCAGCTTTC-3′), which flank the AvaI site. Amplification was performed as follows: 4 min at 94°C; 40 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C; and, finally, 4 min at 72°C. Second-round PCR conditions were identical to the first round except that 2 μl of the first-round amplification product was added to the PCR mixture containing primers P3 (5′-GTGGGAGAAATGAGAAAGGAACAG-3′) and P4 (5′-TGGGGGATCTTCTATTGATGCAC-3′). The second-round PCR product was subjected to digestion with HhaI and analyzed by agarose gel electrophoresis.
MSRE-multiplex PCR on single cells was performed to simultaneously amplify the unrearranged κ DNA in addition to the region flanking the AvaI site (described above). The following primers were used simultaneously: P1, P2, P5 (5′-AGATCCTATGGAAGAGCAGCGAG-3′), and P6 (5′-TTTACCTCCACAAAACCCTCCCC-3′). 50 cycles of amplification were performed using the conditions of the first-round PCR described above. In the second-round PCR, the two distinct regions were amplified in separate tubes in reactions whose conditions were identical (except that each time segment was 45 sec) to the previously described second-round. The following primers (nested within the first-round primers) were used: P7 (5′-CGGGATCCCGGTGGAGGCACCAAGCTG-3′) and P8 (5′-GCTCTAGACCTCCACAAAACCCTCCCTAGG-3′). To visualize products from a larger fraction of cells, a third round (40 cycle) PCR was carried out using primers P7 and P9 (5′-CTGATCTGAGAATGGAATTTGCTAAATCCCTG-3′). This third-round PCR product was subjected to digestion by the restriction enzyme BsmAI and analyzed by agarose gel electrophoresis.
Acknowledgments
We thank K. Rajewsky, S. Takeda, M. Wiegert, and M. Neuberger for providing the trangenic mice used in this study; S. Jung, T. Wirth, and S. Schaal for their collaboration and for providing antibodies; G. Neiman from the Interdepartmental Equipment Unit at the Hebrew University Medical School for technical assistance with the FACS sorting; and G. Paradis of the MIT flow cytometry core facility. We also acknowledge the assistance of G. Hirst in preparing the manuscript. This research was supported by grants from the Israel Science Foundation (Y.B. and H.C.), the German–Israeli Foundation for Scientific Research and Development (Y.B.), the National Institutes of Health (H.C. and A.C.), the Israel Cancer Research Fund (H.C.), the Israel Science Ministry (H.C.), and the Ira W. DeCamp Foundation created under the will of Elizabeth DeCamp-McInery (A.C.). A.C. is a Rita Allen Foundation Scholar and R.M. is the recipient of a fellowship from the Israel Ministry of Science and the Arts.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cedar@md2.huji.ac.il; FAX 972-2 641-5848.
References
- Bertram S, Hufert FT, Haefelin DN, von Laer D. Detection of DNA in single cells using an automated cell deposition unit and PCR. BioTechniques. 1995;19:616–620. [PubMed] [Google Scholar]
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannel R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin κ gene: Critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Bories J-C, Demengeot J, Davidson L, Alt FW. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancer: The role of enhancer elements in controlling V(D)J recombination accessibility. Proc Natl Acad Sci. 1996;93:7871–7876. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Wu L, Scollay R, Ferrier P. TCRβ and TCRα gene enhancers confer tissue- and stage-specificity on V(D)J recombination events. EMBO J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Luning Prak E, Weigert M. Editing disease-associated autoantibodies. Immunity. 1997;6:97–105. doi: 10.1016/s1074-7613(00)80673-1. [DOI] [PubMed] [Google Scholar]
- Chen J, Young F, Bottaro A, Stewart V, Smith RK, Alt FW. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Coleclough C, Perry RP, Karjalainen K, Weigert M. Aberrant rearrangements contribute significantly to the allelic exclusion of immunoglobulin gene expression. Nature. 1981;290:372–378. doi: 10.1038/290372a0. [DOI] [PubMed] [Google Scholar]
- Constantinescu A, Schlissel M. Changes in locus-specific V(D)J recombinase activity induced by immunoglobulin gene products during B cell development. J Exp Med. 1997;185:609–620. doi: 10.1084/jem.185.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demengeot J, Oltz EM, Alt FW. Promotion of V(D)J recombinational accessibility by the intronic Eκ element: Role of the κB motif. Int Immunol. 1995;7:1995–2003. doi: 10.1093/intimm/7.12.1995. [DOI] [PubMed] [Google Scholar]
- Ehlich A, Schaal S, Gu H, Kitamura D, Muller W, Rajewsky K. Immunoglobulin heavy and light chain genes rearrange independently at early stages of B cell development. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- Engler P, Haasch D, Pinkert CA, Doglio L, Glymour M, Brinster R, Storb U. A strain-specific modifier on mouse chromosome 4 controls the methylation of independent transgene loci. Cell. 1991;65:1–20. doi: 10.1016/0092-8674(91)90546-b. [DOI] [PubMed] [Google Scholar]
- Fernex C, Capone M, Ferrier P. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol Cell Biol. 1995;15:3217–3226. doi: 10.1128/mcb.15.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, van-Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin μ gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- Frank D, Keshet I, Shani M, Levin A, Razin A, Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991;351:239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- Gellert M. A new view of V(D)J recombination. Genes Cells. 1996;1:269–275. doi: 10.1046/j.1365-2443.1996.22023.x. [DOI] [PubMed] [Google Scholar]
