Abstract
Many RNAs, proteins, and organelles are present in such low numbers per cell that random segregation of individual copies causes large “partitioning errors” at cell division. Even symmetrically dividing cells can then by chance produce daughters with very different composition. The size of the errors depends on the segregation mechanism: Control systems can reduce low-abundance errors, but the segregation process can also be subject to upstream sources of randomness or spatial heterogeneities that create large errors despite high abundances. Here we mathematically demonstrate how partitioning errors arise for different types of segregation mechanisms and how errors can be greatly increased by upstream heterogeneity but remarkably hard to avoid through controlled partitioning. We also show that seemingly straightforward experiments cannot be straightforwardly interpreted because very different mechanisms produce identical fits and present an approach to deal with this problem by adding binomial counting noise and testing for convexity or concavity in the partitioning error as a function of the binomial thinning parameter. The results lay a conceptual groundwork for more effective studies of heterogeneity among growing and dividing cells, whether in microbes or in differentiating tissues.
At balanced growth the abundances of cellular components are on average doubled during each cell cycle and then halved at cell division. But individual cells can deviate greatly from the average. Stochastic chemical reactions create fluctuations during the cell cycle and stochastic partitioning of components creates further fluctuations at cell division (1–4). This process perturbs concentrations and indirectly shapes molecular mechanisms by placing evolutionary constraints on reaction rates and network topologies.
The variation coming from stochastic production has been closely studied in the last decade (5–13), emphasizing how it could be much greater than expected from Poisson statistics due to upstream sources of randomness (5–7), how it could be controlled (8, 9) or exploited (10, 11) depending on selective pressures, and how fluctuations can reveal properties of the underlying mechanisms (12, 13). Partitioning errors can contribute just as much to the overall heterogeneity and recent results suggest that much of the noise attributed to, e.g., gene expression may in fact originate in stochastic partitioning (14). But not even the basic guiding principles of partitioning have been identified: how partitioning errors depend on upstream spatial heterogeneity or self-control, how they affect the observable cell heterogeneity, or what they can teach us about mechanisms.
The goal of this study is to provide a mathematical understanding of these principles by quantitatively modeling the heterogeneity introduced by various types of segregation mechanisms. We compare simple independent segregation where each segregating unit has a constant and independent probability of ending up in either daughter cell, to disordered segregation where variation in the partitioning machinery or the intracellular milieu further randomizes levels between daughters, or to various types of ordered segregation where copies directly or indirectly interact with each other to create a more even distribution between daughters. We then consider how such partitioning errors can be distinguished experimentally and discuss future directions.
Results
For dividing cells with x copies of a randomly segregating component X, where L and R copies segregate to each daughter cell after division, we can define a statistical partitioning error in several equivalent ways (SI Text),
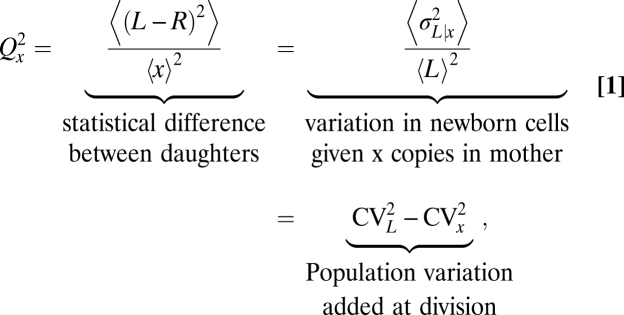 |
where the brackets 〈…〉 denote averages over all dividing cells in the population, regardless of the distribution of x values, and σ and CV denote the standard deviation and the coefficient of variation, respectively. These three perspectives on the same statistical measure show how partitioning errors capture statistical differences between daughters, the perturbation in levels introduced at cell division, and the contributions to population heterogeneity introduced by cell division.
In the simplest example where molecules are statistically independent and each copy goes to either daughter with probability ½, the partitioning error over the whole population simply decreases with the square root of the number of molecules, 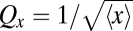 , regardless of fluctuations in x across cells. Because cells are not homogenous well-stirred reactors, for example due to active transport or molecular crowding (15–17), independent segregation may seem unlikely. However, spatial distributions can be uniform even in crowded environments, and even nonuniform spatial distributions can create independent partitioning statistics. Phenotypes consistent with independent segregation have indeed been reported for several cellular components despite complicated molecular mechanisms, e.g., endosomes and lysosomes (18) and symbiotic Chlorella cells (19). For macromolecules, one Escherichia coli study (20) suggested independent protein segregation, and another (6) showed that engineered mRNA transcripts follow independent or weakly disordered segregation, consistent with the observation that some transcripts clustered.
