Abstract
Notch1 regulates gene expression by associating with the DNA-binding factor RBPJ and is oncogenic in murine and human T-cell progenitors. Using ChIP-Seq, we find that in human and murine T-lymphoblastic leukemia (TLL) genomes Notch1 binds preferentially to promoters, to RBPJ binding sites, and near imputed ZNF143, ETS, and RUNX sites. ChIP-Seq confirmed that ZNF143 binds to ∼40% of Notch1 sites. Notch1/ZNF143 sites are characterized by high Notch1 and ZNF143 signals, frequent cobinding of RBPJ (generally through sites embedded within ZNF143 motifs), strong promoter bias, and relatively low mean levels of activating chromatin marks. RBPJ and ZNF143 binding to DNA is mutually exclusive in vitro, suggesting RBPJ/Notch1 and ZNF143 complexes exchange on these sites in cells. K-means clustering of Notch1 binding sites and associated motifs identified conserved Notch1-RUNX, Notch1-ETS, Notch1-RBPJ, Notch1-ZNF143, and Notch1-ZNF143-ETS clusters with different genomic distributions and levels of chromatin marks. Although Notch1 binds mainly to gene promoters, ∼75% of direct target genes lack promoter binding and are presumably regulated by enhancers, which were identified near MYC, DTX1, IGF1R, IL7R, and the GIMAP cluster. Human and murine TLL genomes also have many sites that bind only RBPJ. Murine RBPJ-only sites are highly enriched for imputed REST (a DNA-binding transcriptional repressor) sites, whereas human RPBJ-only sites lack REST motifs and are more highly enriched for imputed CREB sites. Thus, there is a conserved network of cis-regulatory factors that interacts with Notch1 to regulate gene expression in TLL cells, as well as unique classes of divergent RBPJ-only sites that also likely regulate transcription.
Keywords: CSL, lymphoma, transformation
Notch receptors participate in a highly conserved signaling pathway that regulates development and tissue homeostasis. Signaling is mediated through a series of proteolytic cleavages, the last carried out by γ-secretase, that permit the Notch intracellular domain (NICD) to translocate to the nucleus and form a transcriptional activation complex with the DNA-binding factor RBPJ [also known as CSL in vertebrates and Su(H) in flies; for recent review, see ref. 1]. This complex in turn recruits a Mastermind (MAML) family member, other coactivators, and mediator complex components, leading to regulated target gene expression. NICD-containing transcription activation complexes turn over rapidly because of transcription-coupled degradation. In the absence of NICD, RBPJ binds several transcriptional corepressors, supporting models in which RBPJ acts like a transcriptional switch. However, genome-wide studies testing this model have yet to be done in higher organisms.
Notch effects vary with dosage and the chromatin state of the signal-receiving cell. Dysregulated Notch signaling underlies certain developmental disorders and cancers, most notably T-lymphoblastic leukemia/lymphoma (TLL), in which most tumors have somatic Notch1 gain-of-function mutations that increase NICD1 levels (for review, see ref. 2). Dysregulated RBPJ-dependent gene expression is also seen in other cancers, particularly B-cell tumors associated with Epstein-Barr virus (EBV) (3–6). However, activating Notch mutations are virtually unique to TLL, indicating an unusual susceptibility of T-cell progenitors to transformation by Notch.
Recent data indicate that transcription factors bind widely throughout genomes and that individual transcription-factor binding sites show substantial evolutionary divergence (7, 8). To identify conserved relationships of likely fundamental importance, as well as potentially interesting points of evolutionary divergence, we performed ChIP deep-sequencing (ChIP-Seq) for Notch1 and RBPJ in human and murine TLL cell lines.
Results
Notch1 and RBPJ Binding Sites in Human TLL Cell Genomes.
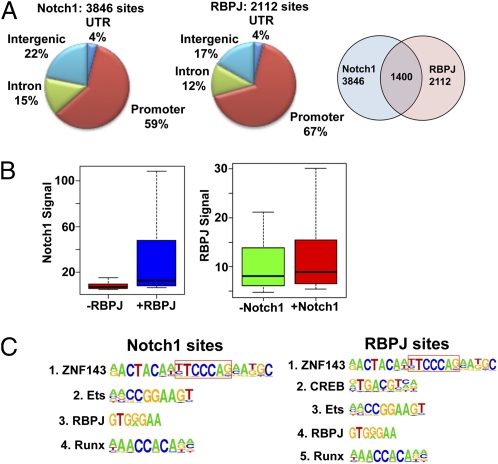
In initial studies, duplicate ChIP-Seq libraries were prepared from the Notch1-dependent human TLL cell line CUTLL1 (9) with RBPJ and Notch1 antisera. Analysis of pooled data identified 3,846 Notch1 binding sites and 2,112 RBPJ binding sites [false-discovery rate (FDR) <0.01] (Fig. 1A). Only 36% of Notch1 peaks overlapped with RBPJ peaks, and 66% of RBPJ peaks overlapped with Notch1 peaks. Most Notch1 and RBPJ binding sites were in promoters [regions within 2 kb of annotated transcriptional start sites (TSS)]. In line with studies in Drosophila suggesting RBPJ is stabilized on genomic DNA by NICD (10), RBPJ and Notch1 ChIP-Seq signals were higher at sites where both bound (Fig. 1B) (P < 10−6 for both comparisons).
