Abstract
The paramyxovirus hemagglutinin-neuraminidase (HN) protein plays multiple roles in viral entry and egress, including binding to sialic acid receptors, activating the fusion (F) protein to activate membrane fusion and viral entry, and cleaving sialic acid from carbohydrate chains. HN is an oligomeric integral membrane protein consisting of an N-terminal transmembrane domain, a stalk region, and an enzymatically active neuraminidase (NA) domain. Structures of the HN NA domains have been solved previously; however, the structure of the stalk region has remained elusive. The stalk region contains specificity determinants for F interactions and activation, underlying the requirement for homotypic F and HN interactions in viral entry. Mutations of the Newcastle disease virus HN stalk region have been shown to affect both F activation and NA activities, but a structural basis for understanding these dual affects on HN functions has been lacking. Here, we report the structure of the Newcastle disease virus HN ectodomain, revealing dimers of NA domain dimers flanking the N-terminal stalk domain. The stalk forms a parallel tetrameric coiled-coil bundle (4HB) that allows classification of extensive mutational data, providing insight into the functional roles of the stalk region. Mutations that affect both F activation and NA activities map predominantly to the 4HB hydrophobic core, whereas mutations that affect only F-protein activation map primarily to the 4HB surface. Two of four NA domains interact with the 4HB stalk, and residues at this interface in both the stalk and NA domain have been implicated in HN function.
The Paramyxoviridae are enveloped, negative-strand RNA viruses that infect both humans and animals (1). The family is divided into two major subfamilies, and it includes, among others, mumps virus, measles virus, parainfluenza viruses 1–5, Newcastle disease virus (NDV), Nipah virus, Hendra virus, respiratory syncytial virus, and metapneumovirus. Although successful vaccines have been developed for some of these pathogens, such as the measles and mumps viruses, it has proven more difficult to develop successful therapeutics for others, particularly respiratory syncytial virus (RSV). RSV and human metapneumovirus (MPV) are second only to influenza virus in causing respiratory infections and are followed by human parainfluenza viruses (1). NDV infection of poultry has been responsible for imposing major economic costs. NDV, Nipah virus, and Hendra virus have been classified in the United States as select agents.
The paramyxovirus RNA genome encodes 6–10 proteins, and virions contain two spike glycoproteins embedded in a lipid membrane that surrounds the matrix protein and nucleocapsid core (1). For infections to proceed, the virion must fuse its lipid envelope with a membrane of the host cell. For nearly all paramyxoviruses, membrane fusion is triggered at the plasma membrane in a receptor-dependent, pH-independent manner (1–3). For most members of the virus family, the two viral glycoproteins mediate this entry process—an attachment protein referred to as HN, H, or G, depending on the virus, and the fusion (F) glycoprotein (1–3). RSV and MPV do not require their attachment protein G for entry or to generate an infectious virus, indicating that the F proteins of these viruses are necessary and sufficient to mediate cell penetration and infection (1). Most of the attachment proteins are type II membrane proteins, with N-terminal transmembrane domains (TM) followed by a stalk region and a C-terminal globular head domain (1). The hemagglutinin-neuraminidase (HN) attachment proteins are thought to form tetramers in their active form and are found in a subset of the paramyxoviruses, including parainfluenza virus 5 (PIV5), mumps virus, NDV, Sendai virus, and human parainfluenza viruses 1–4. HN contributes multiple functions to the virus, including binding to receptor (sialic acid) for virus attachment to cells, receptor-destroying (neuraminidase) activity for virus budding, and fusion protein activation. The C-terminal neuraminidase (NA) domain, obtained by proteolytic cleavage or expression of the NA domain alone, contains the receptor binding site and neuraminidase activity (4–6).
The F proteins form trimers and belong to the class I viral fusion protein group, which includes the influenza virus hemagglutinin, HIV gp160, and Ebola virus GP (1–3, 7–9). However, there is no sequence homology between class I fusion proteins, and they show only limited structural similarities, consisting of helical heptad repeat regions, HRA and HRB in F, and internal hydrophobic fusion peptide sequences that are important in fusion. F initially folds to a metastable, prefusion conformation (1, 10). On activation, a dramatic refolding of the F protein is thought to catalyze the membrane fusion process by juxtaposing the target cell membrane, containing the inserted fusion peptides, with the viral membrane, containing the C-terminal transmembrane domains at the end of HRB. The attachment glycoprotein is thought to activate F refolding, which requires proteins derived from the same paramyxovirus (1, 2, 11, 12).