, regardless of fluctuations in x across cells. Because cells are not homogenous well-stirred reactors, for example due to active transport or molecular crowding (15–17), independent segregation may seem unlikely. However, spatial distributions can be uniform even in crowded environments, and even nonuniform spatial distributions can create independent partitioning statistics. Phenotypes consistent with independent segregation have indeed been reported for several cellular components despite complicated molecular mechanisms, e.g., endosomes and lysosomes (18) and symbiotic Chlorella cells (19). For macromolecules, one Escherichia coli study (20) suggested independent protein segregation, and another (6) showed that engineered mRNA transcripts follow independent or weakly disordered segregation, consistent with the observation that some transcripts clustered.
Many mathematical and computational models of stochastic processes in cells have included independent segregation of molecules (21–26) for example the pioneering gene expression analyses by Berg (22) and Rigney (23), but because independent segregation directly implies binomial partitioning errors, such models are not used to study partitioning itself but rather to provide realistic contexts for studying other mechanisms. Here we analyze stochastic processes that account for nontrivial mechanisms as well as arbitrary intrinsic and extrinsic fluctuations in x (Box 1). Many results are analytically exact (denoted ‘=’), and most approximations (denoted ‘≈’) are highly accurate as shown by comparing with exact simulations.
Box 1.
To increase the applicability of the models, we specify the probabilistic rules of partitioning as broadly as possible. For example, in the trivial case of independent partitioning we assume only that each molecule has an independent probability of being in either cell half. Such independence can be physically realized in many different ways, for example by having a well-mixed cytoplasm, by making immobile molecules with independent probabilities in either cell half, or by active segregation mechanisms that randomly pick molecules and move them to either cell half. However, the derivations of partitioning errors are often more tractable by picking one specific dynamic model that exactly instantiates the general assumptions. The detailed Markov processes (SI Text) used here are thus merely “mock processes” that provably generate the correct partitioning errors corresponding to the assumptions in the main text. In a two-component model where one daughter has L copies of the component of interest (total of x) and n copies of some upstream factor (total of v), one such mock process may include the jumps
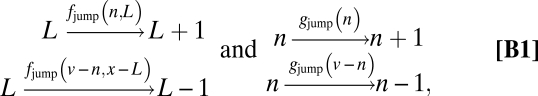 |
and the corresponding Markov process is then solved for the distribution or variance across cells. For independent partitioning we could use fjump = x − L, which gives the expected Qx2 = 1/〈x〉.
To further account for arbitrary heterogeneity in the total numbers x and v between dividing cells, we apply the law of total variance to separate the error given x and v from the error in x and v themselves, i.e., σ2(L) = σ2(〈L | x, v〉) + 〈σ2(L | x, v)〉 = σ2(x)/4 + 〈σ2(L | x, v)〉, where normalizing by 〈L〉2 = 〈x〉2/4 gives Eq. 1 (SI Text).
Disordered Segregation Can Greatly Increase Partitioning Errors.
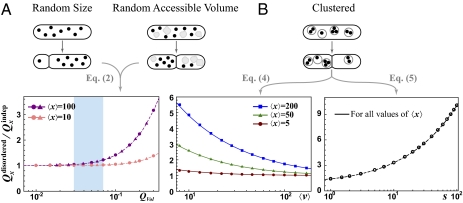
Partitioning errors can be substantial even for components present in high numbers due to upstream sources of randomness. For example, the cytoplasmic volume available to a component can vary between the two daughters either because of errors in the position of the septum or because large cytoplasm-excluding structures such as vacuoles segregate randomly (Fig. 1A). The individual copies then become statistically dependent even if they never interact, increasing the partitioning error. Here we make the single assumption that each molecule independently occupies a cell half with a probability that is proportional to the available volume in that half and that the molecules take up negligible space. The exact partitioning error then follows:
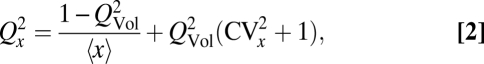 |
where QVol is the partitioning error of relative available volume. The generality of this result allows us to evaluate phenotypic data where mechanisms are unknown. Accounting for the unequal cell division measured for several microbes (27–29) shows that this volume variation should have a minor impact on partitioning errors (Fig. 1A).
Fig. 1.
Disordered segregation can increase partitioning errors greatly. (A) Upper: Variation in division site or random segregation of other large components causes fluctuations in daughter size or in the volume accessible to the segregating component. Lower: In simulations (symbols) x is picked from Poisson distributions and volume variation is generated by assuming either (truncated) Gaussian variation in the size (circles) or random segregation of another large component (triangle), where QVol is determined numerically. The exact analytical curve is dashed. The shaded area corresponds to experimental results for many microbes where 3% < QVol < 7% (27–29). (B) Upper: Segregating molecules (dots) are randomly grouped into vesicles (gray circles) and the vesicles are independently partitioned into daughter cells. Lower: Simulation results (symbols) and analytical expressions that are approximate (solid lines) and exact (dashed line) for cases of Eqs. 4 and 5, respectively. For the case of Eq. 4, x and v are sampled from Poisson distributions. For the case of Eq. 5, the value of v is first sampled from a Poisson distribution and x is then sampled from a Poisson distribution with average sv for each given v, so that q = 1; hence the second term of Eq. 5 is zero in this example. Simulations using different average 〈x〉 produce indistinguishable plots for virtually all values of 〈x〉 (SI Text).