Fig. 1.
Notch1 and RBPJ binding sites in human TLL cells. (A) Distribution and overlap of RBPJ and Notch1 binding peaks in CUTLL1 cells. Promoter regions are defined as sequences within 2 kb of the TSS of annotated genes. (B) Notch1 and RBPJ ChIP-Seq signals (expressed as reads per kilobase per million aligned reads, RPKM) at Notch1-only sites, RBPJ-only sites, and RBPJ/Notch1 cosites. (C) Enriched transcription factor motifs lying ±250 bp of Notch1 and RBPJ binding sites. A RBPJ consensus motif embedded within the extended ZNF143 motif is boxed.
Transcription Factor Motifs Enriched at Sites of Notch1 and RBPJ Binding in Human TLL Cells.
To determine factors that influence genomic Notch1 binding, recombinant RBPJ protein-binding microarray (PBM) data (11) were used to identify the highest affinity RBPJ binding site within 100 bp of the center of each Notch1 binding site. Both Notch1/RBPJ and Notch1-only sites were equally enriched for high-affinity RBPJ binding sequences compared with random genomic sequences (P < 10−10), suggesting that many Notch1-only sites also bind RBPJ. The promoter localizations of Notch1/RBPJ and Notch1-only sites were similar (63% and 59%, respectively). Failure to detect RBPJ at Notch1 sites with RBPJ consensus sequences may stem from shielding of RBPJ epitopes. Other Notch1-only sites may result from Notch1 binding to other chromatin-associated proteins or be an artifact of Notch1 cross-linking via long-range chromatin loops.
Further clues came from a search for transcription factor motifs enriched within 250 bp of RBPJ and Notch1 binding sites. For Notch1 sites, the most enriched motif (compared with overall genomic frequency) was that of ZNF143, followed by those for ETS and RUNX factors (P < 10−50 for each) (Fig. 1C). ZNF143 was also the most enriched motif associated with RBPJ binding sites, followed by the motifs for CREB, ETS factors, and RUNX factors (P < 10−50) (Fig. 1C). Of note, the ZNF143 consensus motif contains an embedded high-affinity RBPJ site. Overall, CREB consensus motifs were found in association with 46% of the RBPJ-only sites, compared with 25% of Notch1/RBPJ cosites (P < 10−6) (Fig. S1A). Compared with RBPJ sites without CREB motifs, the 588 RBPJ/CREB sites had lower-affinity RBPJ binding sites (P < 10−10) (Fig. S1B), but higher RBPJ signals (Fig. S1C) (P < 10−10), suggesting determinants other than DNA binding affinity (e.g., protein–protein interactions) contribute to RBPJ association with imputed CREB sites.
Confirmation of ZNF143 Association with RBPJ and Notch1 Binding Sites.
Western blotting detected NICD1, RBPJ, ZNF143, the ETS factors GABPA and ETS1, RUNX1, and CREB in three human and two murine Notch1-dependent TLL cell lines (Fig. S2A). In local ChIP assays of randomly selected Notch1 binding elements with imputed ZNF143, GABPA, ETS1, or RUNX1 sites, 17 of 17 tested sites showed binding by each factor (Fig. S2B), suggesting most imputed sites are occupied by the imputed transcription factor.
