Abstract
Human apolipoprotein E (apoE) is one of the major determinants in lipid transport, playing a critical role in atherosclerosis and other diseases. Binding to lipid and heparan sulfate proteoglycans (HSPG) induces apoE to adopt active conformations for binding to low-density lipoprotein receptor (LDLR) family. ApoE also interacts with beta amyloid peptide, manifests critical isoform-specific effects on Alzheimer’s disease. Despite the importance of apoE in these major human diseases, the fundamental questions of how apoE adjusts its structure upon binding to regulate its diverse functions remain unsolved. We report the NMR structure of apoE3, displaying a unique topology of three structural domains. The C-terminal domain presents a large exposed hydrophobic surface that likely initiates interactions with lipids, HSPG, and beta amyloid peptides. The unique topology precisely regulates apoE tertiary structure to permit only one possible conformational adaptation upon binding and provides a double security in preventing lipid-free and partially-lipidated apoE from premature binding to apoE receptors during receptor biogenesis. This topology further ensures the optimal receptor-binding activity by the fully lipidated apoE during lipoprotein transport in circulation and in the brain. These findings provide a structural framework for understanding the structural basis of the diverse functions of this important protein in human diseases.
Keywords: apoE-receptor interactions, domain-interactions, lipid transport, lipoproteins
Human apoE is one of the major determinants in lipid transport throughout the body, playing a critical role in atherosclerosis and other metabolic diseases (1). Binding to lipid and heparan sulfate proteoglycans (HSPG) induces apoE to adopt active conformations for low-density lipoprotein receptor (LDLR) and LDLR-related protein (LRP)-binding (2). ApoE functions during lipid transport by binding to the LDLR family (2, 3), with the major LDLR-binding region spanning residues 130–160 (1). Three common isoforms of human apoE have been identified: ApoE3 has C112/R158, apoE4 has R112/R158, and apoE2 has C112/C158 (1). ApoE also interacts with amyloid peptide and manifests critical isoform-specific effects on several major human diseases (4–7). The most pronounced pathological effect attributes to the association of apoE4 with neurodegenerative diseases, including Alzheimer’s disease (2). Clearly, apoE carries out diverse biological functions in several major human diseases by binding to different partners. However, fundamental questions of how apoE adjusts its structure upon binding to regulate its diverse functions remain unsolved. Because apoE isoforms only differ by one amino acid, the structural basis of this isoform-specific effect is another most challenging unanswered question for apoE to date.
It was proposed that apoE contains two domains: an N-terminal (NT) domain (residues 1–191) and a C-terminal (CT) domain (residues 216–299), linked by a flexible hinge region (1). The NT domain is responsible for the LDLR-binding, yet, in isolation, binds weakly to lipids. The CT domain possesses the major lipoprotein-binding sites, but does not bind receptor, possibly resulting in a partially lipidated apoE in which only the CT domain binds to lipids whereas the NT domain remains lipid-free. The X-ray crystal structure of lipid-free apoE-NT (residues 23–165) reveals an elongated four-helix-bundle (8). Although the major LDLR-binding region is partially exposed, only lipid-bound apoE-NT binds the LDLR. Regions beyond residues 23–165 are critical to apoE’s LDLR-binding (9, 10). The crystal structures of apoE2-NT/apoE3-NT/apoE4-NT revealed subtle variations (11, 12). The interchange of C112R in the apoE4-NT leads to a new salt-bridge between E109 and R112, causing the side chain of R61 to adopt an exposed position (13). Mutagenesis study suggests that this change might allow the R61 side chain to interact with E255, whereas apoE3 and apoE2 lack this salt-bridge (14). This salt-bridge in apoE4 is considered to be critical to apoE’s isoform-specific effects on human diseases (2).
The NMR structure of human apoE3 reported here reveals a unique helix-bundle topology of three structural domains that uniquely regulates its diverse biological functions. ApoE3 displays extensive domain interactions of salt-bridges and H-bonds, causing shielding of the major LDLR-binding region by the CT domain from binding to receptors. The CT domain presents a large exposed hydrophobic surface that likely initiates interactions with different binding partners including lipids, HSPG, and amyloid beta peptides. The unique topology of apoE precisely regulates its tertiary structure to permit only one possible conformational adaptation upon binding in a two-step manner. This topology also provides a double security in preventing lipid-free and partially lipidated apoE from premature binding to apoE receptors during receptor biogenesis and further ensures the optimal receptor-binding activity by the fully lipidated apoE during lipoprotein transport in circulation and in the brain. Overall, apoE structure provides a structural framework for the diverse functions of this important protein in human diseases including atherosclerosis and Alzheimer’s diseases.
