Abstract
Neuroblasts in the developing Drosophila CNS asymmetrically localize the cell fate determinants Numb and Prospero as well as prospero RNA to the basal cortex during mitosis. The localization of Prospero requires the function of inscuteable and miranda, whereas prospero RNA localization requires inscuteable and staufen function. We demonstrate that Miranda contains multiple functional domains: an amino-terminal asymmetric localization domain, which interacts with Inscuteable, a central Numb interaction domain, and a more carboxy-terminal Prospero interaction domain. We also show that Miranda and Staufen have similar subcellular localization patterns and interact in vitro. Furthermore, miranda function is required for the asymmetric localization of Staufen. Miranda localization is disrupted by the microfilament disrupting agent latrunculin A. Our results suggest that Miranda directs the basal cortical localization of multiple molecules, including Staufen and prospero RNA, in mitotic neuroblasts in an actin-dependent manner.
Keywords: Miranda, Inscuteable, Prospero, Staufen localization, asymmetric cell division
Asymmetric cell division is fundamentally important to the generation of diverse cell types in many organisms (Horvitz and Herkowitz 1992; Stauger and Doonan 1993; Rhyu and Knoblich 1995; Chang and Drubin 1996; Guo and Kemphues 1996; Shapiro and Losick 1997; Jan and Jan 1998). During development of the Drosophila nervous system, asymmetric division is accomplished at least in part by the asymmetric segregation of the intrinsic cell fate determinants Numb and Prospero (Uemura et al. 1989; Doe et al. 1991; Vaessin et al. 1991; Matsuzaki et al. 1992; Rhyu et al. 1994; Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995). In mitotic neuroblasts of the developing CNS, the membrane-associated protein Numb localizes asymmetrically to form a crescent on the basal cortex and in telophase is segregated exclusively to the basal daughter cell, which becomes a ganglion mother cell (Rhyu et al. 1994; Spana et al. 1995; Spana and Doe 1995).
The homeodomain-containing protein Prospero exhibits a more complex pattern of subcellular localization. Immunostaining of Prospero reveals a faint, transient crescent on the apical cortex of neuroblasts in late interphase and early prophase (Spana and Doe 1995). In late prophase, metaphase, and anaphase, Prospero colocalizes with Numb at the basal cortex and in telophase is similarly segregated into the basal daughter cell (Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995). After the completion of telophase, Prospero enters the nucleus (Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995). The localization patterns of Numb and Prospero are disrupted by treatment with the actin depolymerizing agents latrunculin A and B (Broadus and Doe 1997; Knoblich et al. 1997) but not by treatment with the microtubule depolymerizing agent colcemid (Knoblich et al. 1995; Broadus and Doe 1997). prospero RNA is also asymmetrically localized in neuroblasts and exhibits an apical-then-basal localization pattern similar to Prospero protein (Li et al. 1997). Though prospero RNA localization is not required for Prospero protein localization (Li et al. 1997; Broadus et al. 1998), it may serve as a backup system to ensure a sufficient supply of Prospero protein to the basal daughter cell (Broadus et al. 1998).
Three genes have been implicated in asymmetric localization of proteins and RNA in neuroblasts: inscuteable, miranda, and staufen. inscuteable, which encodes a large novel protein, is the most upstream component identified to date (Kraut and Campos-Ortega 1996). In neuroblasts lacking inscuteable function, Numb and Prospero either fail to form crescents or form crescents that are randomly localized on the cell cortex (Kraut et al. 1996). Furthermore, in inscuteable mutants prospero RNA fails to localize to the basal cortex but localizes to the apical cortex of neuroblasts normally earlier in the cell cycle (Li et al. 1997). Inscuteable itself forms a crescent on the apical cortex of neuroblasts in late interphase, prophase, and metaphase (Kraut et al. 1996). The role of Inscuteable and Numb is not confined to the developing nervous sytem; proper cell fate decisions in the myogenic lineage also require inscuteable and numb function (Burchard et al. 1995; Knirr et al. 1997; Ruiz Gomez and Bate 1997; Carmena et al. 1998).
staufen was originally identified for its role in asymmetric RNA localization during Drosophila oogenesis and encodes a protein that contains five copies of a double-stranded RNA binding motif (St Johnston et al. 1991, 1992). Staufen displays an apical-then-basal localization pattern similar to prospero RNA and is required for the localization of prospero RNA to the basal cortex (Li et al. 1997; Broadus et al. 1998). Staufen is not required for the localization of Prospero protein, however (Li et al. 1997; Broadus et al. 1998). In inscuteable mutant embryos, the apical localization of Staufen is seen less frequently but is not abolished (Li et al. 1997).
Miranda is a novel protein predicted to be rich in coiled–coil structures (Shen et al. 1997; Ikeshima-Kataoka et al. 1997). Miranda interacts with the asymmetric localization domain of Prospero and colocalizes with Prospero in mitotic neuroblasts (Shen et al. 1997; Ikeshima-Kataoka et al. 1997). Loss of miranda function results in the cytoplasmic distribution of Prospero in mitotic neuroblasts and the segregation of Prospero to both daughter cells (Shen et al. 1997; Ikeshima-Kataoka et al. 1997). Miranda also interacts with Numb in vitro (Shen et al. 1997), but its role in the asymmetric localization of Numb is unclear. Miranda is itself asymmetrically localized to the basal cortex in mitotic neuroblasts; proper Miranda localization to the basal cortex requires inscuteable function (Shen et al. 1997). Miranda and Staufen therefore appear to work downstream of Inscuteable to localize Prospero protein and prospero RNA to the basal cortex.
Despite the identification of several genes involved in asymmetric localization, much remains to be learned about the molecular mechanisms of this process. The factors directly responsible for the localization of Miranda and Staufen are yet to be identified. Miranda has been shown to be responsible for the localization of Prospero, but it is unknown whether Miranda is a dedicated adapter for Prospero or is also required for the localization of other factors. Furthermore, it is unclear how the apically localized Inscuteable directs the basal localization of Numb, Prospero, prospero RNA, and Miranda.
To approach these questions, we first examined the distribution of Miranda and Staufen during neuroblast mitosis. We demonstrate that Miranda exhibits an apical-then-basal localization pattern similar to Prospero and that this localization is disrupted by treatment with latrunculin A. We show further that Staufen, like Miranda and prospero RNA, localizes to the apical cortex then the basal cortex and that Miranda interacts with Staufen physically. We show also that miranda function is required for the proper localization of Staufen during neuroblast division. Finally, we show that the amino-terminal domain of Miranda both interacts with a central domain of the Inscuteable protein in vitro and is necessary and sufficient for the complex subcellular localization pattern of Miranda. A central domain of Miranda interacts with Numb, and a more carboxy-terminal domain interacts with Prospero. These results suggest that Miranda occupies a central role during asymmetric cell division and may serve as a link between Inscuteable on the apical cortex and localization of molecules on the basal cortex.
