Abstract
CD4 coreceptor expression is negatively regulated through activity of the Cd4 silencer in CD4–CD8– double-negative (DN) thymocytes and CD8+ cytotoxic lineage T cells. Whereas Cd4 silencing is reversed during transition from DN to CD4+CD8+ double-positive stages, it is maintained through heritable epigenetic processes following its establishment in mature CD8+ T cells. We previously demonstrated that the Runx family of transcription factors is required for Cd4 silencing both in DN thymocytes and CD8+ T cells. However, additional factors that cooperate with Runx proteins in the process of Cd4 silencing remain unknown. To identify collaborating factors, we used microarray and RNAi-based approaches and found the basic helix–loop–helix ZIP transcription factor AP4 to have an important role in Cd4 regulation. AP4 interacts with Runx1 in cells in which Cd4 is silenced, and is required for Cd4 silencing in immature DN thymocytes through binding to the proximal enhancer. Furthermore, although AP4-deficient CD8+ T cells appeared to normally down-regulate CD4 expression, AP4 deficiency significantly increased the frequency of CD4-expressing effector/memory CD8+ T cells in mice harboring point mutations in the Cd4 silencer. Our results suggest that AP4 contributes to Cd4 silencing both in DN and CD8+ T cells by enforcing checkpoints for appropriate timing of CD4 expression and its epigenetic silencing.
Keywords: T-cell development, transcriptional memory, cell fate decision, bipotential precursors
The helper versus cytotoxic T-cell lineage decision in the thymus has long been studied as a model system for binary fate decisions. These two subsets of T cells are selected from a common precursor pool of CD4+CD8+ double-positive (DP) thymocytes through interaction between clonally restricted TCRαβ chains and self-peptides presented on MHC class I or class II molecules expressed by thymic epithelial cells. During differentiation to the cytotoxic lineage, CD4 coreceptor expression is selectively silenced at the transcriptional level (Cd4 silencing), whereas CD8 expression from the Cd8a/Cd8b loci is transiently down-regulated and then restored through activation of postselection stage-specific enhancers (1). The Cd4 silencer, a cis-acting sequence with multiple transcription factor binding sites in the first intron of the Cd4 gene, is required for down-regulation of CD4 during the transition from the DP to CD8 single-positive (SP) stages as well as for repression at the CD4–CD8– double-negative (DN) stage of thymocyte differentiation (2, 3). Runx family transcription factors are required for Cd4 silencing through binding to the silencer, whereas ThPOK, induced following MHC class II-restricted selection, binds to the silencer to maintain active Cd4 expression in CD4SP thymocytes (4, 5). Runx proteins, particularly CD8 lineage-specific Runx3, and ThPOK not only regulate coreceptor expression but are also critically important for the development of the cytotoxic and helper lineages, respectively (5–8).
Genetic studies have shown that the Cd4 silencer is required for the establishment of Cd4 silencing, but not for its maintenance in mature CD8+ effector T cells (3). Conditional deletion of the Cd4 silencer by the Cre-LoxP system before the CD8SP stage of T-cell differentiation results in CD4 derepression similar to that observed in Cd4 silencer-deficient mice. All CD8+ T cells lacking the Cd4 silencer express CD4 at levels similar to CD4+CD8– cells. In contrast, Cd4 silencer deletion in activated proliferating CD8+ T cells using retroviral Cre transduction does not cause CD4 up-regulation. These results indicate that a heritable silenced state of the Cd4 locus is established during development and is propagated through a poorly understood mechanism to prevent Cd4 transcription in mature CD8+ T cells. This well-characterized example of heritable gene silencing provides an opportunity to gain insight into the molecular events required for establishing epigenetic gene regulation.
In this study, we demonstrate that AP4, a basic helix–loop–helix (bHLH) ZIP transcription factor encoded by the Tcfap4 gene, is essential for Cd4 silencing in immature thymocytes. AP4-deficient immature thymocytes derepressed Cd4 beginning at the DN3 stage, before selection of cells with productive rearrangement of Tcrb, and more markedly following β selection. Cd4 derepression caused by AP4 deficiency was further enhanced in the Cd4 silencer-deficient background, suggesting that AP4 functions independently of the Cd4 silencer but cooperates with silencer-binding factors to regulate appropriate timing of CD4 expression. Furthermore, AP4 was also found to complement Cd4 silencer-mediated activity in mature CD8+ T cells, enforcing inactivation of CD4 expression when cells with a mutant silencer and variegated derepression of CD4 acquired a memory phenotype. These results suggest that AP4 is a key negative regulator of the Cd4 gene both early and late in T-cell development.
Results
Identification of AP4 as a Cd4 Gene Regulator.