Results
NMR Structural Determination of apoE3.
ApoE forms a mixture of different oligomers, representing the major challenge that hindered structural determination (1, 15). We generated a monomeric apoE3 by mutations in the CT domain (F257A/W264R/V269A/L279Q/V287E) (15). This monomeric apoE3 retained the properties of the parent protein including domain-domain interactions (15) and allowed us to collect high-quality NMR data for a complete spectral assignment (16). ApoE3 is a 299-residue α-helical protein that displays severe spectral overlap. To solve this problem, segmentally labeled apoE3 samples were prepared (17), allowing spectral simplification and unambiguous NOE assignment (Fig. S1). A total of 3,459 NOEs was assigned, including 572 long-range NOEs (189 unique) and 224 interdomain NOEs (78 unambiguously assigned using special segmentally labeled samples) (Table S1). This NMR data permitted determining the solution structure of apoE3. The 20 best-fit NMR structures of human apoE3 display rmsd of backbone atoms of 0.58 ± 0.05 Å and rmsd for all heavy atoms of 1.13 ± 0.08 Å for all helical regions. ApoE3 displays 68.9% α-helix content, which is consistent with the CD measurements (18). Structural analysis indicates that in addition to 17 Gly and 8 Pro residues, 210 residues (76.8%) are located in the most favored regions, 49 residues (17.7%) are in additional allowed regions, and 12 residues (4.4%) are in the generously allowed region of the Ramachandran plot. Only three residues are in the disallowed region (1.1%) (Table S1).
ApoE3 Structure: Three Domains.
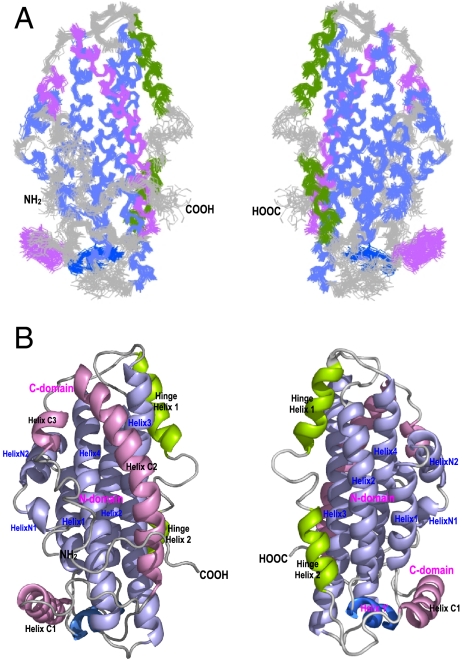
The NMR structure of apoE3 reveals an overall helix-bundle structure (Fig. 1). The helix locations of apoE3 are shown in Table S2. Superposition of the 20 best-fit NMR structures indicates that the helical regions of apoE3 are well defined including the CT helices (Fig. 1A), providing precision and accuracy of backbone and side chain atoms for analysis of H-bonds and salt-bridges. The apoE structure indicates three structural domains: An NT domain (residues 1–167), a hinge domain (residues 168–205), and a CT domain (residues 206–299) (Fig. 1B). The NT domain contains an up-and-down four-helix-bundle (light blue, Fig. 1B), which is similar to the isolated NT domain structures determined by both X-ray crystallography (8) and NMR (19). The major differences between isolated NT domain and the NT domain in full-length apoE3 are in the terminal helices and Helices 1′, 3, and 4 (Fig. S2). In apoE3, HelixN1/N2 and HingeH1 are well defined due to domain interactions. Helices 1′, 3, and 4 are the major helices that interact with the CT domain, causing Helix 1′ to slide down and Helices 3 and 4 to move slightly towards the CT domain (Fig. S2). The hinge domain contains two helices: HingeH1 and HingeH2 (green, Fig. 1B). Previously, we showed that this hinge domain regulates the NT-CT interaction (15). This hinge domain also agrees with the published limited proteolysis data (20). The CT domain contains three helices: Helix C1, C2, and C3 (pink, Fig. 1B). While Helix C1 interacts with Helix 1′, Helices C2 and C3 interact with Helices 3 and 4. These domain-domain-interactions stabilize the CT-domain helices, especially Helix C3 which only contains six residues. Overall, apoE3 form a bundle of six helices. The NT domain separates the hinge domain from the CT domain.