The paramyxovirus HN protein stalk domain carries specificity determinants for F-protein activation, affects neuraminidase activity, and contributes significantly to the oligomerization of the protein (1, 3, 12). Mutational studies of the NDV HN stalk have examined effects on membrane fusion, NA activity, hemadsorption, F-protein complex formation, and oligomerization (13–16). Although mutations in the NDV HN stalk can affect both NA and membrane fusion activities, it has not been clear how these two functions are coupled. Here, we report the crystal structure of the intact NDV HN [Australia–Victoria (AV) strain] ectodomain to ∼3.3 Å. The structure reveals a four-helix bundle (4HB) stalk packed between two NDV NA domain dimers, which provide insight into the structural basis for stalk-dependent HN NA and membrane fusion-promoting activities.
Results and Discussion
Soluble NDV HN Ectodomain Crystallizes as Tetramers and Reveals the Stalk 4HB.
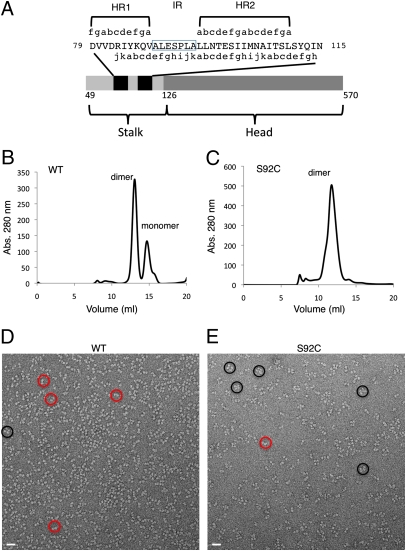
The NDV HN protein forms tetramers on the virion, and similar to the PIV5 HN, the isolated head NA domains exist primarily as monomers (4–6, 16–18). The HN stalk provides a significant driving force for oligomerization, but the TM domains are also thought to play a role (19). We expressed the NDV (AV) HN ectodomain (stalk + head) (Fig. 1) and observed variable amounts of both monomers and dimers in gel filtration chromatography separations (Fig. 1B), with a small number of potential tetramers evident in EM (Fig. 1D), which is consistent with the TM domain stabilizing the tetramer (19).
Fig. 1.
Biochemical and EM analysis of WT and S92C mutant NDV HN proteins. (A) Schematic diagram of the NDV HN ectodomain construct and predicted α-helical regions. The black bars indicate the predicted α-helix positions. (B) WT HN migrates as a mixture of dimers and monomers in gel filtration experiments. (C) The S92C mutant migrates primarily as dimers on gel filtration columns. (D) EM pictures of WT and (E) S92C proteins. The red circles indicate potential dimers, and the black circles indicate potential tetramers.
The NDV HN ectodomain crystallized in space group P43212 and diffracted X-rays to 3.3 Å resolution (Table S1), and the structure was solved by molecular replacement. Two copies of the NA domains are located in the asymmetric unit, with the HN tetramer lying along a crystallographic twofold axis (Fig. 2). Electron density maps showed the presence of a portion of the N-terminal stalk region, forming a 4HB. This helical region of the NDV HN stalk corresponds to 37 aa, representing about one-half of the 79 aa present in the stalk of the construct. No electron density was observed between the stalk helices and the NA domains, indicating that the short (∼7–10 aa) linker segments are disordered in the crystal. The partial stalk region of NDV HN may be stabilized by the packing between two NA dimers, consistent with the observation of dimers rather than tetramers in gel filtration experiments and EM (Fig. 1 B and D). The NDV HN ectodomain, lacking the TM regions, forms a weaker tetramer than the corresponding PIV5 ectodomain, which exists as a homogeneous tetramer in solution (6, 17).
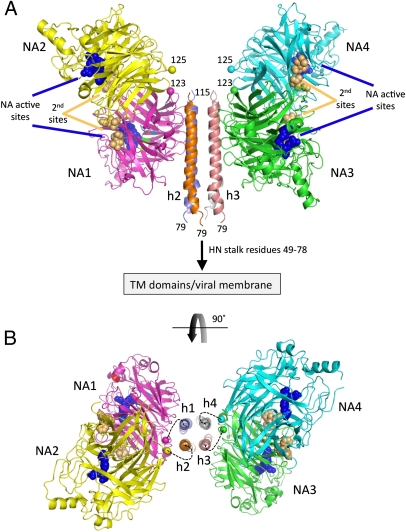
Fig. 2.
Structure of the NDV HN (AV) ectodomaim. (A) Two dimers of the NDV HN NA domains flank the 4HB in the stalk. The four NA domains are labeled NA1–NA4. The active sites are marked by three residues shown as blue CPK spheres (E400, R415, and Y525) and labeled accordingly. The secondary sialic acid binding sites located at the NA domain dimer interfaces are marked by residues shown as orange CPK spheres and labeled (second sites). The N termini of the four NA domains, residues 123 and 125, are labeled and indicated by their CA atoms shown in CPK format colored by chain. The connections of the N-terminal region of the stalk to the HN TM domains and viral membrane are indicated. (B) End-on view of the packing of the HN stalk tetramer between two NA domain dimers rotated through 90° as indicated by the curved arrow. Although no electron density was observed to connect the HN stalk helices with the individual NA domains, the dotted lines indicate possible linkages between these domains, with NA1/NA2 and NA3/NA4 forming covalently linked dimers through C123 and C92 in the S92C mutant. The four-stalk helices are indicated as h1–h4.