Results
Miranda localizes first to the apical, then the basal cortex in an actin-dependent manner
Miranda has been demonstrated to play an important role in the asymmetric localization of Prospero. The localization of Prospero to the apical cortex of neuroblasts during late interphase and early prophase prompted us to examine more closely the localization of Miranda. Miranda forms a crescent on the apical cortex of neuroblasts in late interphase (Fig. 1A). Later in mitosis, Miranda forms a crescent on the basal neuroblast cortex (Shen et al. 1997; Ikeshima-Kataoka 1997).
Figure 1.
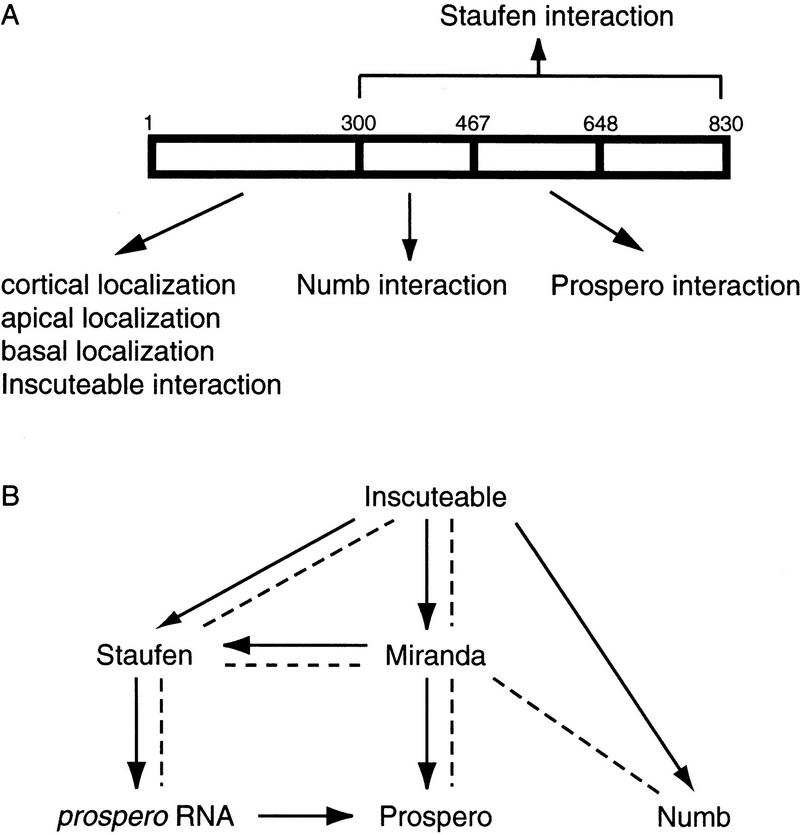
Microfilaments are required for the localization of Miranda to the apical then basal cortex. (A) Wild-type embryos were stained with anti-Miranda antibody (green) and propidium iodide to visualize DNA (red). Apical is toward the top; white dots indicate neuroblast cell borders in this and all subsequent figures. In the neuroblast at right, which is in early prophase, Miranda is localized to the apical cell cortex (arrowhead). The neuroblast at left has nearly completed mitosis. Miranda is present exclusively in the basal daughter cell (arrow). (B) Wild-type embryos were treated with 200 μm latrunculin A for 20 min and stained for Miranda (green) and DNA (red). Under these conditions, phalloidin reactivity is virtually abolished (Knoblich et al. 1997), and Miranda is localized along the entire cortex of mitotic neuroblasts.
Asymmetric localization of both Numb and Prospero has been shown to be dependent on the actin cytoskeleton (Broadus and Doe 1997; Knoblich et al. 1997). We therefore tested the actin dependence of Miranda localization using the actin depolymerizing drug latrunculin A (Spector et al. 1989; Ayscough et al. 1997). After treatment of Drosophila embryos with 200 μm latrunculin A for 20 min, asymmetric localization of Miranda was completely disrupted while membrane association was unperturbed (Fig. 1B). We conclude that the asymmetric localization of Miranda during mitosis is an actin-dependent process.
An amino-terminal fragment of Miranda is localized asymmetrically in neuroblasts during mitosis
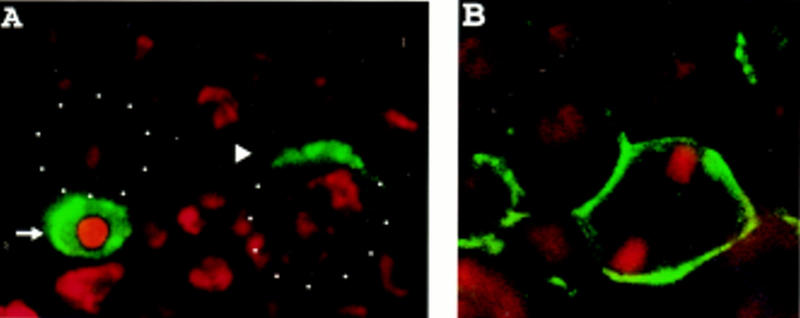
To identify a domain of Miranda sufficient for asymmetric localization, we generated constructs encoding a myc-tagged full length Miranda and a series of myc-tagged Miranda fragments (Fig. 2A). We then used the GAL4–UAS system (Brand and Perrimon 1993) to express these Miranda fragments by introducing these constructs into the pUAST vector, generating transgenic UAS–myc–Miranda flies, and crossing these transgenic flies to the hairy–GAL4 line to express full-length or truncated Miranda in alternate segments of embryos. Immunostaining with an anti-myc antibody was performed to visualize the subcellular localization of the different myc-tagged Miranda proteins in mitotic neuroblasts. All Miranda fragments that contained the amino-terminal 298 amino acids exhibit the same asymmetric localization pattern as wild type Miranda (data not shown). In contrast, a fragment containing amino acids 114–298 localizes to the cytoplasm and fails to segregate preferentially into the basal daughter cell, as does a fragment containing all residues carboxy-terminal to amino acid 300 (data not shown).
Figure 2.
The amino-terminal 298 amino acids of Miranda are sufficient for asymmetric localization even in the absence of endogenous Miranda. (A) Full-length Miranda or Miranda fragments were myc-tagged and expressed in transgenic animals. Staining with an anti-myc antibody was performed to visualize their subcellular localization. The results are summarized at right. Full-length Miranda, the amino-terminal 431 amino acids, and the amino-terminal 298 amino acids of Miranda all localize to the apical cortex in early mitotic neuroblasts and then to the basal cortex later in mitosis (not shown). Amino acids 114–298 and the carboxy-terminal 530 amino acids, however, remain in the cytoplasm of dividing neuroblasts, fail to localize asymmetrically and enter both daughter cells (not shown). (B–E) Embryos expressing myc–MiraN298 but deficient for endogenous miranda were stained for myc (green) and DNA (red). In early prophase, myc–MiraN298 forms a crescent at the apical cortex (arrowhead, B). myc–MiraN298 then localizes to the basal cortex in metaphase (arrowhead, C) and is exclusively localized to the basal daughter in telophase (arrowheads, D,E).