Fig. 1.
NMR Structure of apoE3. (A) Superposition of the 20 best-fit NMR structures of apoE3. (B) Ribbon representations of the average structure of apoE3. The N-terminal helices are colored in light blue, the C-terminal helices are in pink, and the hinge domain is colored in green. The Helix1’ is shown in blue. The helices are labeled respectively. The left and right in (A) and (B) are 180° rotation of apoE3 structure, showing both sides of apoE3.
Extensive and Specific Domain-Domain-Interactions in apoE3.
VADAR analysis (21) indicates that many buried hydrophilic residues are located between apoE domains (Table S3), which is not surprising due to the helix-bundle topology of the NT domain, in which the hydrophilic residues point outside and available for interactions with the other domains. These buried hydrophilic residues form buried H-bonds and salt-bridges between domains (bold, Table 1). In particular, R61 forms an H-bond with T194 and E255 forms a salt-bridge with K95. Both residues are involved in domain interactions, providing a structural explanation of the lipoprotein preference of different apoE isoforms (2). Less buried hydrophobic residues are observed between the NT and CT domains (Table S4). Unlike the NT helix-bundle which are mainly stabilized by hydrophobic interactions, the domain interactions in apoE3 are mainly specific H-bonds and salt-bridges, which are likely critical for the reversibility of lipoprotein-binding activity of apoE.
Table 1.
H-bonds and salt-bridges between apoE domains
| LoopN | HelixN1 | HelixN2 | Helix 1 | Helix 1′ | Helix 2 | Loop | Helix 3 | Loop | Helix 4 | |
| (1–5) |
(6–9) |
(12–22) |
(26–40) |
(45–52) |
(55–79) |
(80–88) |
(89–125) |
(126–130) |
(131–164) |
|
| LoopN | R142-E3 | |||||||||
| K146-Q4 | ||||||||||
| HingeH1 | R172-S94 | |||||||||
| (168–180) | A176-S94, E179-Q98 | |||||||||
| HingeH2 | S54-S197 | R61-T194 | ||||||||
| (190–199) | D65-R191 | |||||||||
| Loop | E49-R206 | R119-Q201 | ||||||||
| (200–209) | ||||||||||
| Helix C1 | Q46-R213 | |||||||||
| 210–223 | E49-R213 | |||||||||
| E50-R217 | ||||||||||
| Loop | K1-T225 | R114-Q235 | Q128-E234 | E131-R226 | ||||||
| (224–235) | E121-Q235 | E132-E234 | ||||||||
| R134-D227 | ||||||||||
| R136-E231 | ||||||||||
| Helix C2 | T83-E266 | T89-K262, T89-E266 | K143-Q246 | |||||||
| (236–266) | E88-K262 | R92-E266,R92-S263 | R147-Q246 | |||||||
| K95-E255, E96-S263 | R147-Q249 | |||||||||
| R103-E245,R114-E238 | R147-E245 | |||||||||
| D110-K242,Q98-R251 | R150-Q253 | |||||||||
| Helix C3 | E13-R274 | K157-D271 | ||||||||
| (271–276) | Q17-R274 | K157-Q275 | ||||||||
| Loop C | Q4-E287 | K143-E287 | ||||||||
| (277–299) | K146-E281 | |||||||||
| K146-E287 | ||||||||||
| R150-E281 |
ApoE3 structure suggests that both the hinge and CT domains cannot fold independently. The NT domain serves as a folding template for the hinge and CT domains. Indeed, the hydrophilic residues located outside of the NT helix-bundle provide ideal candidates for specific H-bonds and salt-bridges with other domains, suggesting a two-step folding mechanism: the NT domain adopts a helix-bundle first and the hinge and CT domains then fold upon the NT domain through specific domain interactions. Experimental evidence supports this two-step folding mechanism, indicating that the isolated NT domain folds independently while the CT domain is more flexible and only marginally stable (15, 18, 20, 22). Chemical denaturation of apoE further reveals this two-step folding process with a clear intermediate stage that is progressively lost as the CT is truncated down to residue 230 (23).