To identify potential Cd4 silencing factors, we used a combination of gene expression analysis and candidate gene approaches. It was previously shown that the mouse Cd4 proximal enhancer (Cd4PE) contains multiple E-protein-binding sequences (E boxes; CD4-1 and CD4-3 in Fig. S1) in motifs identified through DNaseI footprinting assays (9, 10). In vitro reporter assays suggested that, among these, the 5′ E box of the CD4-3 element is essential for enhancer activity. This E box was bound by HEB/E2A complexes in vitro, and DP thymocytes lacking HEB or both HEB and E2A had reduced Cd4 expression (11, 12). In contrast, two E boxes, which are located in CD4-1 and at the 3′ end of CD4-3, are not required for enhancer activity in vitro, and their functions have not been identified (9, 10). In the Cd4 silencer, at least five short sequences or small regions have been shown by targeted mutagenesis to be essential for Cd4 silencing (2). Two of the five sites contain consensus Runx-binding motifs, and mutation of these Runx-binding sequences resulted in complete Cd4 derepression that was similar to that observed in CD8+ T cells lacking the Runx cofactor CBFβ (4, 13). Factors binding to the other sites in the silencer have not been identified. Interestingly, one of the orphan sites, designated site 1, also contains an E-box motif, which is partially conserved between mouse and human (Fig. S2). Based on these results, we hypothesized that additional E proteins may be regulating the Cd4 gene, potentially as repressors.
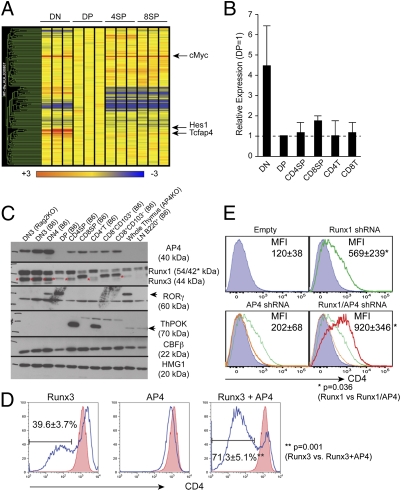
To identify potential E proteins contributing to Cd4 gene regulation in thymocytes, we compared gene expression profiles of cells at the DN, DP, CD4SP, and CD8SP stages of thymocyte differentiation, and focused on factors that contained bHLH domains and whose expression levels correlated with activity of the Cd4 silencer (Fig. 1A). We extracted ∼200 probe sets for known genes containing HLH domains and identified AP4, encoded by Tcfap4, as the only factor that met all criteria (Fig. 1A). We confirmed the stage-specific expression of AP4 by quantitative RT-PCR (Fig. 1B). AP4 mRNA expression was highest in DN thymocytes, and CD8SP thymocytes expressed ∼50% more mRNA than CD4SP thymocytes. At the protein level, AP4 was highly expressed in DN thymocytes, whereas little AP4 protein was detected in DP thymocytes (Fig. 1C). AP4 expression was equally high at the DN3 and DN4 stages, whereas Runx1 expression was down-regulated upon β selection. In mature SP thymocytes and peripheral T cells, AP4 protein expression was higher in CD8+ lineage cells than in CD4+CD8– cells. Consistent with mRNA levels, however, the amount of AP4 was lower in CD8+ cells compared with DN thymocytes.
Fig. 1.
Identification of AP4 as a Cd4 silencing factor. (A) Microarray analysis of genes encoding bHLH domain-containing proteins in different thymocyte subsets. Relative signal intensity is normalized against the expression level in DP thymocytes (shown as yellow). (B) Quantitative RT-PCR analysis of AP4 (Tcfap4) expression. Values were normalized against Hprt1 expression levels and are shown relative to the DP expression level. Data represent the average and SD of two independently prepared samples for each cell type. (C) Western blotting analysis of AP4 expression in different subsets of thymocytes and mature T lymphocytes. Anti-Runx, -RORγt, -ThPOK, and -CBFβ immunoblots serve as controls for population purity, and anti-HMG1 serves as a loading control. (D) Effect of AP4 overexpression in a double-positive thymoma cell line. AP4 and/or Runx3 were retrovirally expressed in AKR1 cells, and CD4 expression was assessed in transduced cells at 72 h postinfection (blue lines). Red-filled histograms indicate CD4 expression in empty virus-transduced controls. Proportions of CD4lo/– cells are shown with averages and SDs from five experiments. (E) AP4 and Runx1 synergistically regulate active Cd4 silencing in the 1200M cell line. CD4 expression in cells transduced with shRNAs targeting AP4 (orange), Runx1 (green), or both (red). Mean fluorescence intensities of CD4 staining are shown with averages and SDs from five experiments. Statistical significance was tested by unpaired two-tailed t test with P values shown.
