Abstract
Legionella pneumophila is a bacterial pathogen of amoebae and humans. Intracellular growth requires a type IVB secretion system that translocates at least 200 different proteins into host cells. To distinguish between proteins necessary for growth in culture and those specifically required for intracellular replication, a screen was performed to identify genes necessary for optimal growth in nutrient-rich medium. Mapping of these genes revealed that the L. pneumophila chromosome has a modular architecture consisting of several large genomic islands that are dispensable for growth in bacteriological culture. Strains lacking six of these regions, and thus 18.5% of the genome, were viable but required secondary point mutations for optimal growth. The simultaneous deletion of five of these genomic loci had no adverse effect on growth of the bacterium in nutrient-rich media. Remarkably, this minimal genome strain, which lacked 31% of the known substrates of the type IVB system, caused only marginal defects in intracellular growth within mouse macrophages. In contrast, deletion of single regions reduced growth within amoebae. The importance of individual islands, however, differed among amoebal species. The host-specific requirements of these genomic islands support a model in which the acquisition of foreign DNA has broadened the L. pneumophila host range.
Keywords: bacterial genomes, transposon site hybridization, essential genes, genome organization, virulence factor
In its natural fresh water environment, Legionella pneumophila is an intracellular parasite of free-living amoebae (1). The host range of L. pneumophila is extensive because the bacterium is capable of replicating in at least 15 different species of amoebae. When contaminated water sources are aerosolized and inhaled by humans, L. pneumophila replicates in alveolar macrophages, causing a potentially fatal form of pneumonia (2, 3). This behavior makes L. pneumophila a generalist because it is able to grow in a diverse array of evolutionarily distinct host cell types. Although the majority of disease-causing organisms are specialists that exhibit a high degree of host tropism, jumping among hosts has been observed for a number of pathogens (4). It is thought that host jumping is followed by adaptation to the new host, which often selects for mutation, inactivation, or loss of virulence factors necessary for growth in the previous host (4). In some cases, this allows escape from immune sensing of the pathogen by the new host (5). This process of host specialization appears to be less prominent in L. pneumophila because it retains the ability to grow in a large assortment of amoebae. What determines the broad host range of this bacterium, how it is maintained, and how it contributes to the fitness of the bacterium in its natural environment are poorly understood.
The intracellular life cycle of L. pneumophila in all host cell types examined is very similar, despite these hosts being phylogenetically distant. The bacterium is internalized into a membrane-bound compartment that avoids delivery to the lysosome (3) and is remodeled through recruitment of host endoplasmic reticulum-derived vesicles into a ribosome-studded replication vacuole (6, 7). Intracellular growth of the bacterium requires a type IVB secretion system encoded by the dot/icm genes (8–10) that delivers bacterial proteins to the host cytoplasm (11), where they manipulate a variety of host cellular processes (12–15). To date, over 200 Dot/Icm translocated substrates (TS) have been identified (16–20). Although the host cell targets and mechanisms of action for a number of these substrates have been elucidated (21–28), the explicit roles of the majority of these proteins remain unclear. This is primarily attributable to the absence of any discernable phenotype of mutants lacking genes encoding these proteins (16, 21, 22, 29), a phenomenon that has been attributed to functional redundancy (16, 30–32).
To understand the nature of the intracellular environment of the host, how the bacterium handles the challenges encountered there, and the differential requirements for growth in different hosts, it is important to distinguish between genes that are essential for viability, basic metabolism, and proliferation and those that are specifically important for growth in a host. To address these issues, a genetic screen using a library of L. pneumophila transposon mutants was performed to identify genes that are essential for growth in nutrient-rich bacteriological media. Clustering of these essential genes to distinct loci defined several genomic islands that were dispensable for growth of the bacterium in rich media and macrophages but not in amoebae. Furthermore, the amoebae-specific requirements of these large genomic segments for intracellular growth demonstrated a modular organization of the L. pneumophila genome distinguishing between regions encoding genes devoted to fundamental biological processes and those dedicated to replication in a host.
Results
Genes Required for L. pneumophila Growth in Nutrient-Rich Bacteriological Media.
A mariner himar1-based transposon consisting of a kanamycin resistance cassette flanked by divergently transcribed T7 promoter elements (Fig. S1) was introduced into L. pneumophila Philadelphia 1 (33) to generate a library of transposon insertion mutants (Materials and Methods) for transposon site hybridization (TraSH) (34). A pool of 100,000 insertion mutants was generated, which, if distributed randomly, was predicted to result in ∼30 insertions per 1-kb gene (the average size of a L. pneumophila ORF). Based on the size of the library, and the fact that randomly chosen insertions showed no sequence specificity, it was judged to be exhaustive and sufficiently random for TraSH (34).
The TraSH library was used to identify genes that are important for growth of L. pneumophila in nutrient-rich bacteriological media (Fig. S1). To do this, an aliquot of the L. pneumophila transposon mutant library was grown to stationary phase in broth culture and plated for single colonies on solid charcoal buffered yeast extract thymidine (CYET) media. Those mutants containing transposon insertions in genes that are important for survival under these conditions will be underrepresented in this pool. To determine the abundance of each mutant in the pool, short regions of the chromosome directly adjacent to the transposon insertion sites were amplified and used to probe a custom gene array representing 94% of L. pneumophila Philadelphia 1 protein encoding genes (Materials and Methods). As a control, a genomic probe was generated by amplifying total DNA from the nonmutagenized parent strain Lp02 using random primers (Materials and Methods), and the relative intensities of each probe were determined at each site on the array. The average insertion/genomic probe ratio for each gene from 10 individual experiments was calculated.