We tested further whether endogenous Miranda is required for the localization of the myc-tagged amino-terminal 298 amino acids of Miranda (myc–MiraN298). Embryos homozygous for a deficiency that removes miranda and expressing myc–MiraN298 under control of the hairy promoter were stained with an anti-myc antibody. Even in the absence of endogenous miranda the myc–MiraN298 fragment displays a wild-type Miranda localization pattern: an apical crescent in late interphase (Fig. 2B) and early prophase, then a basal crescent in metaphase (Fig. 2C), anaphase (Fig. 2D), and telophase (Fig. 2E). The amino-terminal 300 amino acids of Miranda are therefore sufficient for all aspects of Miranda asymmetric localization.
The amino-terminal Miranda fragment interacts physically with Inscuteable
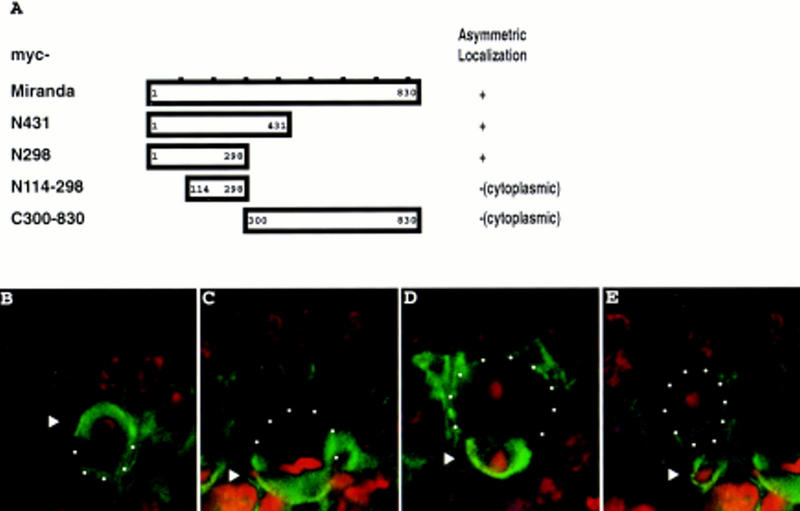
The observation that myc–MiraN298 forms an apical crescent that may coincide with the apical Inscuteable crescent led us to test the possibility that Miranda interacts physically with Inscuteable. In an in vitro binding assay, either GST alone or a protein consisting of GST fused to the amino-terminal 298 amino acids of Miranda was incubated with 35S-labeled full-length Inscuteable. After addition of glutathione beads, Inscuteable coprecipitates with GST–Miranda but not with GST alone (Fig. 3). An Inscuteable fragment from amino acids 252 to 615 also interacts with Miranda in this assay (Fig. 3).
Figure 3.
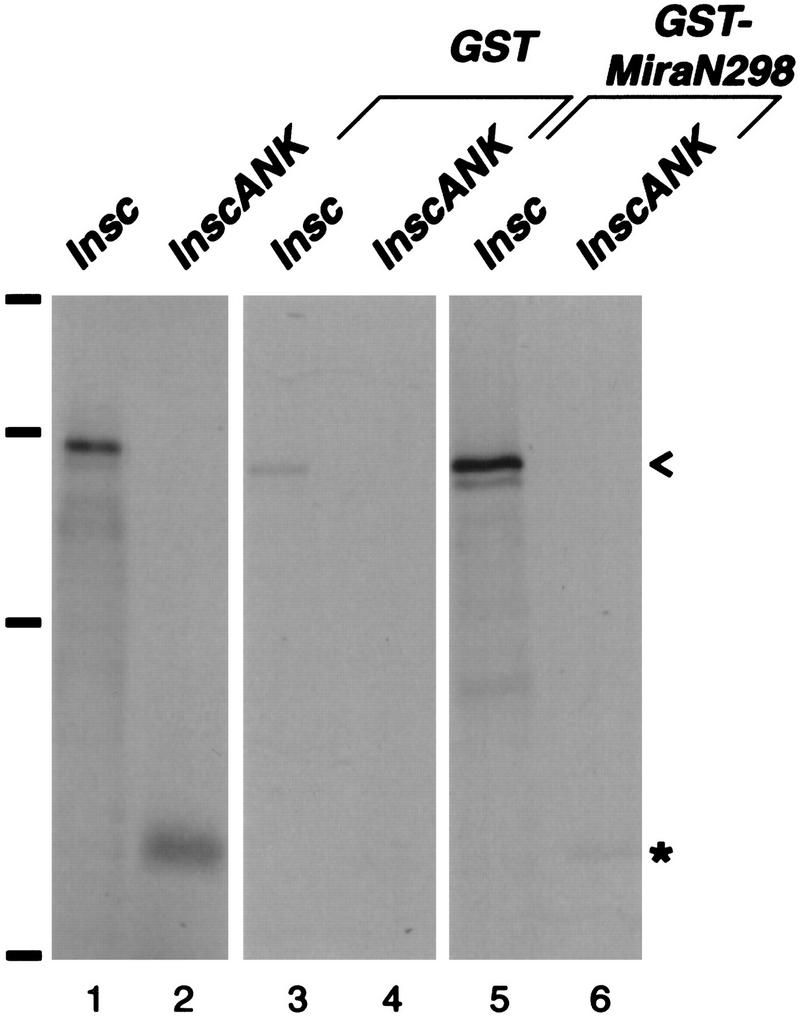
The amino-terminal 298 amino acids of Miranda interact with the ankyrin-like repeat region of Inscuteable. Transcription and translation of full-length inscuteable in vitro produces a major band that migrates at 120 kD (lane 1; <); InscANK, a fragment from amino acids 252 to 615, migrates at 45 kD (lane 2, *). These labeled polypeptides coprecipitate with GST fused to the amino-terminal 298 amino acids of Mira in the presence of glutathione beads (lanes 5,6) but do not coprecipitate with GST alone (lanes 3,4). inscuteable constructs were in vitro translated and transcribed in the presence of [35S]methionine and directly subjected to SDS-PAGE (lanes 1,2) or incubated with bacterially expressed GST or GST–MiraN298, precipitated with glutathione beads, and subjected to SDS-PAGE. Bars at left indicate positions of molecular mass markers (from bottom to top: 42, 79, 130, and 200 kD).
Distinct regions of Miranda interact physically with Numb and Prospero
We studied further the functional domain organization of Miranda with in vitro binding assays. Various 35S-labeled Miranda protein fragments (Fig. 4A) were incubated with GST fused to either the amino-terminal 223 amino acids of Numb, which is sufficient for Numb asymmetric localization (Knoblich et al. 1997), or Prospero amino acids 820–1026, which include the asymmetric localization domain (Hirata et al. 1995). Amino acids 300–467 of Miranda were sufficient to interact with GST–Numb, whereas amino acids 468–648 interact with GST–Prospero (Fig. 4B). Both interaction domains are predicted to adopt a coiled–coil structure (Shen et al. 1997). We conclude that a central 168-amino-acid domain and a more carboxy-terminal 181-amino-acid domain of Miranda interact with Numb and Prospero, respectively.