For the NA domains that directly contact the stalk 4HB, the first observed residue is C123 (Fig. 2A), whereas for the two NA domains positioned above the stalk, the first observed residue is A125. The distances between the end of the stalk helices and each of the four NA domains range from ∼10 to 24 Å, with the longest distances across the observed twofold symmetric arrangement of the four NA domains. The missing linker residues could span distances of 26–38 Å, ranging from 7 to 10 aa in an extended conformation, which is consistent with the distances observed between the individual helices and NA domain residues. Given the twofold symmetry of the tetramer, preferential linkages between the two stalk helices most proximal to a pair of NA domain dimers seem most likely, because linkage distances are shorter and would not lead to steric clashes at the top of the stalk 4HB. The distances are consistent with the number of missing linker residues, but they do not allow an unambiguous assignment of single-chain connectivities. The most likely arrangement is indicated in Fig. 2B.
We generated a panel of cysteine mutations (Fig. S1), including a S92C mutant, in the stalk region to aid our assignment of the sequence register of the 4HB. The S92C mutation stabilizes the dimer of the soluble ectodomain in gel filtration chromatography and produces a leading edge shoulder peak, which could indicate tetramerization (Fig. 1C). An increase in the number of potential tetramers was also observed in EM (Fig. 1E), but quantitation of the oligomer species is complicated by the fact that different projections of the NA domain dimers may seem to be single subunits. NDV HN contains a cysteine at residue 123 that forms an interchain disuflide bond that is not fully oxidized in the expressed ectodomain (Fig. S1). The S92C mutation increases the percentage of covalent dimers on nonreducing SDS gels to near 100% (Fig. S1E), indicating that the mutant forms an additional disulfide bond between the same subunits as C123. The S92C mutant was crystallized (Table S1), and interchain electron density was observed in the stalk region, consistent with a disulfide bond formed by C92 between the same subunits linked through the native disulfide at C123 (Fig. S2). The S92C disulfide also supports the proposed stalk–NA domain linkages (Fig. 2B). The S92C disulfide bond breaks the fourfold symmetry of the stalk 4HB, which is likely a reflection of the predominance of dimers and monomers rather than tetramers for the secreted ectodomain. Similar behavior is observed for a second cysteine mutation at residue Q87C (Fig. S1). The sequence register of the stalk region was, in part, established using the C92 disulfide along with B-factor sharpened electron density maps that provided improved electron density for the HN stalk side chains, such as M104 and Y112 (Fig. S2).
The observed NDV HN stalk spans residues 79–115, and it includes putative heptad repeat regions that have been extensively studied by mutagenesis (13–16). Thirty residues of the ectodomain (49–78) are not observed but would tether the observed stalk to the viral membrane (Figs. 1A and 2A and Fig. S3). The structure reveals the tetramerization of the HN stalk in a helical coiled-coil form, and it complements previously published biophysical studies on the PIV5 stalk (6, 17). The observed contacts between the HN stalk and NA domains are asymmetric, with two of four NA domains packing against the HN stalk and two of four NA domains pointing up and away from the stalk, not making any contacts (Fig. 2).
NDV HN binds sialic acid at the NA domain active site as well as through a secondary site located at NA-domain dimer interfaces (Fig. 2A). The four NA active sites are pointed to the outside of the HN tetramer, with two of these (NA2 and NA4) oriented distal to the stalk and N-terminal TM domains (Fig. 2A). Two of the secondary binding sites point to the top of the HN tetramer, whereas two are oriented below the NA domains. The arrangement of these eight sialic acid binding sites in the NA domain dimers suggests that NA domains might reorient when binding to sialic acid on cells. This orientation might affect NA domain to stalk interactions, and functional studies implicate both NA domain and stalk residues located at the observed NA to stalk interface in membrane fusion, NA activity, and pathogenicity of NDV strains.
NDV HN Stalk 4HB Hydrophobic Core.