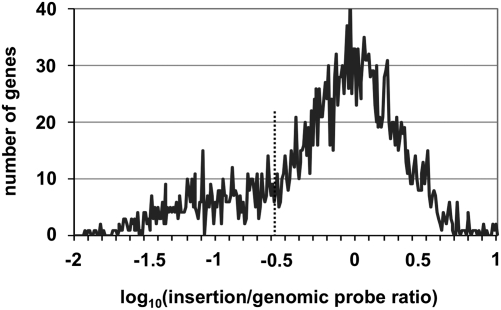
The relative abundance of insertion mutations recovered in each gene was plotted as a histogram of the logarithm of the insertion/genomic probe ratio for each gene analyzed (Fig. 1). The majority of genes exhibited a ratio of ∼1, indicating that these genes are not required for growth in nutrient-rich media. There is a prominent skewing of this plot toward genes in which there was a lower recovery of mutations relative to what was expected based on the distribution of the genomic probe intensities alone, which we interpret as representing insertions in loci resulting in a growth defect. Thus, genes for which the ratio was <0.266 (the point of skewing; Fig. 1, dotted line) and which were significantly depleted relative to the mean population (P < 0.001) were classified as being defective for growth in culture. Based on these criteria, 597 genes required for optimal growth in culture were identified (Dataset S1), including many associated with core metabolic pathways as well as 53 genes encoding proteins of unknown function. Viable mutants have been previously reported in 9 of the genes judged to be required for optimal growth in media (Dataset S1) (35–39); thus, these genes were excluded from further analysis. Of the 588 remaining genes, 98.0% are conserved in the other four sequenced strains of L. pneumophila (40–42) and 78.0% have been described as essential in at least one other bacterial species (Dataset S1).
Fig. 1.
Identification of genes essential for growth of L. pneumophila in nutrient-rich bacteriological media. Histogram plot of the log10(insertion/genomic probe intensity) for all L. pneumophila genes represented on the custom microarray. Skewing of the plot occurs at 10−0.575 (dotted line), corresponding to an insertion/genomic ratio of 0.266, or a 3.8-fold defect in growth. Genes with an insertion/genomic ratio <0.266 and whose value was statistically different from the mean population based on a Student t test P value <0.001 were classified as essential for growth in rich media. Data represent the mean of 10 independent experiments.
Large Contiguous Regions of the L. pneumophila Genome Are Dispensable for Growth in Culture.
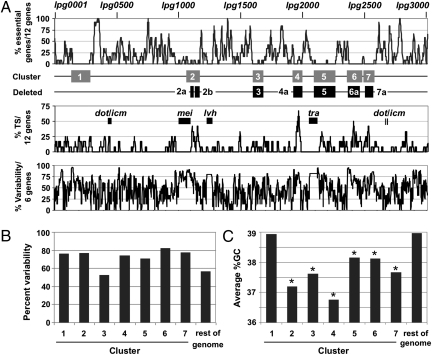
Seven distinct regions of the L. pneumophila chromosome were found to be devoid of genes required for optimal growth (Fig. 2A and Table 1). These clusters, ranging in size from 76 to 171 genes, make up 27.1% of all protein encoding genes. The grouping of these dispensable genes to distinct loci indicates that there is a structured, mosaic organization of the L. pneumophila genome that correlates with function.
Fig. 2.
Mosaic architecture of the L. pneumophila genome. (A) Seven genomic islands are devoid of genes essential for growth in rich media but exhibit a high concentration of Dot/Icm TS. (Top) Running average plot of the percentage of essential genes per 12 genes. The seven gene clusters identified (gray boxes) and the six genomic loci deleted (black boxes) are indicated. (Middle) Running average plot of the percentage of Dot/Icm TS per 12 genes. The locations of the dot/icm, tra, lvh, and mei loci (black boxes) are individually labeled. (Bottom) Running average plot of the percent variability per 6 genes. Pairwise comparison of the L. pneumophila Philadelphia 1 genome to Paris, Lens, Corby, and Alcoy strains was used to identify genes that are conserved or absent across all five species. Percent variability is the number of genes inserted or deleted relative to the total number of genes in that region. (B) Six of the genomic islands have a higher percent variability than the rest of the genome, as defined in A, compared with other sequenced strains of L. pneumophila. (C) Six genomic islands exhibit a lower average %GC for all genes in the cluster compared with the rest of the genome. Clusters exhibiting a statistically significant difference based on a two-tailed Student t test P value <0.05 relative to the rest of the genome are indicated by an asterisk. %GC, percent of guanine and cytosine bases.
Table 1.