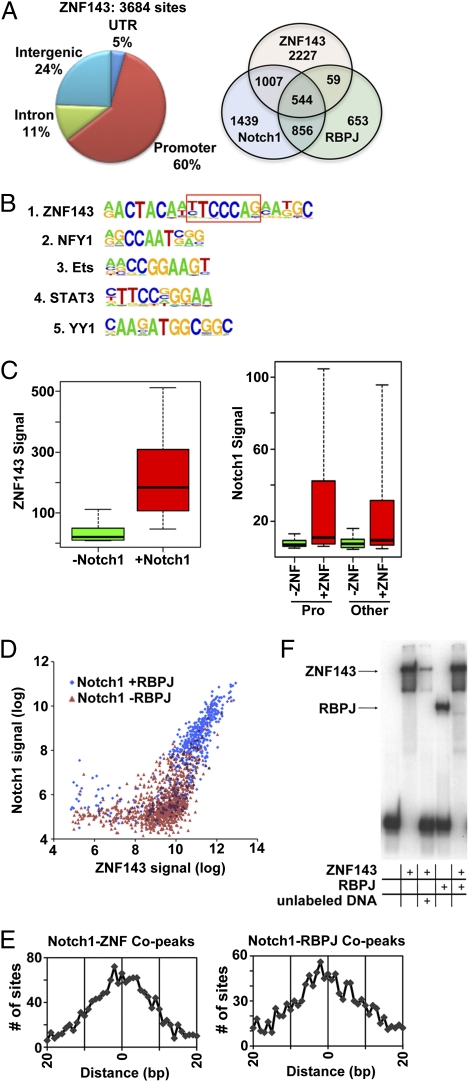
ETS factors and RUNX factors bind widely to Jurkat TLL cell genomes (12, 13), are required during early stages of T-cell development (14), and interact with Notch1 in early hematopoiesis (15), but little is known about ZNF143. ZNF143 ChIP-Seq was thus carried out in CUTLL1 cells, resulting in identification of 3,684 ZNF143 binding sites (FDR < 0.01) (Fig. 2A). ZNF143 also showed a bias for promoters, particularly bidirectional promoters, 285 of which bound ZNF143 in CUTLL1 cells. The most highly enriched motif near ZNF143 peaks was that of ZNF143 (Fig. 2B), followed by NFY1, ETS, and YY1, all of which also often bind bidirectional promoters (16, 17). Remarkably, 40% (n = 1,551) of Notch1 peaks lay within 100 bp of ZNF143 peaks, and 14% (n = 544) overlay RBPJ/Notch “copeaks” (Fig. 2A). Of ZNF143 sites overlapping with Notch1 peaks, 67% (n = 1,044) contained the ZNF143 consensus motif, and of these 57% (n = 591) had an embedded high-affinity RBPJ binding site. ZNF143 signals were higher at sites where Notch1 bound, as were Notch1 signals at sites of ZNF143 binding (Fig. 2C) (P < 10−100 for each). Consistent with these associations, Notch1 and ZNF143 signals correlated at cosites (R2 = 0.66), with cosites with significant RBPJ signals having the highest Notch1/ZNF143 signals (Fig. 2D).
Fig. 2.
Frequent association of ZNF143 and Notch1 binding sites in human TLL cells. (A) Distribution of ZNF143 binding peaks and overlap with RBPJ and Notch1 sites in CUTLL1 cells. (B) Enriched transcription factor motifs lying ±250 bp of ZNF143 binding sites. (C) ZNF143 and Notch1 ChIP-Seq signals (expressed as RPKM) at ZNF143 sites with and without Notch1, and Notch1 promoter (Pro), and nonpromoter sites with and without ZNF143. (D) Correlation of Notch1 and ZNF143 signals on individual cosites sites with or without RBPJ signals. (E) Overlap of Notch1 sites with ZNF143 and RBPJ binding sites in CUTLL1 cells, expressed as the distance in base pairs between the centers of Notch1 and ZNF143 copeaks (Left) and Notch1 and RBPJ copeaks (Right). (F) EMSA done with an oligonucleotide containing the SKP2 promoter ZNF143/Notch1 cosite. ZNF143 and RBPJ were added at concentrations of 280 and 407 nM, respectively. In one lane, an unlabeled probe was added in 200-fold excess over the labeled probe (4 μM:20 nM).
On average the positions of ZNF143 and Notch1 copeaks coincided exactly (within 2 bp), closely approximating the overlap of RBPJ and Notch1 copeaks (Fig. 2E). This finding suggests that Notch1 associates with ZNF143 cosites via RBPJ, which could either co-occupy sites with ZNF143 or exchange with ZNF143 on chromatin. Consistent with the latter possibility, binding of RBPJ and ZNF143 to DNA containing a cosite from the SKP2 promoter was mutually exclusive (Fig. 2F), suggesting that in cells RBPJ/Notch1 and ZNF143 complexes bind sequentially rather than simultaneously. It follows that the correlation of ZNF143 and Notch1 signal strength (Fig. 2D) is a function of binding site accessibility (e.g., chromatin “openness”) rather than cobinding of ZNF143 and Notch1 to these sites.
Identification of Distinct Classes of Notch1 and RBPJ Binding Sites.