Many molecules also form complexes and clusters or segregate in vesicles or organelles (Fig. 1B). The partitioning error then depends both on the distribution of copies among groups and on the segregation of the groups themselves. We assume that (i) each molecule (x copies before division) segregates independently into vesicles (v copies before division) and (ii) each vesicle segregates independently to either daughter (SI Text). The partitioning error then exactly follows:
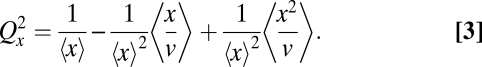 |
Inspecting Eq. 3 shows that the partitioning error further depends on the correlations between x and v across mother cells. We consider two cases to illustrate the effect. First, if x and v are statistically independent, the partitioning error follows
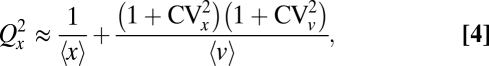 |
which is accurate for a wide range of parameters (Fig. 1B). Second, if the average number of molecules for a given number of vesicles is proportional to the number of vesicles, 〈x | v〉 = sv, so that s corresponds to the average number of molecules per vesicle, the partitioning error exactly follows
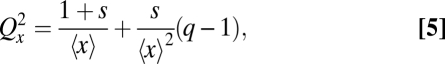 |
where q = 〈σ2(x|v)/〈x|v〉〉, and the second term typically is negligible. In both cases, clustered segregation could increase segregation errors greatly for small v or large s (Fig. 1B). The seemingly structured nature of many cell types—with numerous complexes, clusters, and organelles of varying sizes—can thus have an enormous randomizing effect at cell division. In the extreme where every molecule segregates to the same daughter, Qx2 = 1 + CVx2 (SI Text).
There are virtually no quantitative studies of disordered segregation (the conceptually similar “extrinsic noise” was only recently (7, 30) quantified in genetic networks) but these results suggest a substantial impact on heterogeneity. However, some partitioning mechanisms may also be less disordered than they appear. For example, the Min proteins that help control cell division in E. coli oscillate spatially between cell poles (31), suggesting that the majority of proteins would end up in the same cell half after septation. But the system is also presumably under selection to avoid large fluctuations that could interfere with control, and recent experiments (32, 33) indeed show surprisingly small partitioning errors.
Ordered Segregation Requires Extreme Parameters to Greatly Suppress Partitioning Errors.
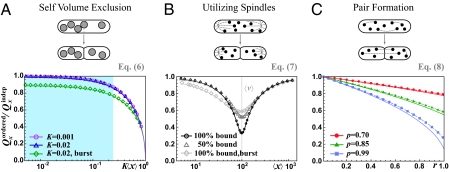
For components present in low numbers cells must either tolerate large partitioning errors or use control mechanisms to reduce them. Because little is known quantitatively about the mechanisms we consider three principles: size exclusion, binding to spindles or other cellular sites, and pair formation.
Cells could reduce partitioning errors passively if the segregating components occupy sufficiently large volume, which would create more available space in the cell half with fewer copies (Fig. 2A) (34). For very large components the error reduction would depend on exact shapes, but because even organelles are fairly small compared to the total cell volume, we assume (i) each individual component occupies a cell half with a probability that is proportional to the available volume, (ii) each molecule takes up a fraction K of the total cell volume, and (iii) the two cell halves have equal available volumes except for the location of component X studied. Such mechanisms produce a partitioning error
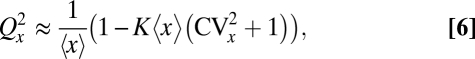 |
where 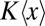 is the average total fraction of cell volume occupied by all segregating X units (SI Text). However, the effect is significant only if the organelles take up a large total fraction (Fig. 2A) and, except for chromosomes in some organisms, Eq. 6 suggests that most cell components occupy too small total volumes for self volume exclusion to substantially affect partitioning errors (34). Eq. 6 also shows that partitioning errors decrease with the variation in organelle numbers, because a larger fraction of cells then have so many organelles that volume exclusion is significant.
is the average total fraction of cell volume occupied by all segregating X units (SI Text). However, the effect is significant only if the organelles take up a large total fraction (Fig. 2A) and, except for chromosomes in some organisms, Eq. 6 suggests that most cell components occupy too small total volumes for self volume exclusion to substantially affect partitioning errors (34). Eq. 6 also shows that partitioning errors decrease with the variation in organelle numbers, because a larger fraction of cells then have so many organelles that volume exclusion is significant.