Cluster deletion mutants
| Intracellular growth |
||||||||||||
| Cluster | Region | Total kilobases | No. genes | Percent total genes* | No. TS | Percent total TS† | Deletion mutant constructed | Growth in vitro‡ | AJ | Dd | Hv | Ac |
| TraSH-identified clusters | ||||||||||||
| 1 | lpg0140–lpg0286 | 172 | 147 | 5.0 | 15 | 6.6 | N | |||||
| 2 | lpg1067–lpg1173 | 133 | 107 | 3.6 | 18 | 8.0 | N | |||||
| 3 | lpg1603–lpg1686 | 109 | 84 | 2.9 | 10 | 4.4 | Y | |||||
| 4 | lpg1924–lpg1999 | 92 | 76 | 2.5 | 20 | 8.8 | N | |||||
| 5 | lpg2097–lpg2267 | 224 | 171 | 5.8 | 17 | 7.5 | Y | |||||
| 6 | lpg2362–lpg2485 | 132 | 124 | 4.2 | 18 | 8.0 | N | |||||
| 7 | lpg2489–lpg2579 | 104 | 91 | 3.1 | 13 | 5.8 | N | |||||
| Total | 966 | 800 | 27.1 | 111 | 49.1 | |||||||
| Single-cluster deletion mutants | ||||||||||||
| 2ab | lpg1104–lpg1128, | 77 | 59 | 2.0 | 15 | 6.6 | Y | + | + | + | + | + |
| lpg1136-–lpg1169 | ||||||||||||
| 3 | lpg1603–lpg1686 | 109 | 84 | 2.9 | 10 | 4.4 | Y | + | + | + | − | − |
| 4a | lpg1933–lpg1999 | 81 | 67 | 2.3 | 19 | 8.4 | Y | + | + | + | + | − |
| 5 | lpg2097–lpg2267 | 224 | 171 | 5.8 | 17 | 7.5 | Y | − | − | − | − | − |
| 6a | lpg2369–lpg2465 | 107 | 97 | 3.3 | 18 | 8.0 | Y | + | + | + | + | + |
| 7a | lpg2508–lpg2573 | 77 | 66 | 2.2 | 9 | 4.0 | Y | + | + | − | + | − |
| Combination-cluster deletion mutants | ||||||||||||
| 3, 2ab | 186 | 143 | 4.9 | 25 | 11.1 | Y | + | + | + | − | − | |
| 3, 2ab, 6a | 293 | 240 | 8.2 | 43 | 19.0 | Y | + | + | + | − | − | |
| 3, 2ab, 6a, 7a | 370 | 306 | 10.4 | 52 | 23.0 | Y | + | + | − | − | − | |
| 3, 2ab, 6a, 7a, 4a | (pentuple) | 451 | 373 | 12.7 | 71 | 31.4 | Y | + | + | − | − | − |
| 3, 2ab, 6a, 7a, 4a, 5 | (hextuple) | 675 | 545 | 18.5 | 88 | 38.9 | Y | − | − | ND | ND | ND |
Ac, Acanthamoeba castellanii; AJ, murine bone marrow-derived A/J macrophages; Dd, Dictyostelium discoideum; Hv, Hartmanella vermiformis; N, no; ND, not determined; Y, yes; +, growth comparable to WT; −, growth defect.
*Total number of L. pneumophila protein encoding genes = 2,942.
†Total number of L. pneumophila TS encoding genes = 226 (14–18) [including only TS with translocation efficiency ≥40% in (18)].
‡In vitro indicates growth in AYE nutrient-rich media.
Closer examination of the seven gene clusters revealed several features that distinguished them from the rest of the genome. First, these regions collectively encoded 49% of the known Dot/Icm TS (Fig. 2A and Table 1). In contrast, the genes encoding the Dot/Icm secretion system did not map to these clusters but, instead, were imbedded in regions required for optimum growth in culture (Fig. 2A). Second, all but one of the seven gene clusters exhibited a high degree of variability in terms of gene conservation and synteny compared with their respective counterparts in the other four sequenced strains of L. pneumophila (Fig. 2B). Third, six of the seven gene clusters had a significantly lower average %GC content relative to the rest of the genome (Fig. 2C), a common characteristic of DNA acquired via horizontal gene transfer (43). Indeed, of the 62 insertion sequence (IS) elements and phage-related genes annotated in the L. pneumophila Lp02 strain, which are characteristic of mobile genetic elements, 60% map to one of the seven gene clusters identified with a heavy concentration at the termini of these loci (Fig. S2).
Because individual genes from the TraSH screen were predicted to be dispensable for growth in bacteriological medium, we tested whether elimination of entire clusters of these genes resulted in viable bacterial strains. Of the seven gene clusters identified, deletion mutations for clusters 3 and 5 were successfully constructed, as were those for clusters 4, 6, and 7 after redefining their outer boundaries (Fig. 2A and Table 1). Of the remaining two clusters, only a partial deletion of cluster 2 consisting of two smaller internal segments could be isolated (Fig. 2A and Table 1), allowing further analysis of strains missing genes in this region. Cluster 1, however, could not be deleted under any circumstance, perhaps because of the lethality of deleting multiple genes simultaneously or an essential role for one or more of the genes from this region that were missing from the custom L. pneumophila arrays (Dataset S2). The correct end points of each deletion were confirmed by microarray analysis (Fig. S3) or by whole-genome sequencing (see below). The isolation of strains lacking large segments of consecutive genes validated the data obtained from the TraSH screen and showed that large contiguous regions of the chromosome are dispensable for growth in culture.
In addition to the five genetic loci deleted here, three other regions of the L. pneumophila Philadelphia 1 genome (Fig. 2A) have been identified, either directly or indirectly, as dispensable for growth in vitro: the lvh gene locus (lpg1228–lpg1271), which is absent from the WT strain used in this work (8); the tra locus (lpg2057–lpg2115) (44); and the metal efflux island (mei) (lpg1006–lpg1096) (45). TraSH data presented here support expendable roles for the tra and mei loci for growth in nutrient-rich media, because none of the genes encoded in these regions passed the essential gene criteria (Dataset S1). Collectively, these loci define an additional 6.6% of protein encoding genes experimentally demonstrated to be dispensable for viability and growth of L. pneumophila in rich media.
Five of the Single Cluster Deletion Mutants Show No Growth Defects During Culture in Nutrient-Rich Media.