To determine whether AP4 could contribute to Cd4 silencing, we overexpressed AP4 in a CD4+CD8+ DP cell line, AKR1 (Fig. 1D). Runx3 overexpression in AKR1 cells induced ectopic Cd4 silencing, resulting in ∼40% of the transduced cells becoming CD4-negative 3 d after retroviral infection. Whereas forced expression of AP4 alone induced uniform, but slight, CD4 down-regulation, coexpression of Runx3 and AP4 resulted in a significant increase in the proportion of CD4-negative cells compared with overexpression of either factor alone (Fig. 1D). This indicates that Runx3 and AP4 synergistically repress CD4 expression, or that AP4 stabilizes Runx3-mediated CD4 repression. To further validate AP4 functions in Cd4 gene regulation, we performed loss-of-function assays in vitro using shRNA-based knockdown (Fig. 1E). In these assays, we used the 1200M cell line that has been used successfully to study Cd4 silencing (2, 14). This cell line is thought to be derived from CD4–CD8+ immature single-positive thymocytes based on high CD24 (HSA) (Fig. S3) as well as Runx1, but not Runx3, expression (6). Upon shRNA knockdown of Runx1 expression, we observed increased CD4 surface expression, consistent with Cd4 silencing in 1200M cells being Runx1-dependent (Fig. 1E, green line). Whereas an shRNA which knocked down ∼50% of endogenous AP4 protein expression induced modest CD4 up-regulation (Fig. 1E, orange line), knockdown of both Runx1 and AP4 resulted in a significant increase in CD4 expression compared with Runx1 or AP4 knockdown alone (Fig. 1E, red line). These results suggest that AP4 and Runx proteins cooperatively regulate Cd4 silencing.
AP4 Is Required for Cd4 Silencing in Immature Thymocytes in Vivo.
To validate AP4 functions in Cd4 gene regulation in vivo, we generated an AP4-null (Tcfap4–) allele using homologous recombination in ES cells (Fig. S4 A–C). Homozygous mice were born at a slightly lower than expected Mendelian frequency upon intercrossing of heterozygous mice (WT 29.3%, heterozygous 54.8%, homozygous 15.9%, 30 litters), but were grossly normal and fertile. Numbers of total thymocytes and frequencies of B and T cells in the spleen were comparable between AP4-deficient mice and littermate controls (Fig. S4 D and E).
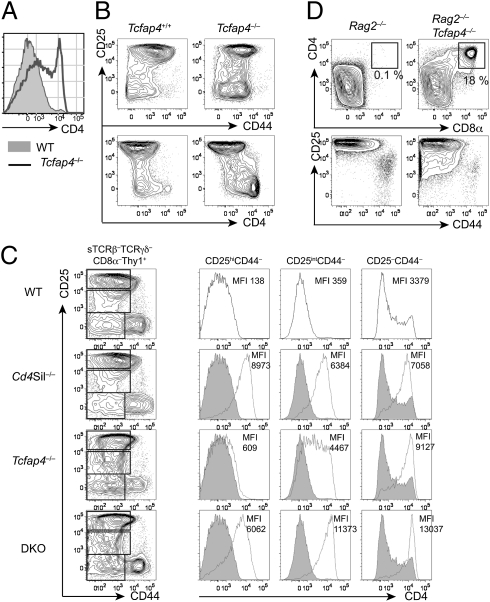
To determine whether AP4 is required for Cd4 silencing, we examined CD4 expression at different stages of thymocyte and T-cell differentiation (Fig. 2). CD4 expression was normal in DP thymocytes and CD4SP thymocytes (Fig. S4F). In contrast, CD4 was up-regulated in immature DN thymocytes, defined as surface TCRβ-negative (sTCRβ–) TCRγδ–CD8α–Thy1+ cells (Fig. 2 A and B). When we subdivided DN thymocytes into subpopulations based on CD25 and CD44 expression, we observed modest CD4 up-regulation in the cells before β selection at the DN3 stage (Fig. 2C, CD25hiCD44–). CD4 up-regulation was further enhanced in DN3 cells from AP4-deficient mice harboring the Cd4 silencer site 3 mutation (Fig. S5A). Interestingly, 15∼20% of AP4/Rag2 doubly deficient thymocytes had the CD4+CD8+CD25int phenotype, suggesting that the modest CD4 expression and/or AP4 deficiency were sufficient to allow Rag2-deficient thymocytes to bypass the β-selection checkpoint (Fig. 2D). Similar phenotypes have been reported in mice ectopically expressing transgenic CD4 in DN3 thymocytes, lacking the Cd4 silencer, or lacking the E2A E-box protein (15–17). In control mice, CD4 remained silenced in intracellular TCRβ (icTCRβ)-expressing thymocytes that down-regulated CD25 following β selection, and only a small fraction of cells started to express CD4 at the DN4 stage (Fig. 2C, CD25intCD44– and CD25–CD44–, and Fig. S5B). In contrast, in the absence of AP4, CD4 was markedly up-regulated in β-selected CD25int cells expressing icTCRβ protein (Fig. 2C and Fig. S5B). CD4 expression increased further in DN4 thymocytes, indicating that AP4 maintains Cd4 silencing in proliferating DN thymocytes following β selection. Postselection DN3 (CD25int) and DN4 thymocytes from mice lacking both AP4 and the Cd4 silencer (Fig. 2C, DKO) expressed CD4 at higher levels than those from AP4-deficient or Cd4 silencer-deficient mice (Fig. 2C and Fig. S5C). These results indicate that AP4 is required for Cd4 silencing in cooperation with silencer-binding factors in double-negative thymocytes and additionally suggest a silencer-independent function for AP4.