The Major LDLR-Binding Region of apoE3 Is Shielded by the CT Domain.
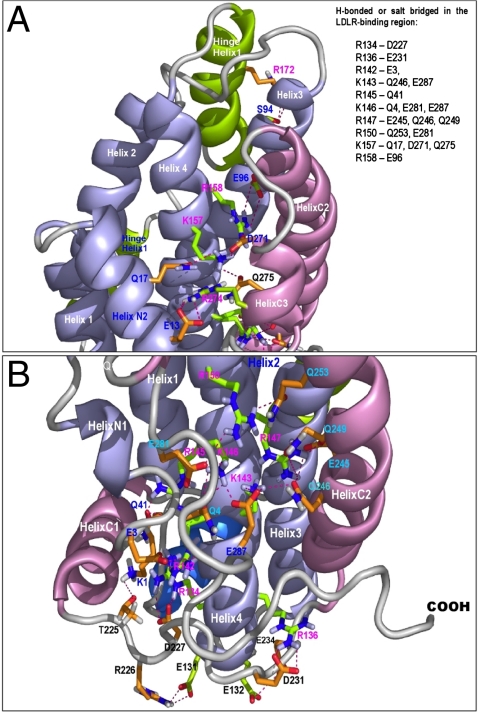
In the major LDLR-binding region, every K and R residues form H-bonds and salt-bridges with the residues in the CT domain (Fig. 2). Two residues in the N terminus (E3 and Q4) are also involved in these H-bonds and salt-bridges (Table 1). These domain interactions significantly stabilizes the CT domain, resulting in a unique structural topology, in which the major LDLR-binding region is shielded (Table 2) and unavailable for receptor/HSPG-binding (3). Furthermore, lipid-free NT helix-bundle also displays an inactive conformation for receptor-binding. These two unique structural properties of apoE3 provide structural rationales for the fact that lipid-free apoE does not bind to LDLR.
Fig. 2.
Buried H-bonds and salt-bridges between apoE3 domains, with a focus on the major LDLR-binding region. The apoE3 is shown in ribbons and the side chains of the interacting residues in stick model. The NT domain is shown in light blue while the CT domain in pink. The side chains of the interacting residues in the major LDLR-binding region are shown in green, whereas the side chains of the other interacting residues are shown in brown. The salt-bridges and H-bonds are indicated with red dashed lines. (A) Detailed interdomain interactions of residues K157 and R158. (B) Detailed interdomain interactions of residues R134, R136, R142, K143, R145, K146, R147, and R150. The interacting residues are summarized in the top box.
Table 2.
Accessible surface area (ASA) of the positively charged residues of the major LDLR-binding region of apoE3 in lipid-free, partially lipidated (Step 1), completely lipidated apoE structures (Step 2)
| Residues | ASA_main | ASA_side | ASA_main | ASA_side | ASA_main | ASA_side |
| Lipid-free apoE3 * | Partially-lipidated apoE3 † | Completely-lipidated apoE3 ‡ | ||||
| R134 | 0.02 | 0.03 | 0.06 | 0.07 | 0.32 | 0.38 |
| R136 | 0.02 | 0.02 | 0.71 | 0.84 | 0.73 | 0.86 |
| H140 | 0.02 | 0.02 | 0.33 | 0.40 | 0.59 | 0.71 |
| R142 | 0.01 | 0.01 | 0.15 | 0.18 | 0.36 | 0.42 |
| K143 | 0.04 | 0.05 | 0.33 | 0.40 | 0.43 | 0.52 |
| R145 | 0.07 | 0.08 | 0.04 | 0.05 | 0.04 | 0.05 |
| R146 | 0.01 | 0.01 | 0.15 | 0.18 | 0.33 | 0.40 |
| R147 | 0.00 | 0.00 | 0.27 | 0.33 | 0.45 | 0.54 |
| R150 | 0.09 | 0.10 | 0.44 | 0.52 | 0.49 | 0.57 |
| K157 | 0.04 | 0.05 | 0.27 | 0.33 | 0.32 | 0.38 |
| R158 | 0.02 | 0.02 | 0.22 | 0.26 | 0.40 | 0.47 |
*The VADAR calculation is based on lipid-free apoE3 NMR structure, showing that the major LDLR-binding region is completely buried due to domain-interaction.