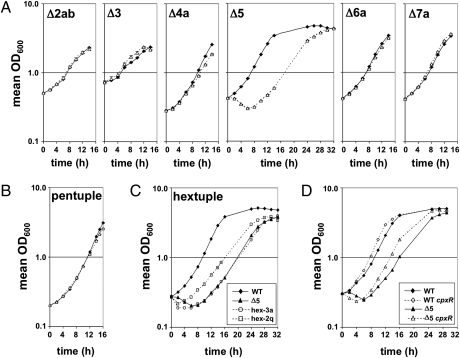
All six cluster mutants constructed were capable of growth on solid medium; however, the cluster 5 mutant produced markedly smaller colonies compared with the WT strain (data not shown). Consistent with these observations, growth of the cluster 2ab, 3, 4a, 6a, and 7a mutants in liquid media was similar to that of the WT strain (Fig. 3A), demonstrating that their absence imparts no adverse effect on the growth of L. pneumophila in culture. In contrast, the cluster 5 mutant exhibited an initial decrease in turbidity at early time points; however, on recovery, it grew at a slower rate than the WT strain (doubling times of 4.4 ± 0.1 h and 3.3 ± 0.1 h, respectively) (Fig. 3A). Despite these defects, upon reaching postexponential phase, the mutant exhibited properties of the WT strain grown to this density, including synthesis of the secreted pigment pyomelanin (46, 47) and enhanced motility relative to exponentially growing bacteria (48).
Fig. 3.
L. pneumophila minimal genome strains exhibit robust growth in vitro. (A) Growth of all but the Cluster 5 single deletion mutants (△) is comparable to that of the WT strain Lp02 (◆) in nutrient-rich media. (B) Growth of the pentuple mutant (△) in rich media is similar to that of the WT strain (◆). (C) Differential growth of the hextuple mutant variants 2q and 3a relative to WT and the Cluster 5 mutant strains. (D) Partial rescue of the cluster 5 mutant in vitro growth defect by the hextuple 2q cpxR point mutation. Data represent the average of two independent experiments of two technical replicates each; error bars indicate ± SD.
Minigenome Strains of L. pneumophila Are Competent for Growth in Culture.
To determine if a highly minimized genome of L. pneumophila was sufficient for growth in culture, the simultaneous deletion of all six clusters was attempted in a single strain (Table 1). The Δ2abΔ3Δ6aΔ7aΔ4a, or pentuple mutant, lacking 12.7% of protein encoding genes in the L. pneumophila genome (Fig. S3 and Table 1), showed similar growth kinetics compared with the WT strain in nutrient-rich liquid media (Fig. 3B). These results demonstrated that a significant portion of the L. pneumophila genome can be deleted without affecting viability or growth of the bacterium in a nutrient-rich environment, indicating that these regions are designated for purposes other than fundamental biological processes.
Deletion of cluster 5 in the pentuple mutant required generating a marked deletion using an antibiotic resistance cassette for selection. By this approach, viable Δ2abΔ3Δ6aΔ7aΔ4aΔ5:cat, or hextuple deletion mutants were obtained, which reduced the genome size by 18.5% compared with the WT strain. Similar to the Cluster 5 mutant, the majority of hextuple mutants exhibited smaller colony sizes than the WT strain when grown on solid media. However, a subset of these mutants generated significantly larger colonies (data not shown). Growth of one of these faster growing variants, 2q, in nutrient-rich liquid media was compared with that of a slower growing isolate, 3a (Fig. 3C). Both hextuple mutants showed a defect in growth at early time points similar to the Cluster 5 mutant, but the larger colony variant recovered in a shorter period, consistent with the acquisition of a suppressor mutation in the larger colony variant.
The entire genomes of both hextuple isolates were sequenced and compared with that of the parental strain, Lp02 (SI Materials and Methods). Each strain had the expected six cluster deletions as well as an unexpected point mutation in tufB that was also present in the Cluster 3 mutant, the progenitor for constructing the hextuple deletion strain. In addition, the slower growing 3a mutant coded for a substitution in RpoC at G195, a residue known to affect RNA polymerase β′ subunit stability (49). In contrast, the faster growing 2q isolate contained three point mutations: one in the intragenic region separating lpg1464 and lpg1465, which encode proteins of unknown function; a mutation in the lysine biosynthesis gene lysCA (lpg1811) that results in substitution of an active site residue of the LysA domain (50); and an insertion mutation in cpxR (lpg1438) that causes premature translational termination of the encoded protein. CpxR is a master two-component response regulator that responds to bacterial envelope stress in Escherichia coli (51) and controls expression of genes encoding components of the Dot/Icm secretion system and its substrates in L. pneumophila (52). The appearance of suppressor mutations in the hextuple strains is consistent with this particular combination of gene deletions approaching the minimal gene set still capable of supporting L. pneumophila growth.
Consistent with the prediction that mutations in the hextuple 2q isolate suppress the effects of the Cluster 5 mutation, introduction of the cpxR insertion mutation into the cluster 5 mutant shortened its early growth defect by roughly 2 h and restored its rate of growth in exponential phase to that of the WT strain (doubling time of 3.2 ± 0.1 h compared with 4.3 ± 0.1 h for the Cluster 5 mutant) (Fig. 3D). Thus, forcing the deletion of cluster 5 in the pentuple mutant through positive selection resulted in the accumulation of mutations that allowed the bacterium to tolerate the loss of cluster 5 in combination with clusters 2ab, 3, 4a, 6a, and 7a.