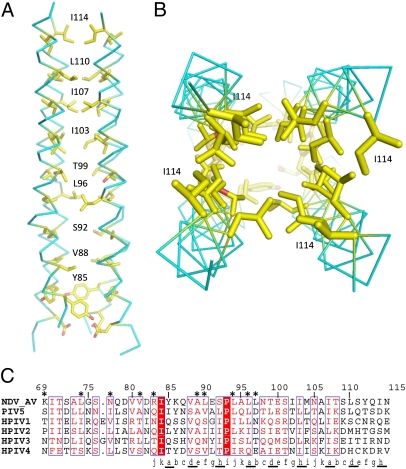
The NDV HN structure reveals that the stalk 4HB hydrophobic core is formed by an 11-residue repeat, not the previously assumed 7-residue (heptad) repeat (Fig. 3). Although electron density is observed for residues 79–115 of the stalk, the well-defined helical region of the stalk begins with residue 83 and ends with residue 114. Residues Y85, V88, S92, L96, T99, I103, I107, L110, and I114, corresponding to the a, d, and h residues of the 11-residue repeat, form the observed core of the 4HB (Fig. 3). This finding generates a repeated packing pattern of (i, i + 3, i + 4, i + 4) rather than the shorter heptad repeat (i, i + 3, i + 4). The 11-residue repeat has been observed in other parallel 4HB bundles from natural and engineered sources (20–22).
Fig. 3.
The hydrophobic core of the stalk 4HB. (A and B) The 4HB is formed by hydrophobic packing of 11-aa (3, 4-4) repeats. The hydrophobic core is formed by nine residues: Y85, V88, S92, L96, T99, I103, I107, L110, and I114. (C) Sequence alignment of the NDV HN stalk region indicating the 11-residue repeat (a–k), with residues a, d, and h underlined to indicate their location in the hydrophobic core of the 4HB. Residues that specifically affect F activation are indicated by an asterisk above the alignment. I84 and P93 are conserved and highlighted with a red background. Residues colored red and surrounded with a blue box are >70% conserved. Residues 69–78 are not present in the structural model.
Previous mutagenesis studies probed an intervening segment between two identifiable heptad repeats in the NDV HN stalk sequence, with HR1 comprising residues 79–88 and HR2 comprising residues 96–110 (Fig. 1A). The predicted HR1 begins four residues upstream of residue 83, with the structure showing partial unraveling of the helices in this N-terminal region of the stalk. The predicted end of HR2 is at residue 110, whereas the structure shows an additional turn of helix through residue 114. The predicted HR1 and HR2 regions are separated by residues 89–95, including a proline at position 93, leading to the suggestion that it might be nonhelical (15), but this is not borne out in the structure. Mutagenesis studies have mapped key functional attributes to residues in the intervening 89–95 segment, and the NDV HN stalk structure shows that these residues adopt the helical 4HB throughout (Fig. 3). Alignment of HN sequences maps the observed a, d, and h residues onto other HN stalk regions (Fig. 3C), revealing partial conservation of the hydrophobic core residues.
Structural Mapping of Stalk Mutations Reveals Two Stripes of Surface Residues Implicated in F-Protein Binding and Activation.
We analyzed previous mutational studies and grouped mutations into distinct categories (Table 1). Two broad categories of stalk mutants emerge from this analysis: mutations that affect only F activation (category I) and mutations that affect both F activation and NA activities (category II). The majority of stalk mutations does not affect hemadsorption (Had) activity or increase this activity relative to WT, and they were considered as WT.
Table 1.
NDV HN mutations
| Category | |
| Category I: Disrupt only fusion activation | |
| A: In structure (surface exposed) | R83 (16), A89 (15), L90 (13, 15), L94 (15), and L97 (13) |
| B: In structure (buried/core) | V81 (13) and L96 (13) |
| C: Not in structure | K69*, L74 (13), and N77* |
| Category II: Disrupt fusion activation and NA activity | |
| A: Buried/core residues or disrupt HN tetramer (bold) | |
| Point mutants | Y85 (16), V88 (13), I103 (13), and L110 (13) |
| CHO mutants* | T99N, E100N-CHO/102T, S101N-CHO/103T, I103N-CHO/N105S, N105-CHO/I107S, and A106N |
| B: Stalk surface residues | P93 (15) |
| C: Mutations located at NA to stalk domain interface | |
| Point mutants | I133 (14) |
| CHO mutants* | I102N-CHO/M104T and M104N-CHO/A106T |
References to mutants are indicated in parentheses.
*N-linked carbohydrate site mutations in HN (16).
Most of the category I mutations map to outer surfaces of the HN stalk 4HB (Fig. 4) and disrupt HN-F interactions, implicating this surface in forming direct contacts with the F protein. The previous mutational studies of the NDV HN stalk show that mutations at residues 69, 74, 77, 81, 83, 89, 90, 94, 96, and 97 specifically affect F-fusion activation, and many have been shown to disrupt F and HN coimmunoprecipitation (Table 1). Interestingly, a subset of these residues map to two opposite faces of the 4HB, defining two accessible surface stripes located at the N-terminal end of the observed 4HB and oriented away from the four NA domains (Fig. 4).
Fig. 4.