Figure 4.
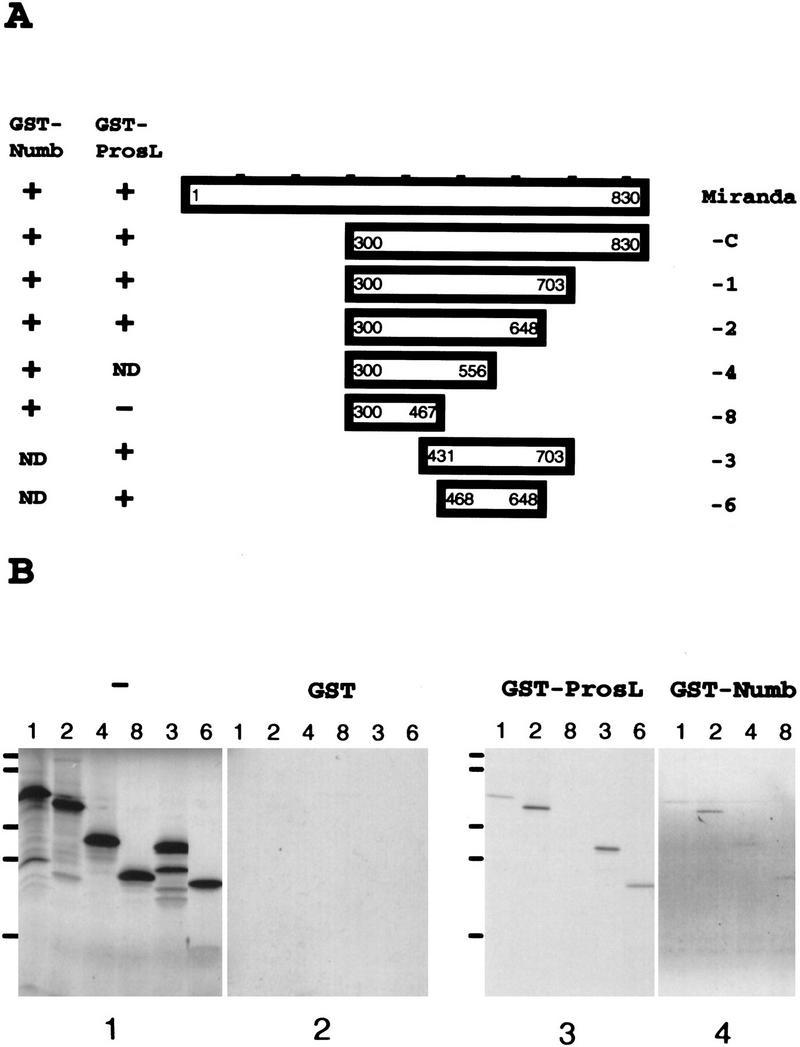
Miranda interacts with Numb and Prospero through distinct domains. (A) Various Miranda constructs were tested for their ability to interact with GST fused to either the Numb amino-terminal 223 amino acids or Prospero amino acids 820–1026 (ProsL), which include the asymmetric localization domain. The results are summarized at left. (B) The labeled Miranda fragments are shown in panel 1. Fragments 1, 2, 3, and 6, which all contain amino acids 468–648 of Miranda, interact with GST–Prospero (panel 3); fragments 1, 2, 4, and 8, which all contain amino acids 300–467, interact with GST–Numb (panel 4). None of the fragments interact with GST alone (panel 2). miranda constructs were in vitro translated and transcribed in the presence of [35S]-methionine and directly subjected to SDS-PAGE (panel 1) or incubated with bacterially expressed GST (panel 2), GST–Prospero (panel 3), or GST–Numb (panel 4) fixed to glutathione beads, which were precipitated, washed, and subjected to SDS-PAGE. Bars at left indicate positions of molecular mass markers (from bottom to top: 17, 32, 41, 71, and 126 kD).
Staufen localizes first to the apical then the basal cortex and interacts physically with Miranda
The report that Staufen is required for the proper localization of prospero RNA to the basal cortex (Li et al. 1997) prompted us to examine the localization of Staufen in mitotic neuroblasts. Staining of wild-type embryos with antiserum against Staufen reveals that Staufen, like Miranda and prospero RNA, is localized to the apical cortex in neuroblasts in late interphase (Fig. 5A) and early prophase. Staufen is then localized to the basal cortex in late prophase, metaphase (Fig. 5B), anaphase (Fig. 5C), and telophase. After mitosis it is segregated into the basal daughter cell (Fig. 5D). Our observations of Staufen localization are consistent with those reported by Broadus et al. (1998).
Figure 5.
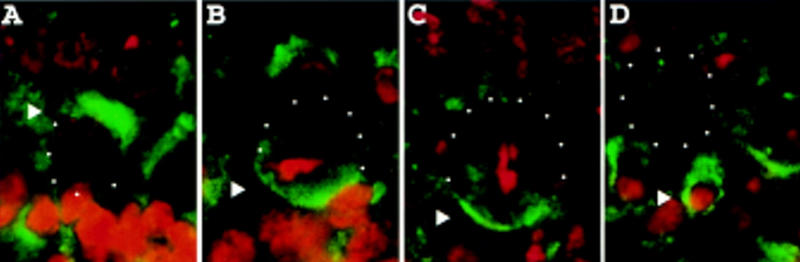
Staufen localizes to the apical and then the basal cortex in dividing neuroblasts. Wild-type embryos were stained for Staufen (green) and DNA (red). Staufen localizes to the apical cortex of neuroblasts in late interphase (arrowhead, A), to the basal cortex in metaphase (arrowhead, B) and anaphase (arrowhead, C), and enters the basal daughter in telophase (arrowhead, D).
The similar localization patterns of Miranda and Staufen suggest that they may interact physically. To test this possibility, we cotranslated Miranda and Staufen in vitro in the presence of [35S]methionine and immunoprecipitated Miranda with anti-Miranda antiserum and protein A beads. Staufen coprecipitates with Miranda under these conditions but not with anti-Miranda antiserum and protein A beads in the absence of Miranda (Fig. 6). We further found that the carboxy-terminal 530 amino acids of Miranda are sufficient for interaction with Staufen (data not shown).
Figure 6.