L. pneumophila Minigenome Strains Are Competent for Growth in Macrophages.
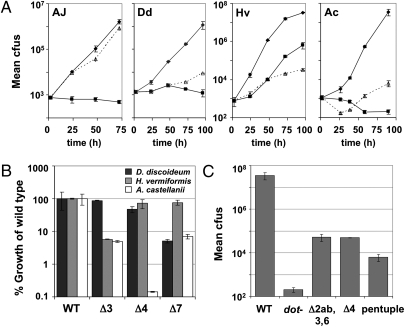
The retarded growth of the hextuple mutants in broth culture, and the ready isolation of fast-growing derivatives that affect Dot/Icm expression, made it difficult to analyze intracellular growth of strains harboring this particular combination of deletions. Therefore, we analyzed the ability of the pentuple mutant (Δ2ab, 3, 4a, 6a, 7a) to grow within bone marrow-derived mouse macrophages. Remarkably, intracellular growth of this mutant was almost indistinguishable from that of the WT strain (Fig. 4A), despite the fact that this mutant was missing 31% of all known Dot/Icm TS (Table 1). As expected, any subcombination of cluster deletions also had no effect on growth within cultured macrophages (Fig. S4, dark bars). Therefore, almost one-third of the proteins targeted for host cells are completely dispensable for intracellular growth in this host cell type.
Fig. 4.
Intracellular growth of the pentuple mutant is severely impaired in amoebae but not macrophage hosts. (A) Growth of WT (◆), dot− (■), and pentuple mutant (△) strains in bone marrow-derived A/J macrophages (AJ) and the amoebal species D. discoideum (Dd), H. vermiformis (Hv), and A. castellanii (Ac). (B) Growth of the Cluster 3, 4, and 7 deletion strains in D. discoideum, H. vermiformis, and A. castellanii as a percentage of the WT strain. (C) Growth of the Δ2ab,3,6a, and 4a mutants in A. castellanii is comparable to that of the pentuple mutant. In each case, bacterial growth based on recovered number of cfus on solid media from lysed host cells was monitored over 3–4 d (encompassing 3 consecutive rounds of infection). Plotted is the total bacterial yield (A and C) over the course of the infection normalized relative to the WT strain by percent uptake at the initial time point tested or the percentage of growth (B) of each mutant relative to the WT strain. Data represent the average of two independent experiments of three technical replicates each; error bars indicate ±SD.
L. pneumophila Minigenome Strains Show Amoebae-Specific Defects in Intracellular Growth.
Because a large number of Dot/Icm TS encoding genes could be removed from the L. pneumophila genome without any clear effect on growth in macrophages, the behavior of the pentuple mutant was analyzed in the amoebal species Acanthamoeba castellanii, Dictyostelium discoideum, and Hartmanella vermiformis (Fig. 4A and Fig. S4), which mimic natural hosts of the bacterium in the environment.
In contrast to its behavior within bone marrow macrophages, the pentuple mutant was severely defective for growth in each of the amoebal species tested, with the most extreme defect observed in A. castellanii (Fig. 4A). To determine if these growth defects were attributable to the simultaneous deletion of all five gene clusters or could be attributed to a single cluster deletion, a variety of cluster deletion derivatives were analyzed for growth in these species (Fig. 4B and Fig. S4). The Cluster 2ab and Cluster 6a mutants grew as well as the WT strain in all three amoeba hosts tested (Fig. S4). In contrast, gene clusters 3 and 7a were differentially important for growth in H. vermiformis and D. discoideum, respectively (Fig. 4B). In A. castellanii, not only were both clusters 3 and 7a required for robust intracellular growth but, unlike in H. vermiformis and D. discoideum, growth of the Cluster 4a mutant was severely impaired in this host (Fig. 4B). Thus, the importance of a cluster in one host does not mean it is critical for growth in all hosts, demonstrating a host-specific requirement for genes in these regions.
In addition to the host-specific requirements observed for a subset of the cluster deletion mutants, three other behavioral trends stood out. First, both the cluster 4 and Δ2abΔ3Δ6a mutants were almost as defective as the pentuple mutant for growth in A. castellanii (Fig. 4C), demonstrating that growth defects of comparable magnitude could be generated in more than one fashion. The simultaneous deletion of multiple functionally redundant virulence genes in different combinations likely accounts for this phenomenon. Second, each deletion mutant generally exhibited a growth defect in A. castellanii that was as great as or greater than that in D. discoideum or H. vermiformis (Fig. S4), consistent with A. castellanii being more restrictive for L. pneumophila growth than these other two species. Third, the growth defect of the pentuple mutant in H. vermiformis could not be explained simply by the absence of critical Dot/Icm TS, because the total yield of bacteria for this mutant was lower than that observed for a dot− translocation deficient strain (Fig. 4A and Fig. S4).
The behavior of the pentuple mutant indicates that a large cohort of genes in the L. pneumophila genome are dispensable for growth within macrophages but are necessary for propagation in the bacterium’s natural environmental host. The importance of some gene clusters for growth in only a subset of amoebal species and their apparent acquisition from an exogenous source indicate that expansion of the bacterial genome through the accumulation of foreign genetic material has broadened the host range of L. pneumophila.
Discussion
We have shown that the L. pneumophila genome is organized in a mosaic fashion, with conserved chromosomal segments encoding genes required for growth in culture. This modular arrangement of the genome allowed its targeted reduction by almost 13%, resulting in a mutant that grew as well as the WT strain in nutrient-rich medium. This series of essential regions defines a core genome that is also sufficient for growth in macrophages, demonstrating that the collection of virulence factors encoded by this core genome can support bacterial replication in a subset of hosts. Dispersed among these essential genomic regions are chromosomal islands of high plasticity that allow replication in a diverse set of amoebal hosts. The impaired growth of mutants lacking individual genomic islands demonstrates a role for these loci in expanding the host range of L. pneumophila.