Fig. 2.
AP4 is required for Cd4 silencing in immature thymocytes. (A) CD4 expression in sTCRβ–TCRγδ–CD8α–Thy1+ thymocytes from AP4-deficient (open histogram) and WT control (filled histogram) mice. (B) CD4 is robustly up-regulated following β selection in AP4-deficient thymocytes. CD4, CD25, and CD44 expression of sTCRβ–TCRγδ–CD8α–Thy1+ thymocytes from AP4-deficient and WT control mice. (C) AP4 synergizes with Cd4 silencer-binding factors to repress CD4 in immature thymocytes. CD4 expression in preselection DN3 cells (CD25hiCD44–), postselection DN3 cells (CD25intCD44–) and DN4 cells (CD25–CD44–) from WT (black lines in the top row and filled histograms), Cd4 silencer-deficient mice (Cd4Sil−/−), AP4-deficient mice (Tcfap4−/−), and mice doubly deficient for the Cd4 silencer and AP4 (DKO) is shown in histograms with mean fluorescence intensity (MFI) indicated. Data shown here are representative of more than three independent experiments. Statistical analysis is shown in Fig. S5C. (D) AP4 deficiency allows Rag2-deficient thymocytes to bypass β selection. CD4, CD8α, CD25, and CD44 expression in Thy1+ thymocytes from Rag2−/− or Rag2−/−Tcfap4−/− mice is shown.
AP4 Modulates Cd4 Silencing in Memory CD8+ T Cells.
Cd4 silencing in CD8+ mature thymocytes or CD8+ T cells appeared to be normal in AP4-deficient mice (Fig. 3 A and B). To further explore the role of AP4 in Cd4 silencing in mature thymocytes and CD8+ T cells, we examined the AP4-deficient mice bred to mice harboring a mutation in the Cd4 silencer. In contrast to its effect on CD4 expression in immature thymocytes, loss of AP4 in CD8SP CD24lo/– mature thymocytes did not exacerbate CD4 repression defects in Cd4 silencer site 1 or site 3 mutant mice, suggesting that AP4 is not required for the establishment of Cd4 silencing in mature CD8+ SP thymocytes (Fig. 3A and Fig. S6). However, in peripheral CD8+ T cells, AP4 inactivation resulted in significantly larger proportions of CD8+ T cells that failed to silence CD4 expression in site 1 or site 3 mutant mice (Figs. 3 B and C and 4). In site 3 mutant mice expressing AP4, ∼30% of CD8+ T cells in the peripheral blood remained CD4-positive, presumably due to inefficient establishment of epigenetic Cd4 silencing as previously reported (2). In the absence of AP4, more than 50% of site 3 mutant CD8+ T cells were CD4-positive, and AP4 heterozygosity caused an intermediate increase in the proportion of silencer mutant CD4+CD8+ T cells (Fig. 3C). Because variegated CD4 expression by CD8+ T cells from site 1 or site 3 mutant mice is considered to be an indication of epigenetic modification of the silenced Cd4 locus (2), our results suggest that AP4 may regulate epigenetic Cd4 silencing after silencer-binding factors, including Runx3 and CBFβ, inactivate transcription of the locus.
Fig. 3.
AP4 modulates variegated CD4 derepression in mice harboring a mutated Cd4 silencer. (A) CD4 and CD8 expression in TCRβhiHSAlo gated mature thymocytes from Tcfap4−/− mice crossed to Cd4 silencer site 3 mutant mice. CD4 expression in site 3 mutant CD8+ mature thymocytes on either AP4-sufficient (Upper) or -deficient (Lower) backgrounds is shown (Right). Filled histograms show CD4 expression in CD8+ mature thymocytes from AP4-sufficient (Upper) or -deficient (Lower) mice with the intact Cd4 silencer. (B and C) CD4 expression in CD8+TCRβ+ lymph node (LN) cells of mice with silencer site 3 mutation, AP4 deficiency, or both. A representative set of data is shown in B and a statistical analysis of frequency of CD4+ T cells within the CD8+ T-cell interval gate in multiple mice is shown in C. Averages and SDs are shown with P values determined by unpaired Student's t test.
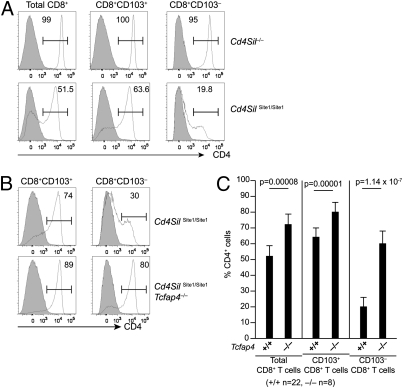
Fig. 4.