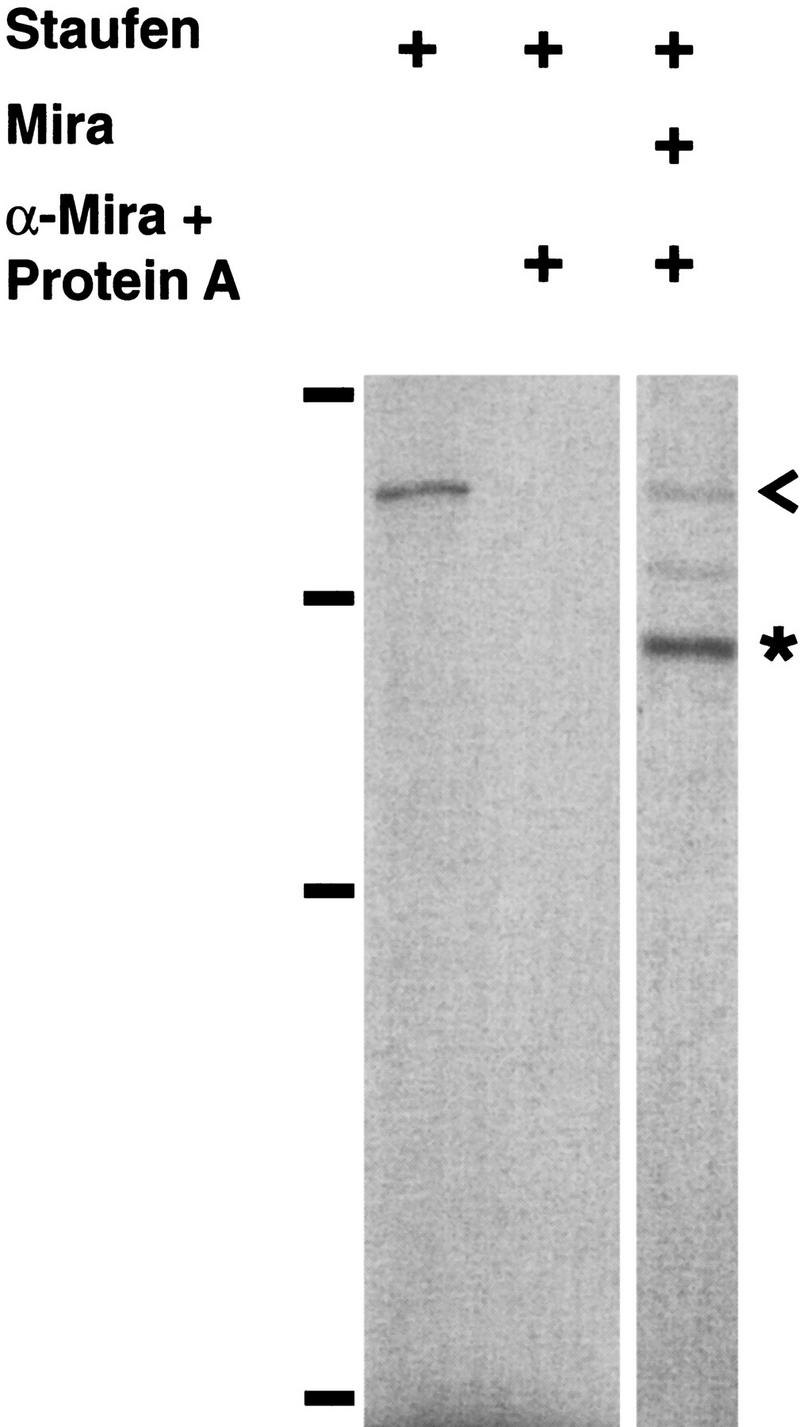
Miranda interacts with Staufen physically. Staufen coimmunoprecipitates with anti-Miranda antiserum and protein A beads in the presence (right lane) but not the absence (middle lane) of Miranda. The left lane shows the position of labeled Staufen (<). A construct encoding full-length Staufen was transcribed and translated in vitro in the presence of [35S]methionine and directly subjected to SDS-PAGE (left lane). Staufen was either translated in vitro alone (middle lane) or cotranslated with Miranda (right lane) and incubated with anti-Miranda antiserum and protein A beads, which were then precipitated, washed, and subjected to SDS-PAGE. Bars at left indicate molecular mass markers (from bottom to top: 42, 79, 130, and 200 kD). (*) The position of Miranda.
Staufen and Prospero localization is disrupted by miraDEB, a protein null mutation in miranda
The physical interaction between Staufen and Miranda and their identical subcellular localization pattern led us to consider whether Staufen, like Prospero, requires miranda for its localization. To test this possibility, we first generated a mira mutant allele, miraDEB, through chemical mutagenesis. miraDEB is recessive lethal, fails to complement Df(3R)oraI9, which removes miranda along with other chromosomal material from 92B2-3 to 92C2-3, but does complement Df(3R)oraK17, which removes material from 92B4-5 to 92B11 but leaves miranda intact (data not shown). miraDEB homozygous embryos do not exhibit Miranda immunoreactivity in immunocytochemical studies (Fig. 7D). Southern blot analysis of miraDEB reveals no gross deletions in a 14-kb genomic region that includes miranda. We conclude that miraDEB is a protein null allele of miranda.
Figure 7.
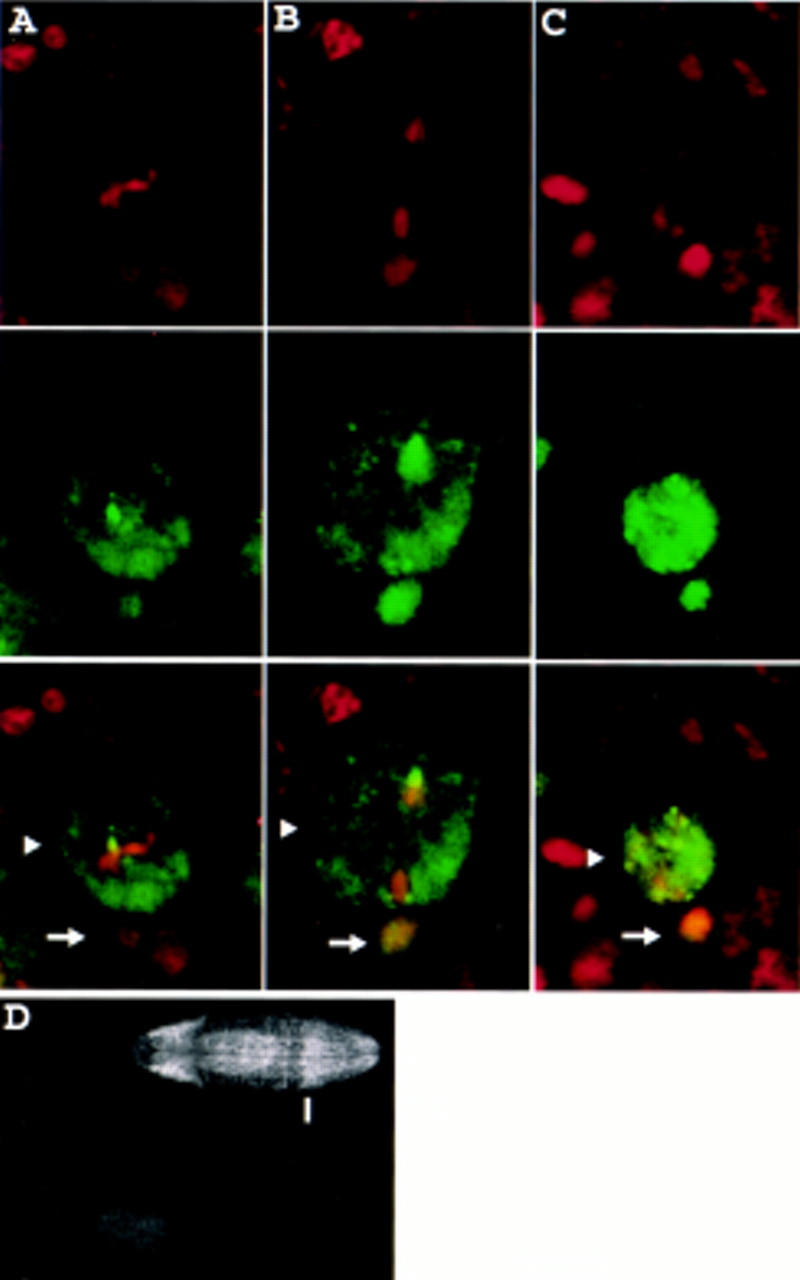
Prospero is mislocalized in embryos homozygous for miraDEB, a protein null allele of miranda. (A–C) Homozygous miraDEB embryos were stained for DNA (red, top row) and Prospero (green, middle row). Prospero is present throughout the neuroblast cytoplasm in metaphase (arrowhead, column A) and anaphase (arrowhead, column B). After cell division Prospero enters the nuclei of both the larger apical daughter (arrowhead) and the smaller basal daughter (arrow, column C). Arrows indicate two small, basally located, Prospero-positive nuclei in column A and one nucleus in column B that belong to ganglion mother cells produced by previous neuroblast divisions. (D) Progeny from males and females of the genotype miraDEB/TM3, Ubx–lacZ were stained with anti-Miranda and anti-β-galactosidase antibodies. Ventral views of two representative embryos are shown; the top and bottom embryos are oriented with anterior to the left and right, respectively. The top embryo expresses β-galactosidase in the Ubx pattern, characterized by a sharp anterior boundary (indicated by the bar) and therefore carries the TM3, Ubx–lacZ “blue balancer” and is not homozygous for miraDEB. This embryo exhibits a wild-type staining pattern of Miranda, with characteristic strong staining in the procephalic neurogenic region and in the midgut. In contrast, the lower embryo, which is identified as homozygous for miraDEB by the lack of β-galactosidase expression, fails to stain with anti-Miranda antibody. Propidium iodide staining shows that both embryos are accessible to staining (not shown).
In neuroblasts of homozygous miraDEB embryos, Prospero is not associated with the cortex in prophase but instead stays in the cytoplasm. Prospero remains in the cytoplasm through metaphase (Fig. 7A) and anaphase (Fig. 7B), is inherited by both daughter cells, and enters both daughter cell nuclei (Fig. 7C). The same defect in Prospero localization is present in homozygous Df(3R)oraI9 embryos (Shen et al. 1997) and in miraDEB/Df(3R)oraI9 embryos (data not shown). Furthermore, embryos homozygous for miraDEB exhibit defects in Even-skipped expression identical to those described for miranda point mutations (Ikeshima-Kataoka et al. 1997; data not shown).