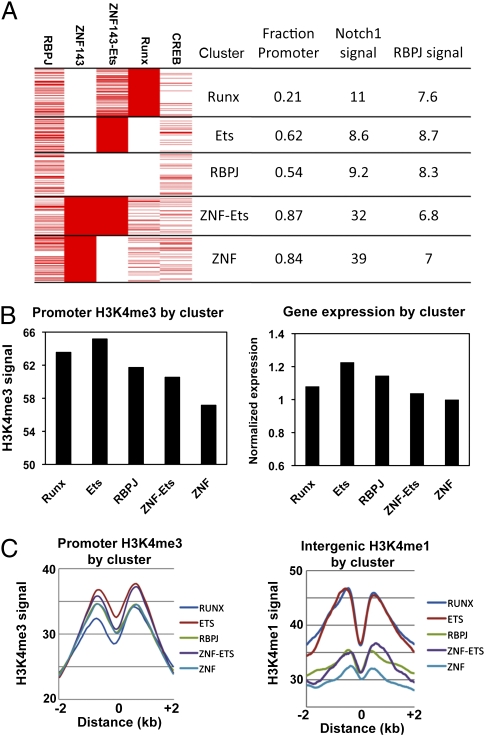
K-means clustering of all 3,846 Notch binding sites revealed five major clusters named RUNX, ETS, ZNF-ETS, ZNF, and RBPJ for the predominant cofactors (Fig. 3A). The RUNX cluster showed a nonpromoter bias, whereas the ZNF143 and ZNF143-ETS clusters had marked promoter biases and high Notch1 signals (Fig. 3A). To further characterize these clusters, Chip-Seq for activating H3K4me1 (enhancer) and H3K4me3 (promoter) histone marks and repressive H3K27me3 marks was done using CUTLL1 cells. The clusters showed subtle but statistically significant differences in H3K4me3 signals at promoters, with the ETS and RUNX clusters having highest marks (Fig. 3B) (P < 0.01) and the greatest nucleosome displacement (Fig. 3C), and the ZNF143 cluster having the lowest. Consistent with these associations, mean-expression levels of genes with Notch1 promoter binding were highest for the ETS cluster and lowest for the ZNF cluster (Fig. 3B) (P < 0.01). More striking differences were observed at nonpromoter Notch1 binding sites, with the ETS and RUNX clusters having the highest H3K4me1 signals and greatest nucleosome displacement and the RBPJ, ZNF143-ETS, and ZNF143 clusters the lowest (Fig. 3C) (P < 0.001).
Fig. 3.
Transcription factors associated with Notch1 binding sites define distinct classes of putative response elements in TLL cells. (A) K-means clustering of Notch1 binding sites in CUTLL1 cells. In the clusters shown at the left, each red line represents a genomic Notch1 binding site where cobinding (RBPJ, Znf143) or a motif (ETS, RUNX, CREB) for the indicated factor is seen. The numbers at the right are the fraction of binding sites within each cluster that are found within promoters of annotated genes and the associated Notch1 and RBPJ ChIP-Seq signal strengths (RPKM). (B) Mean histone H3K4me3 signals (Left) and relative gene expression (Right) for genes with Notch1 promoter binding by cluster designation. Normalized gene expression was determined by expression profiling of CUTLL1 cells. (C) Distribution of H3K4me3 signals in Notch1-binding promoters (Left) and H3K4me1 signals in Notch1-binding intergenic sites (Right) by cluster designation. “0” marks the center of the associated Notch1-binding peaks.
The chromatin marks of ZNF143-only and RBPJ-only (no Notch1) binding sites were also assessed. Promoter and intergenic ZNF143 sites without Notch1 binding had lower active chromatin marks (Fig. S3 A and B) (P < 0.0001 for both comparisons) and higher repressive marks (H3K27me3) (Fig. S3C) (P < 10−100), suggesting ZNF143 associates with repressive complexes. Similarly, RBPJ-only sites had lower intergenic H3K4me1 and promoter H3K4me3 signals (Fig. S3 D and E) (P < 0.05), and of these RBPJ-only sites, those with CREB motifs had lower intergenic H3K4me1 and promoter H3K4me3 signals than those without (Fig. S3 D and E) (P < 0.05). These findings are consistent with a repressive role for RBPJ in the absence of NICD1.
Genomic Notch1 Binding Sites and Target Gene Regulation.
To identify robust direct Notch1 target genes in CUTLL1 cells, we used a γ-secretase-inhibitor (GSI) washout method (18) that permits Notch1 reactivation in the presence of cycloheximide. High-confidence direct canonical Notch1 target genes were defined by a twofold or greater increase in expression within 4 h of GSI washout that was insensitive to cycloheximide and sensitive to dominant-negative MAML1, a specific inhibitor of canonical Notch1 signaling. Two-hundred forty-five genes met these criteria (Dataset S1), including previously identified target genes, such as HES1, HES4, HES5, DTX1, and MYC (18, 19). Notch1 bound the promoters of 61 (25%) of these genes (Fig. S4), an enrichment (P < 10−4, binomial test) over the total fraction of genes with Notch1 binding to their promoters (2,325 of 15,340 genes screened, 15.1%). The remaining target genes are presumably regulated through enhancers, a possibility consistent with the presence of at least one Notch1 binding site within 100 kb of the promoters of 127 of 179 target genes (69%) lacking Notch1 promoter binding.
For each target gene, the Notch binding site closest to the transcriptional start site was designated the most likely regulatory element (summarized in Dataset S1). RBPJ binding was detected by ChIP-Seq at 52% of these sites, an enrichment over the 36% overlap between Notch1 and RBPJ binding genome-wide (P < 10−6, binomial test). The 179 target genes without promoter binding showed larger expression changes in response to Notch1 than the 66 target genes that did (P < 0.05), indicating that Notch1-responsive enhancers are relatively potent inducers of target gene expression.