Fig. 2.
Ordered segregation requires extreme parameters to substantially reduce partitioning errors. (A) Upper: Segregating units with nonnegligible size (gray circles) exclude each other and thereby promote more even segregation. Lower: Simulation results (symbols) and analytical approximation (solid lines) plotted as a function of the average total volume fraction occupied by the segregating units, with x sampled from Poisson distributions for each 〈x〉 (circles and triangles) or assuming that synthesis occurs in a geometric burst with average burst size of 10 (diamonds). All results are truncated to ensure Kx < 1. The shaded area corresponds to the physiological regime for most cellular components (34) and shows that the effect should be minor. (B) Upper: Organelles (dots) compete for available binding sites (ends of astral, gray) and unbound organelles are partitioned independently. Lower: Simulation results (symbols) and analytical approximations (solid lines) when binding sites are distributed evenly between the two daughters. The circles and triangles correspond to v and x sampled from Poisson distributions, using 〈v〉 = 100, and diamonds show x and v sampled from processes where synthesis occurs in a geometrically distributed burst, with average burst sizes of 20 and 10 for v and x, respectively. The results show how modest the effects are when x and v are not exactly matched. (C) Upper: Among (x − δx,odd)/2 possible pairs in each cell, a fraction r of molecules (dots) binomially forms pairs, where δx,odd = 1 if x is odd and zero otherwise. Paired molecules segregate separately with probability p whereas unpaired molecules segregate independently. Under these assumptions, k = r(1 − Po/〈x〉), where Po is the probability that x is an odd number. Lower: Simulations (symbols), exact analytical results (Eq. 8, dashed line), and analytical approximations (k = r in Eq. 8, solid line) when x is sampled from a Poisson distribution of average 10. The results show that both r and p must approach 100% for efficient control. Derivations are given in SI Text.
Even if organelles hitchhiked with chromosomal segregation to reduce partitioning errors (Fig. 2B), for example by binding to mitotic spindles or spindle poles (18, 35), accurate partitioning is still hard to achieve. In cells that by chance have fewer binding sites v than organelles x, the unbound organelles will segregate randomly, and in cells with fewer organelles than binding sites, more organelles could bind to sites on one side than the other (1). To demonstrate the inaccuracy even in an idealized case we assume that (i) free organelles segregate independently and (ii) organelles always bind available sites, with no cells simultaneously having free organelles and free sites. The partitioning error then follows
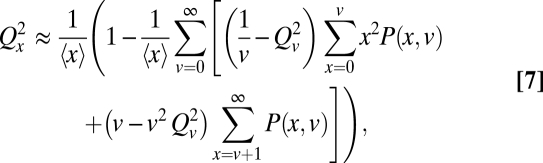 |
where P(x, v) is the probability of x organelles and v binding sites across dividing cells, and Qv is the partitioning error of the binding sites for a fixed v. Analyzing this equation further (Fig. 2B) shows that errors can be reduced even when the numbers are not perfectly matched—more organelles bound to sites in one cell half means relatively more free sites in the other half—but also that the effect is small. For example, even if the averages are perfectly matched, 〈v〉 = 〈x〉 = 100, each daughter cell receives exactly v/2 binding sites, and the only sources of noise in x and v are independent low-abundance Poisson fluctuations, the error Qx would be reduced less than threefold compared to uncontrolled, independent partitioning (Fig. 2B). Control works much worse yet when the averages 〈x〉 and 〈v〉 are lower or unequal, the organelles bind weakly, or the independent fluctuations are larger—with the slight twist that if the averages are unequal, even uncorrelated fluctuations could slightly improve control by making it more likely that the numbers are matched in at least some fraction of the individual cells, i.e., ensuring that sometimes x ≈ v even if 〈x〉 ≠ 〈v〉. With v instead as specific DNA binding sites, so that v may be approximated as fixed and without partitioning error, Eq. 7 simplifies greatly. If we further assume that distribution P(x) is approximately symmetric, the partitioning error can be approximated (SI Text) by
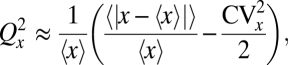 |
where for all symmetric distributions CVx ≤ 1, since x ≥ 0. For example, if both the CV and the normalized absolute deviation are ∼10%, the binding sites can reduce errors only threefold and the effect is smaller yet with increasing fluctuations in x.