The requirements of different genomic regions for L. pneumophila growth in specific hosts indicate that considerable variation exists in the intracellular niches occupied by the bacterium within these hosts, even though the membrane trafficking events that lead to formation of the replication vacuole appear to be similar. Possible sources of this host variation could include allelic differences in, or the absence of, host proteins targeted by the bacterium that render certain Dot/Icm TS ineffective, alterations in the availability of host nutrients, or differences in host antimicrobial defenses. The need to establish a replicative niche in the face of host variation is likely to be the selective pressure driving genome expansion. Therefore, we propose that expansion of the L. pneumophila genome through its acquisition of foreign genetic material has allowed the bacterium to replicate in hosts that are evolutionarily distant from one another. In clinical isolates of L. pneumophila, the large amount of genetic diversity in the regions of the chromosome that are dispensable for growth in vitro likely reflects variation in the amoeba population encountered by these strains in their respective environmental niches. Interaction of the bacterium with any one of multiple hosts in its fresh water habitat could provide the selective pressure for maintaining its diverse repertoire of virulence determinants and contribute to functional redundancy among Dot/Icm TS.
L. pneumophila strains are naturally competent for DNA uptake, allowing acquisition of genetic material from a variety of sources, including eukaryotes (17, 33, 40). Furthermore, the Legionellaceae appear to be capable of actively donating their own DNA to one another, because all sequenced strains encode at least one conjugative DNA apparatus (33, 40–42). The ability to acquire genetic material from multiple sources and reassort DNA among related species likely contributes to increased fitness of the bacterium in a diverse set of hosts.
Continual interaction of L. pneumophila and amoebae over time may have promoted their coevolution in a manner reminiscent of type III effectors and the immune surveillance systems of plants (5). As L. pneumophila evolved effective strategies for promoting intracellular growth, amoebae may have also developed appropriate countermeasures for limiting bacterial replication. This may explain why one amoebal species, such as A. castellanii, is generally more restrictive than others. Unlike plant pathogens, however, which have constrained host ranges and have shed effectors that provide little advantage in their preferred hosts (4, 53), L. pneumophila appears to have retained a broad effector set to facilitate growth in a diverse group of aquatic species. Accumulation of foreign genetic material may enhance the L. pneumophila host range as well as facilitate its adaptation to antimicrobial strategies newly acquired by its hosts.
L. pneumophila is often referred to as an accidental pathogen of humans because its interaction with amoebae provides the selective pressure that allows replication within mammalian macrophages. The pentuple mutant showed robust growth in macrophages despite being severely defective for replication in all three amoebal hosts tested. This behavior supports the existence of an ancestral L. pneumophila strain that encodes a core set of virulence factors sufficient to promote its replication in an unidentified natural host. Further supporting this idea of a reduced genome with a collection of Dot/Icm TS that enables growth in a subset of hosts are the following observations. First, the pentuple mutant has intact copies of the only two Dot/Icm TS whose absence severely impairs intracellular growth, SdhA (13) and DimB/MavN (19, 54). Second, the dot/icm genes do not map to any of the seven gene clusters analyzed here but, instead, are located within the minimized genome (Fig. 2A). Identification of genes in the pentuple mutant required for replication in macrophages should facilitate the discovery of virulence factors specific to this host.
Work on other pathogens has posited the existence of pathogenicity islands, contiguous regions of DNA that provide the bacterium with the potential to colonize a host (55–57). Despite the identification of over 200 Dot/Icm TS, bioinformatic-based approaches to identify such islands in L. pneumophila did not predict the organized clustering of genes required for intracellular growth. The fact that the single most important determinants of disease, the dot/icm genes, are absent from these dispensable regions distinguishes these loci from classically defined pathogenicity islands (58). Instead, the genomic islands identified here by essential gene mapping are regions of host range expansion.
The ordered assembly of the chromosome, in which there are modules of DNA neither associated with a minimal gene set necessary for growth in bacterial culture nor characterized as pathogenicity islands, can be found in other organisms. For example, when we positionally mapped the essential genes of Mycobacterium tuberculosis for growth in rich media, as identified by Rubin and colleagues (59), 11 large gene clusters were devoid of essential genes (Fig. S5). These segments ranged in size from 42 to 112 genes, encompassing 22% of the M. tuberculosis genome. Four of these clusters included genes that encode proteins involved in intracellular survival of the bacterium, demonstrating a correlation between genes in these islands and virulence. Similar information about genome architecture based on experimentally identified regions necessary for optimal growth in bacterial culture should facilitate a more directed approach for genome minimization in other bacteria and provide a strategy for identifying novel virulence genes, including those involved in host range expansion.
This work demonstrates that the L. pneumophila genome was assembled in a modular fashion, building on a core genome that encodes determinants sufficient for growth in vitro as well as replication in selected hosts. Propagation in the environment, however, presumably selected for the ability to grow in multiple hosts and adapt to fluctuations in the host population, rendering the core genome insufficient for survival of the organism in aquatic habitats. Sequestering genes responsible for host range expansion into contiguous regions ensures that the integrity of the core genome is preserved by allowing genetic exchanges resulting in host range alteration being isolated from chromosomal regions encoding functions involved in fundamental cellular processes. Host diversification of L. pneumophila would require that a large fraction of the newly acquired genetic material be maintained to allow growth in multiple hosts. Retention of additional virulence genes would depend on whether they provide an advantage to the bacterium in at least one of its natural hosts without impairing its growth in another. Presumably, the lack of selection against such proteins as Dot/Icm TS would allow L. pneumophila to maintain these proteins, thus accounting for the broader host range of this microorganism. Future studies will focus on distinguishing between factors involved in expanding the host range of L. pneumophila and those encoded by the core genome that play central roles in virulence.
Materials and Methods
Bacterial Strains, Cultures, Cells, and Growth Media.