Changes in CD4 expression patterns in different subsets of CD8+ T cells harboring mutated Cd4 silencers. (A) CD4 expression in total CD8+ T cells, CD8+CD103+ and CD8+CD103– subsets from WT (filled histograms), Cd4 silencer-deficient, and Cd4 silencer site 1 mutant (open histograms) mice. Frequencies of CD4+ cells are shown within the interval gates. (B) CD4 expression in CD8+ T-cell subsets from Cd4 silencer site 1 mutant, AP4-sufficient (Upper), or -deficient (Lower) mice. (C) Statistical analysis by unpaired Student's t test of the percentages of CD4+ cells in different subsets of site 1 mutant CD8+ T cells.
In analyzing the above results, we noticed that in site 1 and site 3 mutants there was “uniform” derepression of CD4 in most CD8SP thymocytes (reminiscent of silencer-mutated DN cells), but there was variegated CD4 expression in mature peripheral CD8 cells. We thus hypothesized that silencer mutant CD8+ T cells that egress the thymus may lose CD4 expression as they populate the peripheral lymphoid compartments and undergo either homeostatic or antigen-driven cell division. To test this hypothesis, we first examined different surface markers to subdivide peripheral CD8+ T cells into “young” versus “aged” subpopulations; we found that CD103 (α E integrin) expression performed this task. CD103 is uniformly expressed in mature CD8SP thymocytes, but the peripheral CD8+ T-cell population contains both CD103+ and CD103– cells (18). To determine whether CD103 expression marks young cells, we examined Rag2 promoter-driven GFP mice in which the GFP transgene is expressed before positive selection and GFP fluorescence is slowly lost as cells age in the periphery or undergo cell division (19, 20). We observed GFP+ cells enriched in the CD103+ subpopulation, but detected only a low frequency of GFP expression in the CD103– population (Fig. S7A), suggesting that CD103– cells are aged cells compared with CD103+ cells. A large proportion of CD103– cells were CD44hi effector or memory T cells but there were few CD44hi cells in the CD103+ population (Fig. S7B), suggesting that CD8+CD103– cells were enriched for aged cells including memory T cells that had undergone cell division driven by antigen stimulation. We next examined CD4 expression in Cd4 silencer mutant CD8+ cells to determine the effects of cell aging or cell division on Cd4 silencing (Fig. 4A). Indeed, failed Cd4 silencing in CD8+CD103+ young cells from site 1 mutant mice resembled that in mature CD8-positive thymocytes: relatively uniform ectopic CD4 expression. In contrast, the majority of CD8+CD103– cells had lost CD4 expression. The marked changes in the CD4 expression patterns were not observed in CD8+ T cells with the 429-bp silencer deleted. These results suggest that CD8+ T cells with suboptimal Cd4 silencer function may establish Cd4 silencing following aging or cell division, probably through propagation of a repressed state, initiated at the Cd4 silencer, to positive regulatory elements, such as the promoter or the proximal enhancer.
Because AP4 deficiency did not result in changes in CD4 expression in CD8SP thymocytes but caused an increased frequency of CD4 expression in CD8+ T cells in site 1 or site 3 silencer mutant mice, we hypothesized that AP4 may be required for inactivation of CD4 expression in silencer-compromised, aged CD8+ T cells. In AP4-deficient mice with the site 1 mutation there was a modest increase in the frequency of CD4+CD8+ T cells in the CD103+ subpopulation compared with control AP4-sufficient mice (Fig. 4 B and C). However, a large proportion of CD103– cells retained CD4 expression in the absence of AP4, thus resulting in a higher overall frequency of CD4+CD8+ T cells in the periphery (Fig. 4 B and C). These results suggest that AP4 may be required to shut off transcription of the Cd4 locus in memory-enriched CD103–CD8+ T cells.
We next determined in vitro whether proliferation per se recapitulates inactivation of CD4 expression in CD8+ T cells in an AP4-dependent manner (Fig. S8). We purified CD4+CD8+CD103+ cells from site 1 mutant mice or site 1/AP4 double-mutant mice and examined CD4 down-regulation following anti-CD3 and -CD28 stimulation (Fig. S8A). About half of activated CD8+ T cells with the site 1 mutation retained expression of CD4 at a high level, whereas the rest of the population expressed low levels of CD4 (Fig. S8B). Interestingly, there were no significant differences between AP4-sufficient and AP4-deficient cells, suggesting that mechanisms of CD4 down-regulation might be different between memory CD8+ T cells generated in vivo and activated CD8+ T cells in vitro.
AP4 Binds to the Cd4 Proximal Enhancer and Physically Interacts with Runx.