The accuracy of chromosomal partitioning instead comes from the fact that the individual molecules form pairs with each other, using spindles to split each pair into separate daughter cells. The underlying molecular mechanisms are highly complex for chromosomes, but they are more tractable for, e.g., plasmid R1, where actin-like ParM proteins are attached to a pair of R1 plasmids via the ParR protein and push each plasmid to the opposite poles of E. coli (36, 37). Here we model the accuracy of such systems in terms of the phenotypic pairing parameters (Fig. 2C). We assume that (i) molecules form pairs by some arbitrary mechanism, with k as the average fraction of paired molecules that may depend on x; (ii) the two units of each pair are separated into different daughters with probability p and into the same daughter with probability 1 – p; and (iii) unpaired molecules (fraction 1 – k) segregate independently. For all such mechanisms—regardless of pairing mechanism—the partitioning error exactly follows
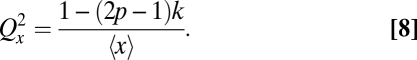 |
As seen from Fig. 2C, substantially reducing the errors is again highly nontrivial. For example, a fivefold reduction of the error requires both pairing and splitting probabilities of at least 90–95%. Chromosomes achieve extremely high k and p, using elaborate control mechanisms, but for most other cellular components such measures are perhaps not cost-effective.
Despite the challenges above, some studies suggest that segregation indeed is moderately ordered for several organelles. For example, during mitosis the chloroplasts in mesophyll protoplasts of Nicotiana tabacum reportedly use actin filaments to segregate more accurately than expected from independent segregation (38), and similar effects have been observed for mitochondrial inheritance in scorpion spermatogenesis (39–41). Studies of Golgi particles in mitotic HeLa cells in turn showed twofold smaller Qx compared with independent segregation for 〈x〉 ≈ 130 (42), whereas chloroplasts in unicellular algae (43) showed a threefold reduction of Qx given the measured 〈x〉 ≈ 20. We know of no quantitative measurement showing ordered partitioning of RNAs or proteins. Some RNAs bind to spindles (44), but also seem to form clusters of random size, with unknown net accuracy. The most precise non-DNA segregation mechanism reported to our knowledge is for carboxysomes in cyanobacteria (45), which achieve a fivefold reduction in Qx compared with independent segregation, for x = 6.
To Suppress Partitioning Errors, Each Source of Randomness Needs Separate Control.
For disordered segregation the task of reducing partitioning errors gets harder yet, not just because of the larger errors but also because fluctuations are introduced at multiple levels. For gene expression, multiple sources of noise can be suppressed by a single feedback loop (46) because deviations in abundances can trigger homeostatic responses regardless of what caused the deviations. Disordered segregation, however, as when proteins are sorted into clusters that then are sorted into the two cell halves, is fundamentally different. Even if the first sorting process is perfectly controlled by a homeostatic feedback system, the randomness of cluster inheritance can still produce large protein partitioning errors. The same is true if the second sorting is perfect but the first is not.
For a quantitative example we assume cooperative control over the sorting process of proteins into clusters and over the sorting of vesicles into each daughter cell. Specifically we assume that each molecule in a vesicle containing x1 copies migrates randomly to any other vesicle with a probability proportional to 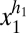 and that each vesicle migrates to the other cell half with a probability proportional to
and that each vesicle migrates to the other cell half with a probability proportional to 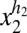 . Further assuming 〈x | v〉= sv as in Fig. 1B, the partitioning error approximately follows
. Further assuming 〈x | v〉= sv as in Fig. 1B, the partitioning error approximately follows
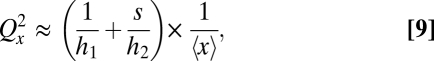 |
where s is the average number of proteins per cluster. Because the two noise terms are inversely proportional to their respective sorting efficiencies, and the partitioning error Qx is proportional to the square root of their sum, both processes must thus be separately and tightly controlled for accurate segregation. The same is true for most combinations of disordered and ordered segregation above: As opposed to negative feedback control of production rates, control of segregation corrects only errors produced by the process being controlled.
Does Segregation Control Serve to Reduce the Risk of Complete Loss?
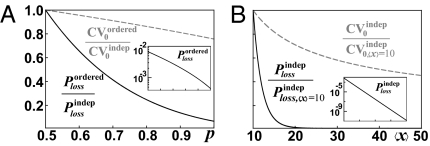
The results above raise the question of why ordered segregation would evolve if its effects on the heterogeneity are marginal anyway. For the many stable components that serve as templates either for their own production (e.g., mitochondria, chloroplasts, and plasmids) or where de novo production is slow [e.g., peroxisomes (47), centrioles (48), or bacterial flagella (49)], the greatest risk may be that all copies segregate to the same daughter. In fact, though ordered partitioning seems to have a small effect on standard deviations—forcing cells to tolerate some fluctuations—such mechanisms can still greatly reduce the risk of complete loss (Fig. 3). Reducing losses at cell division may then be the main selective driving force for ordered partitioning.
Fig. 3.