L. pneumophila strains were grown in liquid N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) buffered yeast extract (AYE) media (60) or on solid charcoal buffered yeast extract (CYE) media (61) containing 0.1 mg/mL CYET (Sigma), 40 μg/mL kanamycin, 5% (wt/vol) sucrose, or 5 μg/mL chloramphenicol when appropriate. E. coli strains were grown in liquid LB or on solid LB plates supplemented with 50 μg/mL ampicillin, 50 μg/mL kanamycin, 50 μg/mL streptomycin, 10 μg/mL tetracycline, or 17 μg/mL chloramphenicol when appropriate. The E. coli strain DH5α λpir (62) was used for all plasmid cloning with the exception of pTO100, which was cloned using a DH5α λpir F+ strain (SI Materials and Methods). Strains are summarized in Table S1.
A/J primary bone marrow-derived mouse macrophages were prepared and cultured as described previously (8). D. discoideum, A. castellanii (ATCC30234; American Type Culture Collection), and H. vermiformis (ATCC50237; American Type Culture Collection), respectively, were cultured as described elsewhere (28, 63, 64). For infections in amoebae, the chromosomal thy− allele in each bacterial strain analyzed was replaced with the thy+ allele by allelic exchange as described (28).
Construction of L. pneumophila Transposon Mutant Library.
Twenty-five individual aliquots of 3 × 109 electrocompetent L. pneumophila Philadelphia 1 strain Lp02 were electroporated with 75 ng of pTO100, recovered at 37 °C for 4 h in AYE, and then plated on CYET containing kanamycin and sucrose. Bacteria were incubated at 37 °C for 4 d. Roughly, 100,000 cfus were harvested into AYE and 20% (vol/vol) glycerol (final concentration of ∼2 × 1010 bacteria/mL), aliquoted into 500-μL volumes, and stored at −80 °C.
TraSH Screen: Growth of L. pneumophila Transposon Library in Vitro.
An aliquot of the L. pneumophila transposon mutant library (∼1010 bacteria) was diluted in AYE media and then cultured overnight at 37 °C to stationary phase. Bacteria were plated for single colonies on solid CYET media containing kanamycin and grown at 37 °C for 4 d. An estimated 100,000–150,000 bacterial colonies were harvested, resuspended in AYE (∼2,500 cfus/mL) and mixed to homogeneity. From this suspension, triplicate aliquots of 2 × 109 bacteria were used to isolate genomic DNA using a Qiagen DNeasy kit, including proteinase K and RNase digestion steps, and then pooled. This procedure was performed with 10 separate aliquots of the library for a total of 10 individual experiments. Array construction, TraSH probe generation, array hybridization, and analysis are detailed in SI Materials and Methods.
Construction of L. pneumophila Deletion Mutants.
Gene clusters were deleted in L. pneumophila Philadelphia 1 strain Lp02 using a double-recombination strategy with the suicide vector pSR47s as described (65) with the following exception: 3,000-bp segments directly upstream and downstream of each cluster were used for homologous recombination. Primer pairs for plasmid construction are listed in Table S1. All plasmids were sequenced before use. For deletion of cluster 5 in the pentuple mutant, a BamHI chloramphenicol resistance cassette was cloned from pM_Cm (pM-Gm was a kind gift from Zhao-Qing Luo, Perdue University, West Lafayette, IN) into the BamHI site of the cluster 5 deletion plasmid, pYA71. For each mutant, 100–600 individual isolates were screened by PCR. For those mutants used in subsequent analyses, their corresponding deletion mutations were verified by microarray analysis (SI Materials and Methods and Fig. S3).
In Vitro Growth Curves.
L. pneumophila strains grown on solid media were diluted in AYE media containing thymidine to 2–3 × 108 bacteria/mL. Bacterial growth at 37 °C was monitored over 16–26 h by diluting culture aliquots 1:5–1:10 in 1× PBS and measuring the absorbance at 600 nm in a Molecular Devices SpectroMax M5 spectrophotometer at regular intervals. Bacterial doubling time, td, was calculated as td = (t2 − t1)/(log102/(log10(q2/q1)), where t1 is time 1, t2 is time 2, and q1 and q2 are the number of bacteria at t1 and t2, respectively.
Intracellular Growth Assays.
Growth of L. pneumophila in A/J macrophages, A. castellanii, and D. discoideum, respectively, was performed as described in elsewhere (8, 28, 66). For growth in H. vermiformis, H. vermiformis was plated at 1 × 105 cells per well in a 96-well tissue culture plate in H. vermiformis media (ATCC1034; American Type Culture Collection) and then immediately challenged with L. pneumophila strains grown to postexponential phase at a multiplicity of infection of 0.05. Cells were incubated for 2 h at 35 °C and then rinsed twice with H. vermiformis media. At 2, 24, 48, 72, and 96 h postinfection, H. vermifomis were lysed with 0.02% saponin (Sigma) and growth of L. pneumophila was determined by plating host cell lysates on solid CYE media and counting the number of bacterial cfus.
Supplementary Material
Acknowledgments
We thank Eric Rubin, Simon Dillon, Jeff Murray, and Chris Sassetti for helpful advice regarding the TraSH protocol, plasmids, and equipment use; Jim Flynn and Yue Shao for helpful advice and equipment use; and Michael Berne and Kip Bodi and Alex Ensminger for helpful advice with genome sequencing protocols and analyses. We thank Eric Rubin, Alex Ensminger, Kerri Sheahan, Greg Crimmins, Eddie Geisinger, Sina Mohammadi, and Liz Creasey for review of the manuscript. This work was supported by a Natalie V. Zucker Fellowship and the Howard Hughes Medical Institute (to T.J.O.). R.R.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111678108/-/DCSupplemental.