†The VADAR calculation is based on partially-lipidated apoE (Structure 5, Fig. 3), showing that the major LDLR-binding region is only partially exposed due to interaction between helices 3 and 4.
‡The VADAR calculation is based on completely-lipidated apoE (Structure 8, Fig. 3), showing that the major LDLR-binding region is completely exposed.
A Unique Exposed Hydrophobic Surface in the apoE-CT.
A striking structural feature of apoE3 was observed: numerous exposed hydrophobic residues in the CT domain (Fig. 3A, left) and few buried hydrophobic residues exist between the CT and NT domains (Fig. 3A, right). As a result, many hydrophilic residues in the CT domain are buried in the domain interface, forming specific H-bonds and salt-bridges with the NT domain (Table 1). While surprising, this arrangement may provide a structural basis for initiation of the CT domain contacting with lipids, HSPG, and amyloid beta peptide. The exposed hydrophobic residues destabilize the CT domain and form a large exposed hydrophobic surface in apoE3 to attract lipid/amyloid beta peptide for initial binding (green arrow, Fig. 3B, right). We found that among the mutations in the monomeric apoE3, V269A/L279Q/ V287E were located in a loop after Helix C3 and F257A/W264R were located in the end of Helix C2. These mutations unlikely change the overall apoE3 structure. However, hydrophobic residues F257/W264/V269/L279/V287 in the wild-type apoE3 enhance the intensity of the exposed hydrophobic surface in the CT domain, facilitating this binding initiation. Initiation of apoE-CT/lipid interaction may trigger a conformational change in the CT domain wherein it moves away from the NT domain, causing exposure of the hydrophilic surface that originally interacts with the NT domain (red arrows, Fig. 3). This conformational change will generate an amphipathic helical conformation in the CT domain in which the hydrophobic surface binds to lipid surface and the opposite hydrophilic surface exposes to the solution, making this partially lipid-bound apoE more stable. Given the increased stability, this conformation will be retained until metabolic processes eliminate the binding sites on lipoprotein surfaces, resulting in apoE release from lipoproteins and recovery of the lipid-free helix-bundle.
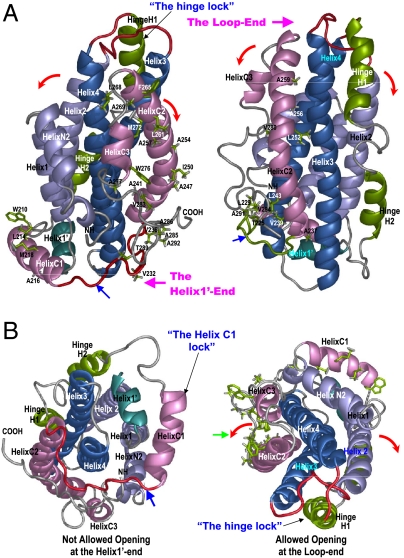
Fig. 3.
Special structural features that regulate initial conformational adaptation of apoE upon lipid binding. ApoE structures are shown in ribbons with the NT helices colored in blue, the CT helices in pink, and hinge domain in green. The Helix1’ is in dark green. (A) Side views of apoE3 structure. Left: ApoE3 structure with the exposed hydrophobic residues of the apoE-CT shown in green sticks. Right: ApoE3 structure with the buried hydrophobic residues of the apoE-CT shown in green sticks. Two ends of the apoE3 helix bundle are named and labeled as: The Loop-end and the Helix1’-end. A hinge lock is formed by hinge domain that prevents the NT bundle from opening at the Loop-end (Helices 2 and 3 moving away from helices 1 and 4) (B) Left: A bottom view of apoE3 structure (the Helix1’-end view). Helix C1 and the flanking loops form a “Helix C1 lock” (Blue arrow) that prevents the CT domain moving away from the NT domain at the Helix1’-end. Right: A top view of apoE3 structure (the Loop-end view) with the exposed hydrophobic residues of the apoE-CT shown in green sticks. Noticeably, most of hydrophobic residues are exposed in the apoE-CT, forming the only major hydrophobic surface in apoE3 structure (green arrow). The directions of the allowed apoE-CT opening are indicated by red arrows.
The Unique Topology of apoE3.