Ordered segregation and increased averages can dramatically reduce the rate of complete loss. (A) Relative reduction of the probability Ploss that all units segregate to the same daughter (solid line), as well as CV0 (dashed line), comparing ordered vs. independent segregation. We assume that 90% of the units form pairs (r = 0.9 model in Fig. 2C), and x is sampled from a Poisson distribution with mean 10. (B) Relative reduction of the loss rate and CV0 with the average number of molecules, again calculated assuming a Poisson distribution for x. Insets: Absolute loss rates Ploss as functions of p and 〈x〉, respectively. Derivations are given in SI Text.
Segregation Mechanisms Are Hard to Infer Experimentally.
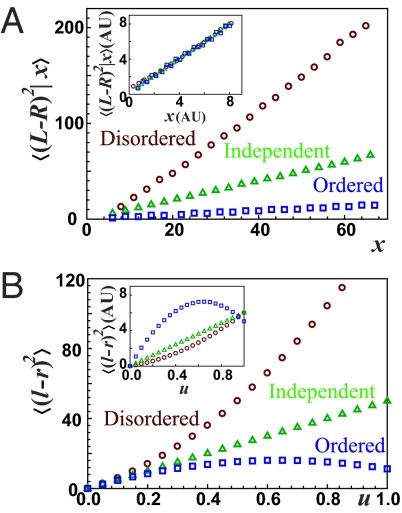
Measurement errors could make precise mechanisms appear imprecise, whereas ignoring legitimate outliers has the opposite effect. In most organelle studies, where it is possible to count individual components, another potential problem is the combination of crowding and low-resolution microscopy, producing systematic tendencies to undercount components in cells that by chance have a high abundance and thereby making mechanisms appear more precise than they actually are. One solution to this problem is to use quantitative microscopy to determine total abundances and then test how the partitioning errors scale with the abundance in the mother cell. However, the error Qx is proportional to the inverse square root of the average for several (but not all) of the mechanisms above, where it is the proportionality constant that represents the accuracy of segregation,
 |
as in Figs. 2 and 3. The problem is that if levels are reported in arbitrary units of fluorescence rather than in integer numbers of molecules, as in most fluorescent protein studies and several measurements of macronuclear DNA in ciliates (50, 51), then A and 〈x〉 are not known separately. Very different partitioning mechanisms—from the very ordered to the very disordered—can then display exactly the same scaling behavior and fit the same experiments (Fig. 4A). For example, the only study to our knowledge that quantified protein segregation statistics (20) showed that the partitioning error for a CI-YFP fusion protein in the λ-cascade of E. coli was inversely proportional to the abundance in the mother cell and assumed independent segregation (A = 1) to infer the average number of proteins. Assuming independent partitioning seems plausible: Most cytoplasmic proteins in E. coli probably follow independent segregation, and even if many CI-YFP molecules would be unspecifically bound to the chromosomes, these copies may also effectively segregate independently. However, it is important to note that such experiments assume rather than demonstrate independent segregation. If proteins formed clusters or were subject to ordered segregation, the fits could be equally good (Eq. 10) and the inferred averages would be incorrect. Both counting and quantification strategies may thus generate qualitatively misleading results, and segregation measurements demand the same attention as, e.g., gene expression noise.
Fig. 4.
Segregation mechanisms are difficult to infer. (A) Examples of ordered, disordered, and independent segregation mechanisms were simulated, and the conditional mean-squared error, given x copies in the mother cell, was plotted as a function of x. This measure is closely related to the partitioning error and was used to interpret experiments for protein segregation in E. coli (20). To illustrate disordered segregation we used the second case of Fig. 1B (Eq. 5) where the number v of vesicles is sampled from a Poisson distribution with average of 25, and x is sampled from a Poisson distribution with average 2v for each given v; hence 〈x | v〉 = 2v. To illustrate ordered segregation we used the pairing mechanism of Fig. 2C, with r = 0.8 and p = 0.99. All three curves are nearly perfectly approximated by 〈(L − R)2 | x〉 = ax. Inset: When levels are measured in arbitrary units, and the three mechanisms are assayed in separate experiments (different units), the curves are indistinguishable and can be laid on top of each other. Each mechanism thus fits the other datasets equally well. (B) Segregation mechanisms can be distinguished without counting the units by utilizing photoconvertible proteins. As the fraction of conversion (u) increases, the partitioning errors of the photoconverted molecules show convex, concave, and straight curves for disordered, ordered, and independent partitioning, respectively. All parameters are the same as in A with 〈x〉 = 50. Inset: The same scaling as in the Inset of A is applied to make the fluorescent level arbitrary, and the shapes of the curves allow us to distinguish the partitioning mechanism.