RBPJ/Notch1 binding sites near some target genes merit comment (Fig. S5). HES1, a gene involved in TLL and normal T-cell development (20), has two high-confidence and one low-confidence binding peaks. HES1 is unusual among target genes in CUTLL1 cells in having high levels of bivalent H3K4me3 and H3K27me3 chromatin marks, a combination that in embryonic stem cells defines genes that are poised to respond to differentiation cues (21); the significance of this chromatin structure near HES1 in TLL cells remains to be determined. Candidate enhancers (defined by high levels of H3K4me1 marks) were identified near other direct Notch1 target genes, including: the clustered GIMAP genes, which have been implicated in TLL cell survival (22); DTX1; and IL7R, IGF1R, and MYC, all of which contribute to Notch1-dependent TLL cell growth (18, 19, 23, 24). Local ChIP confirmed Notch1/RBPJ binding to the candidate MYC enhancer, which also bound p300, CREB binding protein, and RNA polymerase II (Fig. S6A). Although a promoter binding site for RBPJ/Notch1 has been reported in IL7R in CUTLL1 cells (23), we observed no binding to the IL7R promoter in CUTLL1 cells by ChIP-Seq, whereas local ChIP confirmed binding of Notch1 to a putative 3′ IL7R enhancer in three Notch1-dependent human TLL lines (Fig. S6B). Similarly, Notch1 binding to a candidate IGF1R intronic enhancer was also observed in these same lines.
Conservation of Factors Associated with Notch1 Binding Sites and Divergence of Factors Associated with RBPJ-Only Binding Elements.
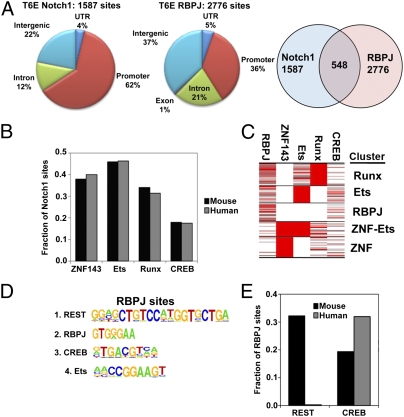
To determine conserved aspects of RBPJ/Notch1 interactions with TLL genomes, ChIP-Seq analyses were done on two Notch1-dependent murine T-acute lymphoblastic leukemia cell lines, T6E (25) and G4A2 (26). T6E cell contained 1,587 Notch1 sites and 2,776 RBPJ sites (FDR < 0.01) (Fig. 4A). The Notch1 binding-site distribution was similar to that in CUTLL1 cells, with a bias for promoters. Furthermore, the transcription factor motifs associated with T6E cell Notch1 binding sites were the same as in CUTLL1 cells and in the same rank order: ZNF143, ETS, and RUNX. The fraction of Notch sites overlapping with each cofactor was almost identical in mouse and human (Fig. 4B), and K-means clustering identified the same five classes of Notch1 binding clusters (Fig. 4C). For unclear reasons, fewer high-confidence Notch1 and RBPJ binding sites (548 and 668, respectively) were identified in G4A2 cells (Fig. S7), but the same associations were found.
Fig. 4.
Conserved and divergent features of Notch1 and RBPJ binding sites in murine and human TLL cells. (A) Distribution and overlap of RBPJ and Notch1 binding sites in the genome of the murine TLL cell line T6E. Promoter regions are defined as sequences within 2 kb of the TSS of annotated genes. (B) Conservation of the proportions of Notch1 binding sites associated with ZNF143, ETS, RUNX, and CREB motifs in the genomes of human CUTLL1 cells and murine T6E cells. The fraction of Notch1 binding sites that contain the indicate motif ±250 bp of Notch1 peaks is shown. (C) Conservation of RUNX, ETS, RBPJ, ZNF143-Ets, and ZNF143 clusters in murine T6E cells. (D) Enriched transcription factor motifs lying ±250 bp of RBPJ peaks in murine T6E cells. (E) Divergence of REST and conservation of CREB motifs near RBPJ peaks in murine T6E cells and human CUTLL1 cells. The fraction of RBPJ binding sites that contain the indicated motif ±250 bp of RBPJ peaks is shown.
In contrast to the conservation of motifs and cofactor combinations, Notch1 binding to orthologous elements was quite divergent. By strict criteria (see Materials and Methods), of the 245 direct Notch1 target genes identified in human CUTLL1 cells, only 50 genes (20.4%) showed Notch1 binding to orthologous elements in murine T6E cells (Dataset S1). Well-characterized target genes with conserved Notch1 binding sites include HES1 and HES5, and divergent Notch1 binding sites were observed in other conserved target genes, such as DTX1. Conserved RBPJ binding to orthologous elements was highly associated with conserved Notch1 binding (Fisher's test, P < 10−16), and conserved imputed ZNF143, ETS, and CREB sites showed smaller significant associations (Table S1).