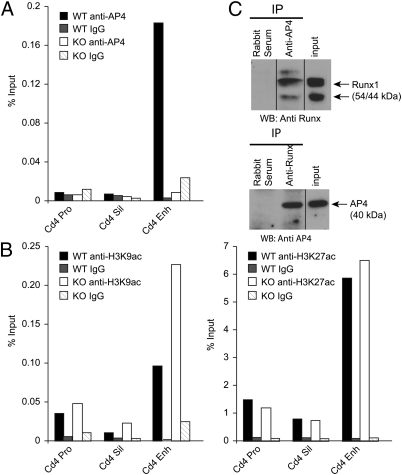
To understand the mechanisms of AP4-mediated Cd4 gene regulation, we examined AP4 binding to known Cd4 regulatory elements by chromatin immunoprecipitation (Fig. 5A). In DN thymocytes, AP4 binding was detected in the Cd4 proximal enhancer located 12 kb upstream of the Cd4 transcription start site, whereas no significant binding was detected in the promoter or silencer regions. In addition, the Cd4 proximal enhancer was hyperacetylated at K9 but not at K27 of histone H3 in the AP4-deficient DN thymocytes (Fig. 5B), which is consistent with previous findings that AP4 represses target gene expression through interaction with histone deacetylases (HDACs) (21, 22).
Fig. 5.
AP4 binding to the Cd4 proximal enhancer and interaction with Runx protein. (A) Chromatin immunoprecipitation analysis showing AP4 binding to the Cd4 proximal enhancer in double-negative thymocytes. Results are representative of four independent experiments. (B) H3K9ac (Left) and H3K27ac (Right) histone modifications at Cd4 regulatory elements in DN thymocytes. Data are representative of two independent experiments. (C) Physical interaction of endogenous AP4 and Runx1 in 1200M cells. Immunoprecipitation (IP) with anti-AP4 (Upper) or anti-Runx (Lower) antibodies was followed by immunoblotting with antibodies specific for the two transcription factors. Results are representative of three independent experiments. WB, Western blot.
Our genetic data suggested that AP4 cooperates with other silencer-binding factors to repress Cd4, which raises the possibility that AP4 and Runx1, the known Cd4 silencer-binding factor, physically interact with each other. To test this hypothesis, we attempted to coimmunoprecipitate endogenous AP4 and Runx1 in 1200M cells, where these factors contribute to Cd4 silencing (Fig. 5C). AP4 and Runx1 were reciprocally coprecipitated by specific antibodies against AP4 or Runx1, suggesting that these two transcription factors interact in cells in which the Cd4 gene is actively silenced in a silencer- and Runx1-dependent manner.
Discussion
We have identified AP4 as a negative regulator of the Cd4 gene. Whereas AP4 was originally identified as an activator of viral gene transcription in cooperation with AP1 (23, 24), recent in vitro studies have suggested that AP4 functions as a transcriptional repressor through interaction with HDACs to silence HIV long-terminal-repeat activity or neuronal gene expression in nonneuronal tissues (21, 22). We have found that AP4 is an enhancer-binding factor required for repression of the Cd4 gene in immature thymocytes in vivo. Repression appears to be dependent on cooperation between AP4 and other factors presumably recruited to the Cd4 silencer, including Runx proteins.
Our analysis of Cd4 gene regulation in immature thymocytes indicates that AP4 is required for Cd4 repression early in thymocyte differentiation. CD4 expression was slightly up-regulated in preselection DN3 cells and robustly up-regulated in DN3 and DN4 cells following β selection in AP4-deficient mice. Before β selection, the Cd4 silencer is fully active and Runx1 expression is high. Upon β selection, silencer function is potentially attenuated in part by Runx1 down-regulation, whereas the Cd4 gene remains inactive in an AP4-dependent manner until developing thymocytes go through the DN4 stage. Premature CD4 expression in DN thymocytes triggers bypassing of the β-selection checkpoint, resulting in generation of double-positive thymocytes even without successful Tcrb rearrangements (15, 16). A similar phenotype was observed in our analysis using AP4/Rag2 compound mutant mice. AP4 thus regulates appropriate timing of CD4 expression during transition from the DN to the DP stage, reinforcing the developmental checkpoint.
AP4-mediated Cd4 repression appears to occur in the absence of its interaction with sequences within the silencer in immature thymocytes. We observed that AP4 deficiency enhances CD4 derepression in DN thymocytes from mice lacking the Cd4 silencer and those harboring the site 1 mutation, indicating that AP4 has at least some function that is independent of the silencer. This repressive influence on Cd4 expression is likely mediated through binding of AP4 to the proximal enhancer. We detected AP4 binding to the Cd4 locus at the proximal enhancer, and there was elevated histone acetylation at this region in AP4-deficient mice. Provided that AP4 and Runx1 interact and that AP4 and Runx1 bind to the enhancer and the silencer, respectively, AP4 may recruit HDAC activity to the Cd4 enhancer by looping of silencer-bound Runx1, and may thus contribute to holding the locus in an off-state.