- Goodhardt M, Cavelier P, Doyen N, Kallenbach S, Babinet C, Rougeon F. Methylation status of immunoglobulin κ gene segments correlates with their recombination potential. Eur J Immunol. 1993;23:1789–1795. doi: 10.1002/eji.1830230809. [DOI] [PubMed] [Google Scholar]
- Gorman JR, van-der-Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Igκ 3′ enhancer influences the ratio of Igκ versus Igλ B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG1 and RAG2 gene expression in pre-B cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Lieber MR. CpG methylated minichromosomes become inaccesible for V(D)J recombination after undergoing replication. EMBO J. 1992;11:315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Qiu RG, Grosschedl R. The immunoglobulin μ enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes & Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- Kelley DE, Pollok BA, Atchison ML, Perry RP. The coupling between enhancer activity and hypomethylation of κ immunoglobulin genes is developmentally regulated. Mol Cell Biol. 1988;8:930–937. doi: 10.1128/mcb.8.2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nature Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- Lewis SM. The mechanism of V(D)J joining: Lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: A novel role for the intronic κ chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes & Dev. 1987;1:751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- Luning-Prak E, Weigert M. Light chain replacement: A new model for antibody gene rearrangement. J Exp Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather EL, Perry RP. Methylation status and DNase I sensitivity of immunoglobulin genes: Changes associated with rearrangement. Proc Natl Acad Sci. 1983;80:4689–4693. doi: 10.1073/pnas.80.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miltenyi S, Muller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Bergman Y. DNA methylation: Regulation of gene expression and role in the immune system. Biochim Biophys Acta. 1997;1333:F29–F50. doi: 10.1016/s0304-419x(97)00010-3. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: Difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes & Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Ohyashiki JH, Ohyashiki K, Kawakubo K, Tauchi T, Nakazawa S, Kimura N, Toyama K. T-cell receptor β chain gene rearrangement in acute myeloid leukemia always occurs at the allele that contains the undermethylated Jβ1 region. Cancer Res. 1992;52:6598–6602. [PubMed] [Google Scholar]
- Okada A, Mendelsohn M, Alt F. Differential activation of transcription versus recombination of transgenic T cell receptor β variable region gene segments in B and T lineage cells. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelanda R, Schaal S, Torres RM, Rajewsky K. A prematurely expressed Igκ transgene, but not a VκJκ gene segment targeted into the Igκ locus, can rescue B cell development in λ5-deficient mice. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Rolink A, Kudo A, Karasuyama H, Kikuchi Y, Melchers F. Long-term proliferating early pre-B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. EMBO J. 1991;10:327–336. doi: 10.1002/j.1460-2075.1991.tb07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F. IL-2 receptor α chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int Immunol. 1994;6:1257–1264. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Serwe M, Sablitzky F. V(D)J recombination in B cells is impaired but not blocked by targetted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 1993;6:2321–2327. doi: 10.1002/j.1460-2075.1993.tb05886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Peng A, Schlissel MS. In vivo occupancy of the κ light chain enhancers in primary Pro- and Pre-B cells: A model for κ locus activation. Immunity. 1997;6:131–143. doi: 10.1016/s1074-7613(00)80420-3. [DOI] [PubMed] [Google Scholar]
- Sharpe MJ, Milstein C, Jarvis JM, Neuberger MS. Somatic hypermutation of immunoglobulin kappa may depend on sequences 3′ of C κ and occurs on passenger transgenes. EMBO J. 1991;10:2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E, Roman CA, Corcoran LM, Schlissel MS, Silver DP, Nemazee D, Nussenzweig MC, Shinton SA, Hardy RR, Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiaton in Rag-1-deficient mice. Genes & Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel M. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- Storb U, Arp B. Methylation patterns of immunoglobulin genes in lymphoid cells: Correlation of expression and differentiation with undermethylation. Proc Natl Acad Sci. 1983;80:6642–6646. doi: 10.1073/pnas.80.21.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Zou Y-R, Bluethmann H, Kitamura D, Muller U, Rajewsky K. Deletion of the immunoglobulin κ chain intron enhancer abolishes κ chain gene rearrangement in cis but not λ chain gene rearrangement in trans. EMBO J. 1993;12:2329–2336. doi: 10.1002/j.1460-2075.1993.tb05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Boekel E, Melchers F, Rolink A. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7:1013–1019. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]