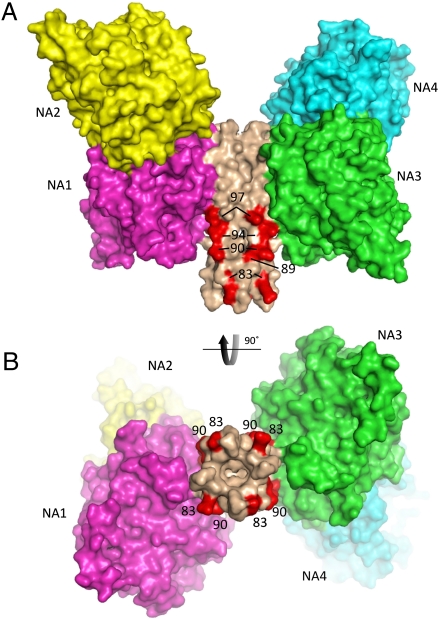
Stalk residues implicated in direct interactions with the NDV F protein. (A and B) Mutations of NDV HN stalk residues R83, A89, L90, L94, and L97 decrease F activation specifically and are implicated in forming direct contacts with the F protein. These residues are colored red and labeled on a surface representation of the HN tetramer model. B is a view rotated by 90°, which is indicated by the arrow. NA domains are colored as in Fig. 2, and the observed packing arrangement against the stalk would sterically restrict access to two sides of the proposed interaction site for F.
Residues R83, A89, L90, L94, and L97 are within the 4HB model, forming an exposed, predominantly hydrophobic surface on the 4HB. These exposed residues correspond to the j, e, f, and b positions of the 11-residue repeat of the 4HB (Fig. 3C). R83 was previously predicted to lie at position c of HR1, whereas residues A89, L90, and L94 were predicted to fall within the HR1/HR2 intervening region (Fig. 1A). Residues V81 and L96 are also in the structure, but they are buried at intersubunit interfaces; they may indirectly affect the local conformation of the stalk but do not perturb other HN functions. Residues K69, L74, and N77 are not within our modeled 4HB, but they are within the immediately N-terminal segment for which there was no observed electron density. Extrapolation of the 11-mer repeat from the observed 4HB to these residues places L74 and N77 at a and d positions of the bundle and residue K69 at a g position. From the current data, it is not possible to determine if these residues form a 4HB and if so, whether there is a register shift in the helical packing that would reposition these amino acids relative to the 4HB hydrophobic core. The alignment of the sequences of NDV HN with other paramyxovirus HN proteins (Fig. 3C) shows that these F-activation residues are only partially conserved and may contribute to the specificity of HN-F interactions.
NDV HN Stalk Mutants Can Affect both NA and Fusion-Promoting Functions by Destabilization of the Stalk Structure.
It has been unclear how HN stalk mutations could affect both NA and fusion-promoting activities and how these two functions might be linked. Structural analysis of NDV HN stalk mutants (Table 1) not only reveals a local surface patch of residues that specifically affects F activation (Fig. 4), but it also suggests that many of the mutations in the HN stalk that affect both F activation and NA activity likely disrupt the HN stalk structure (Table 1). The majority of the mutations (9/13) that have this dual affect on HN activities involves residues that are buried in the hydrophobic core of the 4HB (Table 1, category II A and B) or have been shown experimentally to disrupt the native HN tetramer (Table 1, category IIB, bold). Two mutants, engineered to introduce N-linked carbohydrate sites into the NDV HN stalk (E100N-CHO/102T and A106N), do not change hydrophobic core residues but destabilize the HN tetramer (16). E100 and A106 are located at the interface between stalk helices, and the introduction of a bulky carbohydrate modification could explain the experimentally observed destabilization. In addition, mutation of P93 may affect the 4HB stalk structure, although this residue is primarily surface-exposed. A subset of mutations map to the observed NA to stalk domain interface (Table 1, category IIC).
Disruption of the HN 4HB structure would disrupt the underlying oligomer scaffold that forms the putative F-interaction surface involving two adjacent helices of the 4HB (Fig. 4), thereby reducing F-protein interactions and activation. It is less clear how disruption of the HN 4HB stalk would affect NA activity. NA activity may be disrupted in these mutants, because loss of the stalk structure and disassembly of the HN tetramer may lead to a loss of cooperativity between the HN NA domains, which has been suggested previously (23). It is also possible that full NA activity is dependent on activating interactions with the stalk, potentially exemplified by the interactions observed in the crystal structure. The structural mapping of most of these mutations indicates that the observed linkage between the NA and fusion-promoting activities of the HN stalk mutants is likely caused by perturbations of the stalk structure that, therefore, affect both functions.
Specific Interactions Between the NDV HN NA and Stalk Domains May Define Functional Surfaces Important for HN Fusion-Promoting Activities.