In our previous studies, we proposed that Cd4 silencing is differentially regulated between DN thymocytes and CD8+ mature T cells based on observations that mutations in the Cd4 silencer resulted in uniform derepression in DN cells and variegated derepression in CD8+ T cells (2). This hypothesis was also supported by the finding that, in contrast to reversible repression of CD4 in DN cells, silencing in mature CD8+ T cells is heritable and no longer requires the presence of the silencer sequence once it is established (3). CD8SP thymocytes from mice with mutations within the Cd4 silencer appear to have substantially less variegation of CD4 expression compared with mature CD8+ T cells in the periphery. This finding suggests that Cd4 silencing in DN thymocytes may be mechanistically similar to that in CD8SP thymocytes before establishment of epigenetic memory in the latter. It remains unclear at which stage epigenetic maintenance is established, as it is not possible to distinguish cells with and without heritable Cd4 silencing based on loss of surface CD4 expression. By using Cd4 silencer mutant mice, we observed distinct patterns of CD4 derepression between naive and effector/memory CD8+ T cells. In the former subset, a large proportion of naive CD8+ T cells (CD103+CD44lo) from site 1 mutant mice consisted of CD4-positive cells. In contrast, in the CD103–CD44hi memory subset, a large proportion of site 1 mutant CD8+ T cells lost CD4 expression. Such a difference in CD4 expression patterns between naive and memory cells was not observed in CD8+ T cells lacking the entire silencer, suggesting that this may be due to the continued activity of silencer-binding factors, for example, Runx3, and may reflect imposition of heritable silencing, which then no longer requires persistence of the silencer-binding factors. Because memory T cells are thought to experience antigen-driven cell division and are long-lived in peripheral tissues after thymic egress, imposition of heritable silencing may require cell-cycle progression or aging. DNA replication may facilitate efficient recruitment of epigenetic modifiers to the locus.
AP4-deficient CD8+ T cells were able to repress CD4 expression during the transition from the DP to CD8SP stages, indicating that AP4 is not required to down-regulate Cd4 after positive selection. This is consistent with our observation that AP4 does not affect Cd4 silencing in site 1 or site 3 mutant CD8+ SP thymocytes. In contrast, we observed a slightly larger fraction of naive CD8+ T cells retaining CD4 expression in AP4-deficient mice. More significantly, inactivation of Cd4 in CD103–CD8+ peripheral T cells from silencer-attenuated mice was markedly compromised in the absence of AP4. In animals with a mutation in site 1, CD4 was repressed in CD8+ T cells after they acquired markers of memory cells, likely due to the action of silencer-binding proteins such as Runx3. However, this compensatory late establishment of silencing was largely abrogated in the absence of AP4. These findings suggest that AP4 may enforce reimposition of Cd4 repression, possibly by mediating interaction of its cis-binding sequences in the enhancer with the Runx3-containing silencer. An alternative, but not mutually exclusive, explanation is that AP4 may mediate a fail-safe mechanism for heritable stable gene repression, should there be compromised recruitment of the heritable silencing machinery to the Cd4 locus. Either full Cd4 silencer activity or AP4-mediated repression of the enhancer may be required to shut off the Cd4 locus. AP4 may facilitate a stable interaction of the Cd4 locus with the heritable silencing machinery during memory cell differentiation. However, we cannot rule out the possibility that the change of the Cd4 repression pattern observed in AP4-deficient mice could be through an indirect mechanism. Further studies are needed to determine precise binding sites for AP4 and whether AP4 acts as a repressor through chromosomal looping. Although it will be necessary to examine more rigorously whether mutations in site 1 or 3 or the Cd4 silencer result in delayed imposition of heritable Cd4 silencing/repression in memory CD8+ T cells, our findings raise the possibility that AP4-dependent establishment of heritable silencing may be relevant to differentiation of memory CD8+ T cells.
In summary, we provide genetic evidence for the role of the AP4 transcription factor in Cd4 gene regulation in vivo. Our results strongly suggest that Cd4 silencing both in immature thymocytes and CD8+ T cells is not solely dependent on the silencer but requires cooperation of silencer-binding factors and distal factors that potentially bind to the enhancer. Future studies, including purification of AP4/Runx-containing complexes and genome-wide mapping of AP4 binding sites, will contribute to elucidation of molecular mechanisms of epigenetic silencing and to the identification of other target genes regulated by AP4 during T-cell development.
Materials and Methods
Mice.
AP4-deficient mice were generated by deleting exons 2, 3, and 4 by gene targeting (Fig. S4). Refer to SI Materials and Methods for details. Mice were analyzed in the 129/B6 mixed genetic background. Rag2-deficient mice were purchased from Taconic. Rag2p-GFP mice were purchased from Jackson Laboratory. Cd4 silencer mutant mice were described previously (2). All experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees at the New York University School of Medicine and Washington University School of Medicine.
Retroviral Overexpression and shRNA Knockdown.
AP4 and Runx3 cDNAs were inserted into murine stem cell virus (MSCV)-based retroviral backbones containing ires hCD2 (tailless) or ires EGFP, respectively. shRNA targeting AP4 was purchased from Open Biosystems (TRCN0000082124). For Runx1 knockdown, a 5′-ATAGATGCCAAACAGGATGGG-3′ sequence complementary to the 3′ UTR of mouse Runx1 was inserted into an miR-30-based hairpin of the pMSCV-LMP vector (Open Biosystems) according to the manufacturer's instruction. Retroviruses were packaged in PlatE cells (25) by transient transfection using TransIT 293 (Mirus Bio). Lentiviruses were packaged by cotransfection of viral vectors and CMV-Δ8.9 along with a vector encoding vesicular stomatitis virus glycoprotein (VSV-G). Cells were transduced by spin infection at 1,200 × g at 30 °C for 90 min in the presence of 10 μg/mL polybrene (Sigma). For sequential knockdown of AP4 and Runx1, 1200M cells were initially infected with AP4 shRNA or control lentivirus, selected for puromycin resistance for 48 h, and subsequently infected with retrovirus expression Runx1 shRNA.