Unexpectedly, the distribution of RBPJ binding sites was highly divergent in human and murine TLL cells. Only 36% of RBPJ sites were in promoters and only 35% of RBPJ sites overlapped with Notch1 sites in murine T6E cells (Fig. 4A); similar distributions were observed in murine G4A2 cells (Fig. S7). The motif most highly enriched near RBPJ binding sites in T6E cells (Fig. 4D) and G4A2 cells was that of REST, a DNA-binding transcriptional repressor (27), followed by the motifs for CREB and ETS. Overall, 895 of 2,776 (32%) RBPJ binding sites in T6E cells were associated with imputed REST sites. Most RBPJ/REST sites were outside of promoters and none bound Notch1; thus, these sites explain the divergence in RBPJ binding distributions between human and murine TLL genomes. Analyses using PBM data showed that RBPJ/REST sites generally lack high-affinity RBPJ binding motifs (Fig. S8A), suggesting that RBPJ binding to these sites involves protein-protein interactions. In contrast, only 6 of 2,112 (0.3%) of the RBPJ sites in human CUTLL1 cells were associated with REST motifs. To confirm REST binds CUTLL1 cell genomes, local ChIP for REST was done on four conserved RBPJ/REST sites identified in T6E cells. All four sites showed significant REST binding in both species (Fig. S8B). Furthermore, of 2,319 REST and 10,529 RBPJ binding sites identified by ChIP-seq [FDR <0.01 (28)] in EBV-transformed lymphoblastoid cell lines, only 12 (0.1%) overlapped. Thus, although REST binding sites may be conserved, RBPJ/REST colocalization in genomes may be restricted to murine cells.
A second interspecies difference in RBPJ-only sites was the presence of a higher fraction of RBPJ/CREB sites in human TLL cells (Fig. 4E) (Fisher's test P < 10−16). Conservation of CREB motifs was strongly associated with conserved RBPJ binding to orthologous elements (Table S2) (Fisher's test P < 10−16). Thus, unlike RBPJ/REST sites, there appears to be selective pressure favoring conservation of RBPJ-only sites associated with CREB motifs.
Discussion
Analysis of RBPJ/Notch1 interactions with TLL cell genomes identified conserved features with implications for transcriptional regulation as well as unexpected points of evolutionary divergence. Although Notch1 binds predominantly to promoters, promoter binding is a poor predictor of Notch1 responsiveness; all told, 97% of genes with Notch1 promoter binding do not respond robustly when Notch1 is activated. This finding is in line with studies in yeast, which found only 3% overlap between promoter occupancy and response to transcription factor perturbations (29). Similar findings have been noted in Drosophila (30). In nonresponsive genes with promoters bound by Notch1, loss of Notch1 may be compensated by other transcription factors, or Notch1 might only affect gene expression when other signaling pathways are also perturbed. The only positive predictor of Notch1 responsiveness in TLL cells was RBPJ cobinding, but only a small subset of genes with RBPJ/Notch1 promoter binding are Notch1 responsive, and other determinants must exist. The ChIP-Seq studies described here also revealed a large number of candidate Notch1-binding enhancers, which presumably regulate target genes that lack Notch1 promoter binding. Additional studies using methods such as chromosome conformation capture will be necessary to link these elements to target gene regulation.
Another variable likely to influence the Notch1 responsiveness of genes are cofactors that bind nearby. We identified a significant enrichment for overlapping ZNF143 binding sites and ETS and RUNX factor motifs near Notch1 binding sites in human and murine TLL cells. These cofactors occur in nonrandom combinations, pointing to a conserved cis-regulatory transcription factor network that forms the basis for a new understanding of Notch1 function in this cellular context. ETS factors are involved in T-cell specification and development (31, 32). Similarly, RUNX factors interact with Notch1 during hematopoietic stem cell development (15, 33), act upstream of Notch1 in early T-cell development (14), and (when deficient) predispose mice to TLL (34). An open question in understanding Notch1 function in T-cell progenitors is the identity of factors that “condition” the epigenome, allowing NICD1 access to regulatory elements that control T-cell development or, which when overactive, contribute to TLL. ETS and RUNX family members, by virtue of their association with Notch1 binding sites in TLL genomes, are candidates for such roles.
Unlike ETS and RUNX factors, ZNF143 has no known role in T-cell development, TLL, or Notch signaling. ZNF143 is a ubiquitously expressed zinc finger protein of unknown function originally identified as a regulator of selenomethionine tRNA gene expression (35). Our ChIP-Seq analyses confirm prior studies suggesting that ZNF143 localizes to promoters (36), particularly those driving transcription bidirectionally (17). Gene pairs regulated by bidirectional promoters are highly conserved in vertebrates (37); whether this is a consequence of convergent functions necessitating coordinate regulation or chance insertion of a coding sequence adjacent to a promoter with bidirectional activity during early vertebrate evolution is uncertain. ZNF143 motifs also appear to be enriched near hepatocyte SREBP-1 binding sites (38), suggesting ZNF143 is a general factor with broad functions. An issue emerging from our work is whether sites where RBPJ/Notch1/ZNF143 colocalize load ternary complexes or are instead occupied sequentially by RBPJ/Notch1 and ZNF143 complexes. In vitro studies suggest that binding of these factors to ZNF143 motifs is mutually exclusive, supporting a complex exchange model, but further work is needed to confirm this relationship in cells and to understand its basis.