When the integer numbers of molecules cannot be directly counted, the type of segregation mechanism can be alternatively inferred by introducing an additional binomial source of noise (Fig. 4B). For example, a protein could be translationally fused to a photoconvertible fluorescent protein (PC-FP). If each PC-FP has an independent probability 0 < u < 1 of photoconversion for a certain amount of light (which should be demonstrated), the partitioning error of the photoconverted proteins (l and r copies in each daughter cell) follows (SI Text)
The partitioning error of the fluorescence from the photoconverted PC-FP, which is proportional to 〈(l − r)2〉, is thus a parabolic function of u, controllable by light. For disordered protein segregation the curve will be convex (〈(L − R)2〉 > 〈x〉), for ordered segregation it will be concave (〈(L − R)2〉 < 〈x〉), and for independent segregation it will be a straight line (〈(L − R)2〉 = 〈x〉). It is not necessary to measure the conversion fraction u: The shape of the curve is sufficient, as long as u is not too small. If u is also measured, which is possible with PC-FP but difficult with photoactivatable proteins, we can further fit the curve to a parabola and calculate 〈(L − R)2〉/〈x〉 by umin,max = (2 − 2〈(L − R)2〉/〈x〉)−1, which gives the value of A for the cases in Eq. 10. The intuition behind this effect is that for ordered partitioning, binomial sampling introduces significant noise relative to the original fluctuations when u is small so that 〈(l − r)2〉 overestimates the real partitioning error, but as u approaches 1 this additional noise is eliminated, and hence Eq. 11 becomes concave. For disordered partitioning, the dominant binomial sampling errors at low u instead hides a large natural error and hence 〈(l − r)2〉 underestimates the actual partitioning error, an effect that again disappears as u approaches 1. The curve of the partitioning error as a function of u then becomes convex. In practice, the highest value of u that can be achieved with many current photoconvertible proteins may be in the range where high-quality data are required to practically use this approach. However, more such proteins are being engineered and higher u can be achieved by photobleaching the FPs.
Outlook
Appropriate segregation of components at cell division is key to cell proliferation. However, although the heterogeneity caused by random births and deaths of molecules during the cell cycle has received enormous attention in recent years, the heterogeneity caused by partitioning errors has been virtually ignored in comparison. Many theoretical studies consider independent segregation and binomial errors. But just as gene expression is poorly approximated by a homogenous Poisson process due to feedback control or upstream sources of heterogeneity, cells are not bags of enzymes where all individual molecules segregate independently, but rather contain complexes, vesicles, cytoskeletons, and organelles. By considering these features we show how surprisingly hard it is for cells to eliminate partitioning errors through controlled segregation, how easily such errors are amplified by upstream factors, and how difficult it is to interpret seemingly direct partitioning measurements.
In addition to identifying these effects, we systematically investigate what patterns to expect from partitioning and how to more effectively design experiments. So far, measurements have been possible for only a handful of systems—often in obscure and poorly characterized processes and often with too low accuracy to discriminate among different types of segregation mechanisms—whereas the partitioning errors of crucial proteins and RNAs in model organisms have barely been measured at all. Although these studies are very interesting in their own right, not all components that have been counted thus necessarily count so much toward our general understanding of segregation, whereas the components that count cannot necessarily be counted (to paraphrase an alleged Einstein quote). This problem is likely to change in the near future. Several of the major experimental challenges have recently been overcome (6, 12, 52) and much of the necessary mathematical theory is now in place to address the central questions—How random is segregation and how much does it contribute to cellular heterogeneity?—quantifying one of the most central aspects of life at the level of individual cells.
Supplementary Material
Acknowledgments
We thank A. Hilfinger for discussions and comments on the manuscripts. J.P. gratefully acknowledges support from the Division of Mathematical Sciences of the National Science Foundation (Grant 0748760) and support from the US National Institutes of Health (Grant GM081563-01A).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013171108/-/DCSupplemental.