Antibodies.
Anti-AP4 antibody was generated by immunizing rabbits with a GST-fusion protein containing amino acids 180–338 of AP4 protein (Covance Research). Rabbit bleeds were initially depleted of reactivity against GST using immobilized GST agarose (Pierce) and subsequently affinity-purified with GST-AP4 (180–338). Anti-H3K9ac and anti-H3K27ac antibodies were purchased from Millipore. Anti-Runx, -ThPOK, -HMG1, and -RORγt antibodies were described previously (26, 27).
Western Blotting and Immunoprecipitation.
Whole-cell extract was prepared by lysing cells in nondenaturing buffer containing 150 mM NaCl, 1% Nonidet P-40, and protease inhibitor mixture (Sigma). For immunoprecipitation, 500 μg of whole-cell extract of 1200M cells, which actively silence CD4 and express endogenous Runx1 and AP4, was incubated with preimmune serum, anti-Runx antibody, or anti-AP4 overnight. Immune complexes were precipitated by protein-G agarose (Roche), separated by SDS/PAGE, and subjected to Western blotting.
Chromatin Immunoprecipitation.
Pro-T cells from Rag2-deficient or Tcfap4/Rag2 double-deficient thymocytes were fixed with 1% paraformaldehyde and sonicated using a Bioruptor (Diagenode). Immunoprecipitation was performed using Dynabeads protein G (Invitrogen). Precipitated DNA was quantitated by quantitative PCR using PerfeCTa SYBR Green FastMix (Quanta BioSciences) and a LightCycler 480 (Roche). Percentages of input were calculated using ΔΔCt (28). Primer sequences are available upon request.
Supplementary Material
Acknowledgments
The authors thank Mary Jean Sunshine and the New York University ES cell core for microinjection, John Hirst for cell sorting of microarray samples, the Memorial Sloan-Kettering Cancer Center genomics core for microarray, Jonathan Chow and Elke Kurz for technical assistance, and Gretchen Diehl and MacLean Sellars for comments on the manuscript. T.E. was a Special Fellow of the Leukemia and Lymphoma Society. This work was supported in part by the start-up fund of the Department of Pathology and Immunology, Washington University (T.E.). D.R.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE31082).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112293108/-/DCSupplemental.
References
- 1.Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat Rev Immunol. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- 2.Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell. 2002;10:1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- 3.Zou YR, et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 4.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 5.Setoguchi R, Taniuchi I, Bevan MJ. ThPOK derepression is required for robust CD8 T cell responses to viral infection. J Immunol. 2009;183:4467–4474. doi: 10.4049/jimmunol.0901428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 8.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 9.Sawada S, Littman DR. Identification and characterization of a T-cell-specific enhancer adjacent to the murine CD4 gene. Mol Cell Biol. 1991;11:5506–5515. doi: 10.1128/mcb.11.11.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naoe Y, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf β binding to the Il4 silencer. J Exp Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 15.Norment AM, Forbush KA, Nguyen N, Malissen M, Perlmutter RM. Replacement of pre-T cell receptor signaling functions by the CD4 coreceptor. J Exp Med. 1997;185:121–130. doi: 10.1084/jem.185.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu M, et al. Nucleoprotein structure of the CD4 locus: Implications for the mechanisms underlying CD4 regulation during T cell development. Proc Natl Acad Sci USA. 2008;105:3873–3878. doi: 10.1073/pnas.0800810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grueter B, et al. Runx3 regulates integrin α E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 19.McCaughtry TM, Wilken MS, Hogquist KA. Thymic emigration revisited. J Exp Med. 2007;204:2513–2520. doi: 10.1084/jem.20070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, et al. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 1999;400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 21.Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J Biol Chem. 2006;281:12495–12505. doi: 10.1074/jbc.M511773200. [DOI] [PubMed] [Google Scholar]
- 22.Kim MY, et al. A repressor complex, AP4 transcription factor and geminin, negatively regulates expression of target genes in nonneuronal cells. Proc Natl Acad Sci USA. 2006;103:13074–13079. doi: 10.1073/pnas.0601915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu YF, Lüscher B, Admon A, Mermod N, Tjian R. Transcription factor AP-4 contains multiple dimerization domains that regulate dimer specificity. Genes Dev. 1990;4:1741–1752. doi: 10.1101/gad.4.10.1741. [DOI] [PubMed] [Google Scholar]
- 24.Mermod N, Williams TJ, Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature. 1988;332:557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- 25.Morita S, Kojima T, Kitamura T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 26.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberl G, et al. An essential function for the nuclear receptor RORγ(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.