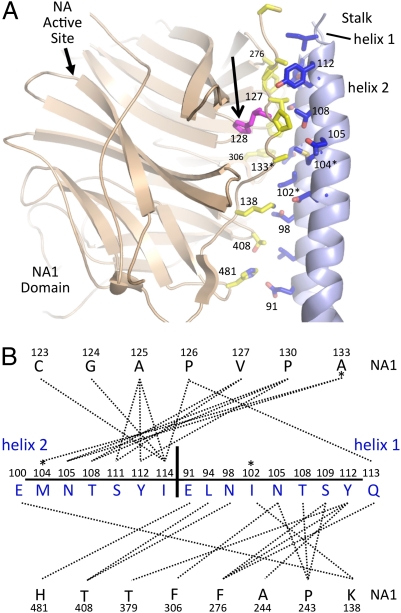
In the NDV HN structure, two of the NA domains pack against the observed 4HB (Fig. 2, NA1 and NA3), creating a twofold symmetry of the overall structure. These two NA domains individually form extensive interactions with helical pairs of the stalk 4HB (Fig. 5). Although the observed contacts between the NA1/NA3 domains and the stalk may be the result of crystallization, four features point to their potential functional significance. First, the arrangement is symmetrical, with the lower NA domains forming identical interactions with opposite sides of the 4HB. Second, mutations collected in Table 1 (category IIC) that are located at this interface interfere with both NA and fusion activities. Third, functional studies have implicated HN stalk residues through approximately residue 141 (12–16) in determining F-protein specificity, and five residues between 125 and 138 form part of the observed interface. Fourth, analysis of pathogenic mutants of NDV indicates that substitution of residue 128 from proline to histidine increases NA activity and may influence virulence (24). Residue 128 is adjacent to residues in the NA domain that pack against the stalk, and substitution may affect the local structure of the protein and stalk interactions (Fig. 5).
Fig. 5.
Functional mutations map to the NA to stalk interface. (A) An expanded view of a single NA domain to stalk interface is shown between the NA1 domain (beige) and stalk helices from two subunits (h1 and h2; light blue). Interface residues are shown in stick format, with oxygen atoms colored red and nitrogen atoms colored dark blue. Interface residues in the NA1 domain have carbon atoms colored yellow. Residues in the stalk domain have carbon atoms colored blue. Residue H128, implicated in NDV virulence, is shown in magenta and highlighted with an arrow. (B) Contact map between the NA1 domain and h1/h2 helices generated with the program MONSTER (36). NA1 domain residues are shown above and below a line of h1/h2 stalk residues. Residues in the stalk helices from the two subunits (h1/h2) are separated by a vertical line and indicated. Mutations of residues in this interface (102, 104, and 133) that affect HN NA and fusion activities (Table 1, category IIC) are indicated by asterisks in both A and B. I133 and L244 were truncated to alanine in the model.
Overall, the NA domain to stalk interface buries 928 Å2 surface area and involves contacts between 14 NA domain residues and 13 stalk residues (Fig. 5), spanning two of the 4HB helices. The overall character of the interface is hydrophobic, with 88 hydrophobic contacts and 3 electrostatic contacts. The size and nature of the interface are compatible with other functional protein interfaces.
The contact map between the NA domain and stalk (Fig. 5B) shows that residues throughout the NA domain contribute to the contact surface of the two stalk helices. The N-terminal region of the NA domain (residues 123–138) forms 16 contacts with the stalk helices, with V127 making contacts at the interface of the helices. A mutation of His128 to proline, which is observed in low pathogenicity strains of NDV (24), would likely disrupt the V127 interactions. A hydrophobic patch, including residues Y112 of the stalk and F276 of the NA domain, lies adjacent to the V127 site. Y112 forms three contacts with the NA domain (A244, F276, and T379), whereas F276 makes three reciprocal contacts with a single stalk helix (S109, Y112, and Q115). Finally, the category IIC mutants (Table 1), affecting both NA and F activities, map to this interface. Residue 133 and two carbohydrate mutants, affecting residues 102/104 and 104/106, fall into this group of mutants and are located at the observed NA to stalk domain interface (Fig. 5).
Conclusions
The membrane fusion and entry process in most paramyxoviruses is dependent on a poorly understood activation step that couples receptor recognition by the attachment protein to F-protein activation and membrane fusion. Key steps in this process remain to be fully understood, including potential receptor-triggered changes in HN, the nature, energetics, and specificity of the HN-F interaction, and the mechanism by which HN structural changes could promote F-mediated fusion. It seems more likely that HN proteins promote F conformational changes by actively destabilizing the prefusion conformation (a provocateur model) after receptor binding (11). An alternative clamp model, in which HN would stabilize the prefusion state to be released on receptor binding, seems less consistent with experimental evidence. Both models posit direct contacts between the F and HN proteins, and the HN stalk is thought to contain specificity determinants for this interaction.