Finally, we have identified major classes of genomic RBPJ sites that fail to bind Notch1 or do so very inefficiently. Both murine and human TLL genomes contain numerous RBPJ-only binding sites enriched for CREB motif that have relatively high levels of repressive chromatin marks, tend to flank genes with low expression levels, and often lack high-affinity RBPJ binding motifs. A second RBPJ-only site is associated with motifs for REST, a corepressor best known for its role in regulating neurogenesis (27) but which also has many other functions, such as regulating herpes simplex virus latency (39). RBPJ/REST sites are common in murine TLL cells, but essentially undetectable in human TLL cells or EBV-transformed human B cells, presumably because of divergence in factors that mediate recruitment of RBPJ to REST sites. One important Notch-independent RBPJ-dependent function in mammals involves the association of RBPJ with the transcription factor Ptf1a (40). The identification of CREB/RBPJ-only and REST/RBPJ-only sites in TLL cells suggests that other Notch-independent RBPJ functions exist and that the RBPJ “transcriptional switch” model of gene regulation is overly simplistic, at least in higher vertebrates.
Materials and Methods
Cell Culture.
Cell lines were cultured in RPMI medium 1640 (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, penicillin, and streptomycin; T6E cell medium was additionally supplemented with 1 mM sodium pyruvate and 5 μM 2-mercaptoethanol.
ChIP-Seq.
ChIP was done using the ChIP assay kit protocol (Millipore). ChIP-seq libraries were constructed per the Illumnia ChIP DNA library preparation kit. High-throughput sequencing was performed using the Illumina Genome Analyzer II. Reads were mapped to human genome hg18 or mouse genome mm9. Mapped reads ranged from 1 × 107 to 3.4 × 107 per library. ChIP-seq data (accession# GSE29600) are available through the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/).
Local ChIP.
Antibodies used for local ChIP assays are listed below. After DNA purification, real-time PCR was performed in triplicate on the CFX96 Real-Time PCR System (Bio-Rad), using input DNA as a standard and preimmune or nonimmune Ig as a control. Enrichment was estimated using the ΔΔCT method. Primer sequences used in local ChIP assays are available on request.
RNA Preparation and Gene-Expression Profiling.
Induction of Notch1 signaling through GSI washout was performed as previously described (18); additional details are available in SI Materials and Methods. Total RNA prepared with RNeasy Mini kit (Qiagen) was subjected to expression profiling in triplicate on Affymetrix human genome U133 plus 2.0 arrays. Affymetrix expression data were normalized using the robust multichip average package in R. Gene-expression profiling data (accession number GSE29544) are available through the Gene Expression Omnibus.
Analysis of ChIP-Seq Data
ChIP-Seq data were processed using QUEST (41). High-confidence peaks were defined by ChIP enrichment of ≥ threefold and FDR < 0.01. The FDR was determined by running the same peak calling analysis on a randomly selected subset of the input ChIP-seq data. Peaks for two transcription factors were called overlapping when the centers of the peaks were within 100 bp of each other. All binding signals were normalized to reads per kilobase per million mapped. Binding sites in the mouse genome (version mm9) were mapped to orthologous sequences in the human genome (version hg18) using the University of California Santa Cruz Liftover tool. A site was called conserved if Notch1 binding was present in both species within 100 bp of the center of the binding peak in CUTLL1 cells.
Motif Analysis and Clustering.
Enriched motifs within 250 bp of the center of each binding peak were determined using HOMER (42) in the default setting. Controls were genomic sequences selected at random with DNA percent GC content similar to the binding peaks. The hypergeometric test was used to quantify enrichment of motifs in DNA regions adjacent to binding peaks. For cluster analysis, each binding site was represented as a vector defined by the presence (i) or absence (ii) of a given cofactor (based on ChIP-Seq) or binding motif. Clusters were identified by K-means clustering of all of the binding sites.
PBM Analysis.
PBM data were obtained from Del Bianco et al. (11). Each 8-mer sequence was associated with a z-score based on RBPJ affinity. The 100-bp region around each Notch1 and RBPJ binding site were scanned for the motif with the highest z-score.
Antibodies.
Antibodies used in these studies are given in SI Material and Methods.
Statistical Methods.
Unless otherwise specified, statistical comparisons were performed and P values were calculated using the Mann-Whitney U test.
Electrophoretic Mobility Shift Assay.