References
- 1.Warren G. Membrane partitioning during cell division. Annu Rev Biochem. 1993;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- 2.Marshall WF. Stability and robustness of an organelle number control system: Modeling and measuring homeostatic regulation of centriole abundance. Biophys J. 2007;93:1818–1833. doi: 10.1529/biophysj.107.107052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birky CW., Jr The partitioning of cytoplasmic organelles at cell division. Int Rev Cytol Suppl. 1983;15:49–89. doi: 10.1016/b978-0-12-364376-6.50009-0. [DOI] [PubMed] [Google Scholar]
- 4.Nordström K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 5.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 6.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 7.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 8.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 10.Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 11.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 13.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 14.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet. 2011;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss M, Elsner M, Kartberg F, Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys J. 2004;87:3518–3524. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni RP, Castelino K, Majumdar A, Fraser SE. Intracellular transport dynamics of endosomes containing DNA polyplexes along the microtubule network. Biophys J. 2006;90:L42–L44. doi: 10.1529/biophysj.105.077941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golding I, Cox EC. Physical nature of bacterial cytoplasm. Phys Rev Lett. 2006;96:098102. doi: 10.1103/PhysRevLett.96.098102. [DOI] [PubMed] [Google Scholar]
- 18.Bergeland T, Widerberg J, Bakke O, Nordeng TW. Mitotic partitioning of endosomes and lysosomes. Curr Biol. 2001;11:644–651. doi: 10.1016/s0960-9822(01)00177-4. [DOI] [PubMed] [Google Scholar]
- 19.McAuley PJ. Partitioning of symbiotic Chlorella at host cell telophase in the green hydra symbiosis. Philos Trans R Soc Lond B Biol Sci. 1990;329:47–53. [Google Scholar]
- 20.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 21.Brenner N, Farkash K, Braun E. Dynamics of protein distributions in cell populations. Phys Biol. 2006;3:172–182. doi: 10.1088/1478-3975/3/3/002. [DOI] [PubMed] [Google Scholar]
- 22.Berg OG. A model for the statistical fluctuations of protein numbers in a microbial population. J Theor Biol. 1978;71:587–603. doi: 10.1016/0022-5193(78)90326-0. [DOI] [PubMed] [Google Scholar]
- 23.Rigney DR. Stochastic model of constitutive protein levels in growing and dividing bacterial cells. J Theor Biol. 1979;76:453–480. doi: 10.1016/0022-5193(79)90013-4. [DOI] [PubMed] [Google Scholar]
- 24.Paulsson J, Ehrenberg M. Noise in a minimal regulatory network: Plasmid copy number control. Q Rev Biophys. 2001;34:1–59. doi: 10.1017/s0033583501003663. [DOI] [PubMed] [Google Scholar]
- 25.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rausenberger J, Kollmann M. Quantifying origins of cell-to-cell variations in gene expression. Biophys J. 2008;95:4523–4528. doi: 10.1529/biophysj.107.127035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyson JJ. Effects of asymmetric division on a stochastic model of the cell division cycle. Math Biosci. 1989;96:165–184. doi: 10.1016/0025-5564(89)90057-6. [DOI] [PubMed] [Google Scholar]
- 28.Nanninga N, Koppes LJH, de Vries-Tijssen FC. The cell cycle of Bacillus subtilis as studied by electron microscopy. Arch Microbiol. 1979;123:173–181. doi: 10.1007/BF00446817. [DOI] [PubMed] [Google Scholar]
- 29.Trueba FJ. On the precision and accuracy achieved by Escherichia coli cells at fission about their middle. Arch Microbiol. 1982;131:55–59. doi: 10.1007/BF00451499. [DOI] [PubMed] [Google Scholar]
- 30.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juarez JR, Margolin W. Changes in the Min oscillation pattern before and after cell birth. J Bacteriol. 2010;192:4134–4142. doi: 10.1128/JB.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Ventura B, Sourjik V. Self-organized partitioning of dynamically localized proteins in bacterial cell division. Mol Syst Biol. 2011;7:457. doi: 10.1038/msb.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birky CW, Skavaril RV. Random partitioning of cytoplasmic organelles at cell division: The effect of organelle and cell volume. J Theor Biol. 1984;106:441–447. doi: 10.1016/0022-5193(84)90001-8. [DOI] [PubMed] [Google Scholar]
- 35.Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: Visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Møller-Jensen J, et al. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 37.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheahan MB, Rose RJ, McCurdy DW. Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 2004;37:379–390. doi: 10.1046/j.1365-313x.2003.01967.x. [DOI] [PubMed] [Google Scholar]
- 39.Wilson EB. The distribution of the chondriosomes to the spermatozoa in scorpions. Proc Natl Acad Sci USA. 1916;2:321–324. doi: 10.1073/pnas.2.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson EB. The distribution of sperm-forming materials in scorpions. J Morphol Physiol. 1931;52:429–483. [Google Scholar]
- 41.Hood RD, Watson OF, Deason TR, Benton CLB., Jr Ultrastructure of scorpion spermatozoa with atypical axonemes. Cytobios. 1972;5:167–177. [PubMed] [Google Scholar]
- 42.Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hennis AS, Birky CW., Jr Stochastic partitioning of chloroplasts at cell division in the alga Olisthodiscus, and compensating control of chloroplast replication. J Cell Sci. 1984;70:1–15. doi: 10.1242/jcs.70.1.1. [DOI] [PubMed] [Google Scholar]
- 44.Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179:1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 46.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 47.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- 49.Neidhardt FC. Escherichia coli and Salmonella: Cellular and Molecular Biology. DC: American Society for Microbiology, Washington; 1996. p. 123. [Google Scholar]
- 50.Doerder FP. Regulation of macronuclear DNA content in Tetrahymena thermophila. J Protozool. 1979;26:28–35. doi: 10.1007/BF00327377. [DOI] [PubMed] [Google Scholar]
- 51.Berger JD. Regulation of macronuclear DNA content in Paramecium tetraurelia. J Protozool. 1979;26:18–28. doi: 10.1083/jcb.76.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang B, et al. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.