References
- 1.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen TM, et al. A community-wide outbreak of legionnaires disease linked to industrial cooling towers—How far can contaminated aerosols spread? J Infect Dis. 2006;193:102–111. doi: 10.1086/498575. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz MA. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindeberg M, Cunnac S, Collmer A. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol Plant Pathol. 2009;10:767–775. doi: 10.1111/j.1364-3703.2009.00587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma W, Dong FF, Stavrinides J, Guttman DS. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2:e209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 8.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 9.Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 12.Derré I, Isberg RR. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect Immun. 2005;73:4370–4380. doi: 10.1128/IAI.73.7.4370-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci USA. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belyi Y, Tabakova I, Stahl M, Aktories K. Lgt: A family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol. 2008;190:3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Losick VP, Haenssler E, Moy MY, Isberg RR. LnaB: A Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 2010;12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Felipe KS, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burstein D, et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L, et al. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2011;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 22.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 24.Banga S, et al. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci USA. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X, et al. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 27.Xu L, et al. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ensminger AW, Isberg RR. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun. 2010;78:3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conover GM, Derré I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: A translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 30.Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol. 2005;56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 31.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: Exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e4. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanRheenen SM, Luo ZQ, O’Connor TJ, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74:3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chien M, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 34.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachman MA, Swanson MS. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol. 2001;40:1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 36.Samrakandi MM, Cirillo SL, Ridenour DA, Bermudez LE, Cirillo JD. Genetic and phenotypic differences between Legionella pneumophila strains. J Clin Microbiol. 2002;40:1352–1362. doi: 10.1128/JCM.40.4.1352-1362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyake M, et al. Characterization of Legionella pneumophila pmiA, a gene essential for infectivity of protozoa and macrophages. Infect Immun. 2005;73:6272–6282. doi: 10.1128/IAI.73.10.6272-6282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent CD, et al. Identification of non-dot/icm suppressors of the Legionella pneumophila DeltadotL lethality phenotype. J Bacteriol. 2006;188:8231–8243. doi: 10.1128/JB.00937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ninio S, Celli J, Roy CR. A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 2009;5:e1000278. doi: 10.1371/journal.ppat.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 41.Glöckner G, et al. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol. 2008;298:411–428. doi: 10.1016/j.ijmm.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 42.D’Auria G, Jiménez-Hernández N, Peris-Bondia F, Moya A, Latorre A. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics. 2010;11:181–193. doi: 10.1186/1471-2164-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann S, Chen YP. Bacterial genomic G+C composition-eliciting environmental adaptation. Genomics. 2010;95:7–15. doi: 10.1016/j.ygeno.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Brassinga AK, et al. A 65-kilobase pathogenicity island is unique to Philadelphia-1 strains of Legionella pneumophila. J Bacteriol. 2003;185:4630–4637. doi: 10.1128/JB.185.15.4630-4637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim E-H, Charpentier X, Torres-Urquidy O, McEvoy MM, Rensing C. The metal efflux island of Legionella pneumophila is not required for survival in macrophages and amoebas. FEMS Microbiol Lett. 2009;301:164–170. doi: 10.1111/j.1574-6968.2009.01813.x. [DOI] [PubMed] [Google Scholar]
- 46.Ristroph JD, Hedlund KW, Gowda S. Chemically defined medium for Legionella pneumophila growth. J Clin Microbiol. 1981;13:115–119. doi: 10.1128/jcm.13.1.115-119.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wintermeyer E, et al. Sequence determination and mutational analysis of the lly locus of Legionella pneumophila. Infect Immun. 1994;62:1109–1117. doi: 10.1128/iai.62.3.1109-1117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albert-Weissenberger C, et al. Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J Bacteriol. 2010;192:446–455. doi: 10.1128/JB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nedea EC, Markov D, Naryshkina T, Severinov K. Localization of Escherichia coli rpoC mutations that affect RNA polymerase assembly and activity at high temperature. J Bacteriol. 1999;181:2663–2665. doi: 10.1128/jb.181.8.2663-2665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gokulan K, Rupp B, Pavelka MS, Jr, Jacobs WR, Jr, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis. J Biol Chem. 2003;278:18588–18596. doi: 10.1074/jbc.M301549200. [DOI] [PubMed] [Google Scholar]
- 51.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 52.Altman E, Segal G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol. 2008;190:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi DY, Tamaki SJ, Keen NT. Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc Natl Acad Sci USA. 1989;86:157–161. doi: 10.1073/pnas.86.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Losick VP, Isberg RR. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morschhäuser J, Vetter V, Emödy L, Hacker J. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: Cross-talk between different adhesin gene clusters. Mol Microbiol. 1994;11:555–566. doi: 10.1111/j.1365-2958.1994.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 56.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochman H, Soncini FC, Solomon F, Groisman EA. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hacker J, et al. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 59.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 60.Horwitz MA, Silverstein SC. Intracellular multiplication of Legionnaires’ disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J Clin Invest. 1983;71:15–26. doi: 10.1172/JCI110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feeley JC, et al. Charcoal-yeast extract agar: Primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolter R, Inuzuka M, Helinski DR. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 63.Solomon JM, Rupper A, Cardelli JA, Isberg RR. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun. 2000;68:2939–2947. doi: 10.1128/iai.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moffat JF, Tompkins LS. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect Immun. 1992;60:296–301. doi: 10.1128/iai.60.1.296-301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. Analysis of the Legionella pneumophila fliI gene: Intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Solomon JM, Isberg RR. Dictyostelium discoideum strains lacking the RtoA protein are defective for maturation of the Legionella pneumophila replication vacuole. Cell Microbiol. 2005;7:431–442. doi: 10.1111/j.1462-5822.2004.00472.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.