The structure of the NDV HN ectodomain reveals a 4HB, consistent with the predicted oligomeric arrangement for native, full-length HN (19, 25). The 4HB structure is formed by hydrophobic core residues, with an 11-residue repeat that maps stalk residues to surface vs. exposed positions differently from prior heptad repeat predictions. Mutations in the hydrophobic core of the 4HB disrupt both NA and fusion-promoting activities, consistent with the interpretation that both functions depend on the integrity of the tetramer structure. Mutations that affect only F-protein activation and F-protein binding map to the surface of the 4HB, defining a potential binding site for F. NDV HN and F interactions are not of high affinity (15, 16, 26), and the structural organization of tetrameric HN proteins with trimeric F proteins is unclear. The fourfold symmetry of the stalk is not maintained through the native disulfide at C123, and the NDV HN structure reveals an overall twofold symmetry for the ectodomain, suggesting a possible stoichiometry of one HN with two F trimers. Whether HN-F assemblies would be limited to discrete oligomers or larger HN-F arrays at the membrane surface remains to be established.
The recruitment of F proteins to HN through the stalk region would provide a mechanism for colocalization of the two proteins in a manner that could prime the fusion apparatus before the engagement of sialic acid receptors. The missing N-terminal residues of the NDV HN stalk (49–78) would form a ∼45-Å-long helix, defining the position of the F-protein binding site relative to the viral membrane (Fig. S3A). However, these residues were not observed in the structure, suggesting that they do not form a stable 4HB. If the N-terminal residues adopt a fully extended conformation, the HN stalk could lengthen to ∼105 Å (Fig. S3). By comparison, the head of the prefusion F trimer is likely to be restricted to be ∼45–60 Å from the viral membrane. HN-F interactions before receptor binding could constrain the HN stalk to a compact conformation (Fig. S3). Receptor binding by the HN NA domains tethers virus to cells and could potentially provide a driving force for the extension of the HN stalk through the independent Brownian motions of the virus particle and cell. The preformed interactions of HN with F could transmit this force to F and initiate F conformational changes (Fig. S3), potentially driving the unraveling and extension of the F prefusion stalk and leading to an early intermediate that has been characterized in the PIV5 F-fusion mechanism (10, 27). Other important HN-F interactions or conformational changes, such as rearrangements of the NA domains, could also play a role in F activation (6). For measles virus H, functional and structural evidence for such domain rearrangements has been reported (28, 29). For NDV HN, disulfide bond cross-linking between NA1/NA2 and NA3/NA4 dimers enhances membrane fusion (30), but dimer of dimer arrangements and NA to stalk domain interactions might contribute to fusion activation. However, a stalk extension model for HN-dependent activation of F protein-mediated fusion has many features that are consistent with current experimental observations and provides an interesting foundation for future investigation of paramyxovirus entry mechanisms.
Materials and Methods
Cloning and Mutagenesis.
The NDV (AV) HN ectodomain, including residues 49–570, was cloned into pBACgus-11 (EMD; Novagen), and recombinant baculovirus stocks were generated. The N terminus of the purified HN protein contains additional residues introduced from the vector, including three amino acids (Ala, Met, and Ala), a 6His tag, and a thrombin cleavage site. The cysteine mutants, K86C, Q87C, S92C, S101C, and T108C, were generated using the QuikChange mutagenesis kit (Agilent).
Protein Expression and Purification.
The WT HN protein was expressed in both Hi5 and SF+ insect cell lines, whereas the cysteine mutants were only expressed in SF+ cells. Cell cultures were harvested within 72 h after infection with virus stocks. The proteins were purified from the supernatants using Nickel affinity chromatography (Qiagen) followed by size exclusion chromatography (GE Healthcare). The purified proteins were analyzed by SDS/PAGE and detected by Coomassie brilliant blue staining. Protein molecular weights and oligomerization were assessed by SDS/PAGE and size exclusion chromatography.
Crystallization.
The NDV HN was concentrated to ∼8 mg/mL in 25 mM Tris, pH 7.5, and 50 mM NaCl and was crystallized at room temperature by the hanging drop vapor diffusion method. The drops contained protein and precipitant of 12% PEG 4000, 200 mM Li2SO4, and 100 mM N-(2-Acetamido) Iminodiacetic Acid (ADA), pH 6.5, at 1:1 ratio. The mutant S92C protein was crystallized similarly, except that the PEG 4000 concentration was raised to 17% and 1 mM 2-mercaptoethanol was included. The crystals were harvested in 2–3% higher PEG 4000 concentrations, and for WT HN, the pH was also increased to pH 7.0.
Data Collection, Structure Determination, and Refinement.