Similarly, in neuroblasts of miraDEB embryos, Staufen is no longer asymmetrically localized; it resides either in the cytoplasm or diffusely along the cell cortex throughout prophase (Fig. 8A), metaphase (Fig. 8B), anaphase, and telophase (Fig. 8C). Staufen also fails to localize asymmetrically in embryos heterozygous for miraDEB and miraZZ176, a point mutation in Miranda (data not shown). We conclude that miranda function is required for the asymmetric localization of Staufen in mitotic neuroblasts.
Figure 8.
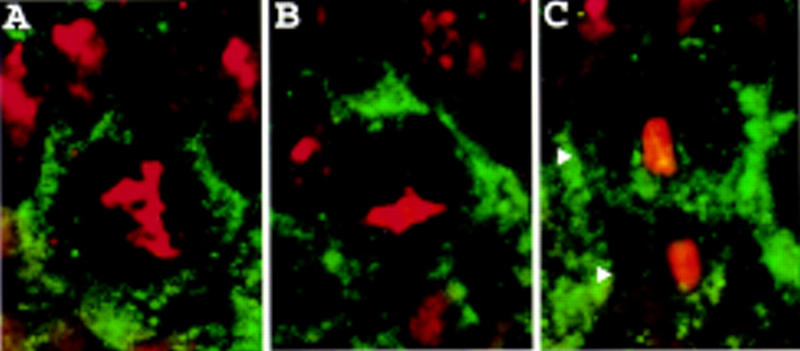
Staufen is mislocalized in homozygous miraDEB embryos. Homozygous miraDEB embryos were stained for Staufen (green) and DNA (red). Staufen is diffusely localized to the cell cortex in neuroblasts in prophase (A) and metaphase (B) and in telophase is present in both daughter cells (arrowheads, C).
Discussion
We have shown that Miranda localizes to the apical cortex in late interphase and early prophase, prior to its localization to the basal cortex. This complex apical-then-basal localization pattern requires microfilaments. Furthermore, Miranda consists of three distinct domains. The asymmetric localization domain lies in the amino terminus of Miranda, is necessary and sufficient for asymmetric localization, and interacts with a central domain of Inscuteable. The Numb interaction domain lies in a central 168-amino-acid region, and the Prospero interaction domain in a more carboxy-terminal 181-amino-acid region. We showed further that Staufen displays the same localization pattern as Miranda in mitotic neuroblasts, that this localization pattern requires miranda function, and that Miranda interacts with Staufen in vitro. The domain composition of Miranda is schematized in Figure 9A, and a summary of the known functional and physical interactions among asymmetrically localized factors is shown in Figure 9B.
Figure 9.
(A) Miranda contains several independent domains. The amino-terminal 298 amino acids of Miranda is sufficient for physical interaction with Inscuteable, localization to the neuroblast cortex, apical localization in late interphase and early prophase, and basal localization later in mitosis. Amino acids 300–467 are sufficient for Numb interaction, and amino acids 468–648 for Prospero interaction. The carboxy-terminal 530 amino acids are sufficient for interaction with Staufen. (B) Summary of known interactions. Arrows indicate functional interactions. Broken lines indicate physical interactions.
The Miranda asymmetric localization domain lies in the amino-terminal 298 amino acids
Identifying a minimal region of a protein required for asymmetric localization can provide useful tools for studying asymmetric localization. For instance, the identification of a discrete asymmetric localization domain of Prospero led to the finding that Miranda localizes Prospero (Hirata et al. 1995; Ikeshima-Kataoka et al. 1997; Shen et al. 1997). Our analysis of fragments of Miranda show that the amino-terminal 113 amino acids are required while the carboxy-terminal 530 amino acids of Miranda are dispensable for asymmetric localization and cortical localization (Fig. 2). At the levels of expression achieved in our experiments, we were able to observe asymmetric localization of exogenous Miranda in the presence of endogenous Miranda; the factors involved in the localization of Miranda are therefore not saturated under wild-type conditions.