Recombinant RBPJ was prepared as previously described (43). Human ZNF143 was expressed in Escherichia coli Rosetta(de3)pLys cells and purified by sequential chromatography on Ni-NTA affinity matrix followed by a MonoQ ion exchange column. An oligonucleotide containing the SKP2 promoter ZNF143/Notch1 cobinding site (GGAAAACTACAATTCCCAGCTTGGGCCTAGC) (ZNF143 site underlined, embedded RBPJ site in bold) was end-labeled with 32P and mixed at a final concentration of 20 nM with 200 ng of ZNF143 (280 nM) or 200 ng RBPJ (407 nM) in 20 mM Hepes, pH 7.9, containing 60 mM KCl, 10 mM DTT, 5 mM MgCl2, 10 μg/mL poly-dIdC, and 0.2 mg/mL albumin. After 30 min at 30 °C, the mixtures were loaded onto a 10% native Tris-glycine gel, separated by electrophoresis, and analyzed by autoradiography.
Supplementary Material
Acknowledgments
We thank Jeremiah Huang for technical assistance and the University of Pennsylvania Diabetes and Endocrinology Center for the use of the Functional Genomics Core (P30-DK19525). This work was supported in part by National Institutes of Health Grant P01CA119070 and a Specialized Center of Research grant from the Leukemia and Lymphoma Society (to J.C.A., S.C.B., and W.S.P.); The Howard Hughes Medical Institute, the National Human Genome Research Institute, and the National Institutes of Health Epigenome Mapping Centers Consortium, Encyclopedia Of DNA Elements Grant U54 HG004570, and Roadmap Epigenomics Grant U01 ES017155 (to B.E.B.); National Institutes of Health Grants R01CA047006, R01CA131354, and R01 CA085180 (to E.K.); and a National Science Foundation Graduate Student Fellowship (to J.Z.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE29544 and GSE29600).
See Commentary on page 14715.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109023108/-/DCSupplemental.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223:262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 5.Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, et al. Epstein-Barr virus nuclear protein 3C domains necessary for lymphoblastoid cell growth: Interaction with RBP-Jkappa regulates TCL1. J Virol. 2009;83:12368–12377. doi: 10.1128/JVI.01403-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowell RD. Transcription factor binding variation in the evolution of gene regulation. Trends Genet. 2010;26:468–475. doi: 10.1016/j.tig.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palomero T, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 10.Krejcí A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Bianco C, et al. Notch and MAML-1 complexation do not detectably alter the DNA binding specificity of the transcription factor CSL. PLoS ONE. 2010;5:e15034. doi: 10.1371/journal.pone.0015034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollenhorst PC, et al. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenberg EV. Regulatory factors for initial T lymphocyte lineage specification. Curr Opin Hematol. 2007;14:322–329. doi: 10.1097/MOH.0b013e3281de72a8. [DOI] [PubMed] [Google Scholar]
- 15.Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin JM, et al. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res. 2007;17:818–827. doi: 10.1101/gr.5623407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anno YN, et al. Genome-wide evidence for an essential role of the human Staf/ZNF143 transcription factor in bidirectional transcription. Nucleic Acids Res. 2010;39:3116–3117. doi: 10.1093/nar/gkq1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wendorff AA, et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010;33:671–684. doi: 10.1016/j.immuni.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 22.Chadwick N, et al. Identification of novel Notch target genes in T cell leukaemia. Mol Cancer. 2009;8:35. doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-García S, et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7Ralpha gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206:779–791. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haluska P, et al. In vitro and in vivo antitumor effects of the dual insulin-like growth factor-I/insulin receptor inhibitor, BMS-554417. Cancer Res. 2006;66:362–371. doi: 10.1158/0008-5472.CAN-05-1107. [DOI] [PubMed] [Google Scholar]
- 25.Pear WS, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashworth T, et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Zhao B, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA. 2011;108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 30.Li XY, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 32.Yu S, Zhao DM, Jothi R, Xue HH. Critical requirement of GABPalpha for normal T cell development. J Biol Chem. 2010;285:10179–10188. doi: 10.1074/jbc.M109.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns CE, et al. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 2009;113:5776–5782. doi: 10.1182/blood-2008-12-193607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundu M, et al. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood. 2005;106:3621–3624. doi: 10.1182/blood-2005-04-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J Biol Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 36.Myslinski E, Gérard MA, Krol A, Carbon P. A genome scale location analysis of human Staf/ZNF143-binding sites suggests a widespread role for human Staf/ZNF143 in mammalian promoters. J Biol Chem. 2006;281:39953–39962. doi: 10.1074/jbc.M608507200. [DOI] [PubMed] [Google Scholar]
- 37.Yang MQ, Taylor J, Elnitski L. Comparative analyses of bidirectional promoters in vertebrates. BMC Bioinformatics. 2008;9(Suppl 6):S9. doi: 10.1186/1471-2105-9-S6-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo Y-K, et al. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proc Natl Acad Sci USA. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA. 2005;102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hori K, et al. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.