The native dataset was collected at the Advanced Photon Source L and processed to 3.3 Å using HKL2000 (31). The S92C mutant dataset was collected at the Stanford Synchrontron Radiation Lightsource beamline 11–1 and processed to 4.3 Å. The NDV HN Kansas (4) structure (Protein Data Bank ID code 1E8T) was used as the search model to determine initial phases for the native dataset in both P43 and P43212 space groups, with four and two NA domains found in the asymmetric unit, respectively. Initial refinement of the structure was carried out in the P43 space group with four copies of the NA domains and stalk helices to allow for possible deviations in the symmetry of the stalk 4HB. The rfree set was transferred from the P43 to P43212 space group at the final stages, providing similar model statistics for two copies of the HN subunits in P43212 to confirm the correct space group. Model building and structure refinement were performed with Coot (32), Refmac (33), and Phenix (34, 35). Phenix was used in the final stages of refinement, applying Ramachandran restraints and automated rotamer building while refining coordinates, TLS groups, and restrained individual B factors. Use of noncrystallographic restraints or group B factors yielded higher Rfree values. The data collection and final refinement statistics are collected in Table S1.
Supplementary Material
Acknowledgments
We thank past and present members of the T.S.J. and R.A.L. laboratories. G.P.L. is a Research Specialist and R.A.L. is an Investigator of the Howard Hughes Medical Institute. This research was supported in part by National Institute of Health Research Grants AI-23173 (to R.A.L.) and GM-61050 (to T.S.J.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3T1E).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111691108/-/DCSupplemental.
References
- 1.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 2.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: The increasing diversity of paramyxovirus entry. FEBS J. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 5.Takimoto T, Taylor GL, Crennell SJ, Scroggs RA, Portner A. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology. 2000;270:208–214. doi: 10.1006/viro.2000.0263. [DOI] [PubMed] [Google Scholar]
- 6.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 7.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell CJ, Luque LE. The structural basis of paramyxovirus invasion. Trends Microbiol. 2006;14:243–246. doi: 10.1016/j.tim.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi SB, Dutch RE, Lamb RA. A core trimer of the paramyxovirus fusion protein: Parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 10.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: Implications for the mechanism of fusion triggering. J Virol. 2009;83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio RM, Melanson VR, Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 2009;4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone-Hulslander J, Morrison TG. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J Virol. 1999;73:3630–3637. doi: 10.1128/jvi.73.5.3630-3637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravel KA, Morrison TG. Interacting domains of the HN and F proteins of newcastle disease virus. J Virol. 2003;77:11040–11049. doi: 10.1128/JVI.77.20.11040-11049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 2006;80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378:282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bousse TL, et al. Biological significance of the second receptor binding site of Newcastle disease virus hemagglutinin-neuraminidase protein. J Virol. 2004;78:13351–13355. doi: 10.1128/JVI.78.23.13351-13355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinnes L, Sergel T, Morrison T. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology. 1993;196:101–110. doi: 10.1006/viro.1993.1458. [DOI] [PubMed] [Google Scholar]
- 20.Sales M, Plecs JJ, Holton JM, Alber T. Structure of a designed, right-handed coiled-coil tetramer containing all biological amino acids. Protein Sci. 2007;16:2224–2232. doi: 10.1110/ps.062702907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol. 2000;7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- 22.Stetefeld J, et al. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat Struct Biol. 2000;7:772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- 23.Mahon PJ, Deng R, Mirza AM, Iorio RM. Cooperative neuraminidase activity in a paramyxovirus. Virology. 1995;213:241–244. doi: 10.1006/viro.1995.1564. [DOI] [PubMed] [Google Scholar]
- 24.Tsunekun R, Ito H, Kida H, Otsuki K, Ito T. Increase in the neuraminidase activity of a nonpathogenic Newcastle disease virus isolate during passaging in chickens. J Vet Med Sci. 2010;72:453–457. doi: 10.1292/jvms.09-0474. [DOI] [PubMed] [Google Scholar]
- 25.Parks GD, Lamb RA. Folding and oligomerization properties of a soluble and secreted form of the paramyxovirus hemagglutinin-neuraminidase glycoprotein. Virology. 1990;178:498–508. doi: 10.1016/0042-6822(90)90347-t. [DOI] [PubMed] [Google Scholar]
- 26.Deng R, et al. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology. 1999;253:43–54. doi: 10.1006/viro.1998.9501. [DOI] [PubMed] [Google Scholar]
- 27.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: Capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navaratnarajah CK, et al. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashiguchi T, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 30.Mahon PJ, Mirza AM, Musich TA, Iorio RM. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: Implications for the mechanism of fusion promotion. J Virol. 2008;82:10386–10396. doi: 10.1128/JVI.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology: Macromolecular Crystallography, part A. Vol 276. London: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Salerno WJ, Seaver SM, Armstrong BR, Radhakrishnan I. MONSTER: Inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–W568. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.