A similar deletion analysis of Numb, which like Miranda resides at the cell cortex, suggested that cortical localization is strictly required for asymmetric localization (Knoblich et al. 1997). We cannot determine, therefore, whether the amino-terminal 113 amino acids are required solely for cortical localization or whether they are required to direct asymmetric localization of Miranda as well. Similarly, we cannot exclude the possibility that the carboxy-terminal 530 amino acids contain an asymmetric localization domain that cannot function in the absence of cortical localization. Nevertheless, it is clear that the amino-terminal 298 amino acids of Miranda are sufficient to direct a complex pattern of asymmetric localization at the neuroblast cortex.
Miranda is an adapter for multiple asymmetrically localized factors
We proposed previously that Miranda is an adapter protein that allows Prospero to interface with the asymmetric localization machinery. We have shown that Miranda interacts with Staufen, is required for the asymmetric localization of Staufen, and therefore plays a role in Staufen asymmetric localization similar to its role in Prospero asymmetric localization. Thus, Miranda is not an adapter solely dedicated to Prospero protein but is involved in the localization of Staufen as well.
The multidomain structure of Miranda allows it to interact with several different proteins and thereby serve as a multifunctional adapter. The existence of a distinct Numb interaction domain in Miranda suggests that Miranda may also serve as an adapter for Numb. Loss of zygotic miranda function does not abolish Numb localization, however. It is possible that Numb interacts with molecules other than or redundant with Miranda for its asymmetric localization. It is also possible that maternally supplied miranda is sufficient for Numb localization. Miranda may also be required for the asymmetric localization of as yet unknown molecules important for asymmetric cell division. These molecules could potentially be identified on the basis of their interaction with Miranda.
Staufen localizes prospero RNA by linking it to Miranda
We have observed a localization pattern of Staufen similar to that described for prospero RNA (Li et al. 1997) and consistent with a recent report (Broadus et al. 1998). It has also been shown that Staufen binds prospero RNA and that staufen function is required for the basal localization of prospero RNA (Li et al. 1997). Taken together with the requirement of miranda function for Staufen localization, these findings suggest that Staufen localizes prospero RNA by directly binding both to prospero RNA and to Miranda. The asymmetric localization of Miranda to the basal cortex during mitosis then results in the basal localization of both Staufen and prospero RNA as well.
Potential mechanisms of Miranda localization
The asymmetric localization of Miranda requires both inscuteable function and microfilaments. Our experiment does not allow us to distinguish whether the actin cytoskeleton is required for establishment or maintenance of asymmetric localization, or both. Actin-based mechanisms for Miranda localization might involve actin-based motors or rapid reorganization of the cortical cytoskeleton. Alternatively, actin may only be involved in the anchoring of Miranda after its localization by an actin-independent mechanism.
The localization of Miranda to the apical neuroblast cortex in late interphase and early prophase and the physical interaction between Miranda and Inscuteable in vitro suggest the simple model that Miranda is recruited to the apical cortex by Inscuteable. Such an association between Miranda and Inscuteable would necessarily be transient and regulated during the cell cycle, because in late prophase Inscuteable and Miranda form distinct crescents opposite each other on the apical and basal cortex, respectively.
Alternatively or additionally, Inscuteable may facilitate the assembly of a Staufen–Miranda complex. Staufen has been demonstrated to interact with Inscuteable by yeast two-hybrid and in vitro biochemical analyses (Li et al. 1997). Yet another possible function for Inscuteable is to load Miranda onto factors that transport Miranda to the basal cortex. Miranda crescents can still be seen in inscuteable mutant embryos but are no longer localized to the basal cortex (Shen et al. 1997).
An apical-then-basal localization pattern has now been described for Prospero, prospero RNA, Staufen, and Miranda, but the importance of apical localization is still unknown. During delamination of the neuroblast from the ectodermal epithelium, the neuroblast retains contact with the apical epithelial surface through an extended cell process. This apical process may provide the sole apical–basal polarity cue for the neuroblast. The role of Inscuteable would then be to interpret these cues and to use them to orient the mitotic spindle and localize protein and RNA crescents along the apical–basal axis. The early expression of Inscuteable in the apical process of the delaminating neuroblast (Kraut et al. 1996) is consistent with such a role.
In summary, the localization of Miranda to the apical cortex, its interaction with Inscuteable in vitro and its role in localizing several downstream factors suggest that Miranda occupies a central link between Inscuteable at the apical cortex and the localization of Prospero, Staufen, and prospero RNA to the basal cortex. How the apically localized Inscuteable early in mitosis dictates basal localization of intrinsic factors for asymmetric cell division may be elucidated by further studies on the genetic and cell biological mechanisms of the asymmetric localization of Miranda.
Materials and methods
Drosophila stocks and transgenics
The deficiency Df(3R)oraI9 deletes chromosome regions 92B1-3 to 92C1-3 and uncovers miranda (Shen et al. 1997). The deficiency Df(3R)oraK17 deletes chromosome regions 92B4-5 to 92B11 and does not uncover miranda (C.-P. Shen, unpubl.). Both stocks were kindly provided by Dr. William Pak (Purdue University, West Lafayette, IN). The miranda point mutant miraZZ176 (Ikeshima-Kataoka et al. 1997) was kindly provided by Dr. Chris Doe (University of Illinois, Urbana–Champaign).
Fragments of the miranda cDNA (diagrammed in Fig. 2A) were cloned into the pSK–N-myc vector (J.A. Knoblich, unpubl.) and subcloned into the pUAST vector (Brand and Perrimon 1993). The resulting constructs were individually injected into w− embryos to generate transgenic flies (Spradling and Rubin 1982). Flies carrying the transgene on the second chromosome were balanced with the CyO balancer and crossed into a Df(3R)oraI9/TM3, Sb Ubx–lacZ background. hairy–GAL4 (Brand and Perrimon 1993) was recombined onto the Df(3R)oraI9 chromosome. Flies of the genotype UAS–myc–miranda/CyO; Df(3R)oraI9/TM3, Sb Ubx–lacZ were crossed to hairy–GAL4 Df(3R)oraI9/TM3, Sb Ubx–lacZ flies to produce progeny of the genotype UAS–myc–miranda/+; hairy–GAL4 Df(3R)oraI9/Df(3R)oraI9. These flies, which are deficient for endogenous miranda and express exogenous Miranda in odd stripes in the embryo, were distinguished from siblings carrying TM3, Sb Ubx–lacZ by the absence of β-galactosidase expression.
DEB mutagenesis
Newly hatched male flies isogenic for the third chromosome were collected, starved for 5 hr, transferred to bottles containing filter paper (Whatman) soaked in 1% glucose and 7.5 mm diepoxybutane (DEB; Sigma), and kept at room temperature for 20 hr. The male flies were then mated en masse to female flies of the genotype TM3, Sb/TM6B, Tubby (Tb) in bottles. Two thousand sixty-eight Tubby male progeny from this cross were mated singly to female flies of the genotype Df(3R)oraI9/TM6B, Tb. Vials were scored for the absence of Tb+ pupae. Three such vials were identified, and progeny from these vials were used for complementation tests. One line was found to complement Df(3R)ora17 and was then balanced over TM3, Sb Ubx–lacZ. Homozygous embryos from this line failed to react with anti-Miranda antiserum. We conclude that this line carries a protein null mutant allele called mirandaDEB.
Drug treatments and antibody staining
Treatment with latrunculin A and antibody staining of embryos was performed as described in Knoblich et al. (1997) and St Johnston et al. (1991). Rabbit anti-Miranda antibody (Shen et al. 1997) was used at 1:1000 dilution. Monoclonal anti-myc antibody (9E10, Santa Cruz Biotechnology) was used at 1:300 dilution. Monoclonal anti-Prospero antibody MR1A (kindly provided by Dr. Chris Doe) was used at 1:5 dilution. Rabbit anti-Staufen antibody (kindly provided by Dr. Daniel St Johnston, Wellcome/CRC Institute, University of Cambridge, UK) was used at 1:2500 dilution. Rabbit anti-β-galactosidase antibody (Cappel) was used at 1:1000 dilution.
In vitro binding assay
cDNA fragments encoding amino acids 1–298 of Miranda, amino acids 1–223 of Numb, and amino acids 820–1026 of Prospero were cloned into the GST gene fusion vector pGEX-4T1 (Pharmacia). These constructs as well as pGEX-4T1 were transformed into DH5α Escherichia coli. IPTG was added to 100 μm to log phase bacteria to induce expression of fusion proteins, and the culture was incubated for an additional 3 hr before harvesting. Following sonication of the bacterial culture in 1× PBS, Triton X-100 was added to 1% and the lysate was then centrifuged to remove insoluble debris. The supernatant was mixed with glutathione–Sepharose-4B beads (Pharmacia) at 4°C for 30 min. The beads were then washed six times with 1× PBS/0.1% NP-40. The washed beads were kept at 4°C as a 50% suspension until use.
Fragments of the miranda cDNA (diagrammed in Fig. 4A) were cloned into the pNAC vector (kindly provided by Dr. J.P. O’Connor, University of Pennsylvania, Philadelphia). Full-length inscuteable and a fragment encoding amino acids 252–615 of Inscuteable were cloned into pSK–N-myc. These constructs were transcribed and translated in vitro in the presence of [35S]methionine with the TNT-coupled lysate system (Promega).
To test binding in vitro, 15–20 μl of the 50% suspension of beads bound to the GST fusion protein were mixed with 10–20 μl of lysate containing 35S-labeled polypeptides and incubated in 150 μl of 1× PBS/0.1% NP-40 for 30 min at 4°C. The beads were then washed six times with PBS/0.1% NP-40. After washing, loading buffer was added and the samples analyzed by SDS-PAGE.
Coimmunoprecipitation of in vitro-translated proteins
cDNAs encoding full-length Miranda and full-length Staufen (kindly provided by Dr. St Johnston) or staufen cDNA alone were transcribed and translated in vitro in the presence of [35S]methionine with the TNT-coupled lysate system (Promega). The lysate (30–50 μl) was mixed with 15–20 μl of packed protein A–agarose (Bio-Rad) and 2 μl of anti-Miranda antiserum and incubated in 150 μl of 1× PBS/0.1% NP-40 at 4°C for 30 min. The beads were then washed six times with PBS/0.1% NP-40. After washing, loading buffer was added, and the samples analyzed by SDS-PAGE.
Acknowledgments
We thank S. Abdelilah for comments on the manuscript, S. Younger-Shepherd for advice on the mutagenesis, and all members of the Jan laboratory for helpful discussions; Dr. William L. Pak for the Df(3R)oraI9 and Df(3R)oraK17 stocks; Dr. Chris Doe for the anti-Prospero antibody and the miraZZ176 stock; and Dr. Daniel St Johnston for the anti-Staufen antibody. C.-P.S is supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship DRG-1410. J.A.K. was supported by a European Molecular Biology Organization postdoctoral fellowship and by the Howard Hughes Medical Institute. Y.M.C. has received support from a Medical Scientist Training Grant, the Program in Biological Sciences Markey Grant, the Sussman Fund, and the Herbert Boyer Fund. L.Y.J. and Y.N.J. are Howard Hughes investigators.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ynjan@itsa.ucsf.edu; FAX (415) 476-5774.
References
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Broadus J, Doe CQ. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr Biol. 1997;7:827–835. doi: 10.1016/s0960-9822(06)00370-8. [DOI] [PubMed] [Google Scholar]
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;39:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- Burchard S, Paululat A, Hinz U, Renkawitz-Pohl R. The mutant not enough muscles (nem) reveals reduction of the Drosophila embryonic muscle pattern. J Cell Sci. 1995;108:1443–1454. doi: 10.1242/jcs.108.4.1443. [DOI] [PubMed] [Google Scholar]
- Carmena A, Murugasu-Oei B, Menon D, Jiminez F, Chia W. inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes & Dev. 1998;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin DG. Cell division: Why daughters cannot be like their mothers. Curr Biol. 1996;6:651–654. doi: 10.1016/s0960-9822(09)00440-0. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–465. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of a homeoprotein, Prospero, during cell divisions in neural and endodermal development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath J, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Asymmetric cell division. Nature. 1998;392:775–778. doi: 10.1038/33854. [DOI] [PubMed] [Google Scholar]
- Knirr S, Breuer S, Paululat A, Renkawitz-Pohl R. Somatic mesoderm differentiation and the development of a subset of pericardial cells depend on the not enough muscles (nem) locus, which contains the inscuteable gene and the intron located gene, skittles. Mech Dev. 1997;67:69–81. doi: 10.1016/s0925-4773(97)00109-3. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- ————— The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc Natl Acad Sci. 1997;94:13005–13010. doi: 10.1073/pnas.94.24.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R, Campos-Ortega JA. inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell division in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Li P, Yang X, Wasser M, Cai Y, Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Koisumi K, Hama C, Yoshioka T, Nabeshima T. Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem Biophys Res Commun. 1992;182:1326–1332. doi: 10.1016/0006-291x(92)91878-t. [DOI] [PubMed] [Google Scholar]
- Rhyu M, Knoblich J. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–526. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of Numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Ruiz Gomez M, Bate M. Segregation of myogenic lineages in Drosophila requires Numb. Development. 1997;124:4857–4866. doi: 10.1242/dev.124.23.4857. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- Shen C-P, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The Prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of Numb autonomously determines sibling neuron identity in Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Blasberger D, Kashman Y. Latrunculins—Novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauger C, Doonan J. Cell decision in plants. Curr Opin Cell Biol. 1993;5:226–231. doi: 10.1016/0955-0674(93)90107-2. [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:942–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]