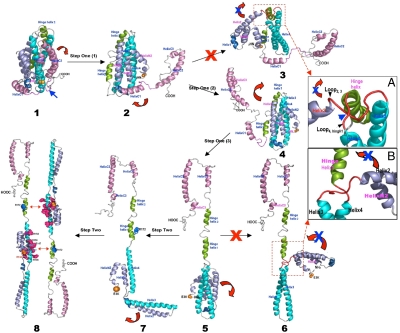
Compelling evidence indicates that apoE may undergo major conformational changes upon lipoprotein-association (2). However, controversy arises as to how apoE adjusts its conformation upon lipid binding, due to lack of structural information of the CT domain (2). Using surface plasmon resonance, Lund-Katz and coworkers proposed a two-step mechanism for apoE-binding to HDL and very low density lipoprotein (VLDL) (24) and a similar mechanism has been also proposed by others (2, 25). ApoE3 structure supports this model and further reveals a structural rationale for a unique conformational adaptation upon lipoprotein association. The first step involves lipid binding induced interruption of domain interactions, promoting sequential dissociation of the CT and the hinge domains from the NT domain (1–3, Fig. 4). Experimental data suggests that this step is a fast reversible step (24). The apoE3 tertiary structure prevents the CT domain from opening at the Helix 1′-end due to a “Helix C1 lock” (left, Fig. 3B). As such, the CT domain can only open at the opposite loop-end (right, Fig. 3B) and the loop between Helices C1 and C2 serves as a hinge (blue arrow, Fig. 3). Once the CT domain moves away from the NT domain, the lipid-associated CT domain causes Helix C1 to dissociate from Helix 1′ (2, Fig. 4). The NT helix-bundle opening is prohibited at this stage due to a “hinge lock” that prohibits the NT domain opening from the loop-end (Fig. 3A). Similarly, the red loops will be tangled if the NT domain opens at the Helix1’-end (3, inset, Fig. 4A). Only one possibility is allowed: the hinge domain swings the CT domain to the top of the NT domain (2, Fig. 4), unwinding the loops and releasing the hinge lock to generate a partially lipidated intermediate wherein only the CT domain binds to the lipid surface (3, Fig. 4). The major LDLR-binding region is only partially exposed in the lipid-free NT helix-bundle due to interactions between helices 3 and 4 (Table 2), thus displaying an inactive receptor-binding intermediate.
Fig. 4.
A two-step conformational adaptation of apoE3 upon lipoprotein binding. ApoE3 structures are shown in ribbons and the colors are coded the same as Fig. 2. The helices are labeled respectively. This two-step conformation adaptation of apoE3 upon lipoprotein association follows a specific and precise self-modulation pathway, which is regulated by the special topology of lipid-free apoE3 structure. We assume that there is no major change of the helix locations in apoE3 during lipid-binding.
The second step involves the NT helix-bundle opening. Experimental data suggests that this step is a slow reversible step (24). The tertiary structure of apoE3 again precisely modulates this opening to only allow for one possibility: the NT helix-bundle opening at the loop-end. Indeed, the hinge domain imposes severe stereo hindrance that restricts the helix-bundle opening at the Helix 1′-end (6, inset, Fig. 4B). Additionally, the lipoprotein-bound CT domain is located at the loop-end, preventing lipid surface from inducing the helix-bundle opening at the opposite Helix 1′-end. We previously showed that the NT helix-bundle opening at Helix 1′ is not preferred (Table S5) (19). The only allowed NT helix-bundle opening is at the loop-end and involves Helices 2 and 3 moving away from Helices N1, N2, 1, and 4 (4, Fig. 4). The CT-bound lipid surface is located at the same loop end that is able to induce this helix-bundle opening. Many buried hydrophilic residues are located between Helices 1 and 2 and between Helices 3 and 4, making their helix-helix interfaces much less stable (Table S5). The final lipoprotein-associated structure of apoE3 is a completely opened conformation, containing two lobes connected by the hinge domain (5, Fig. 4): the CT lobe and NT lobe. Both lobes interact with the lipoprotein surface, providing the maximum lipoprotein-binding surface possible. The major LDLR-binding region in this conformation is fully exposed due to the removal of interactions between Helices 3 and 4 (Table 2), resulting in an enhanced positive charge potential for the optimized receptor-binding activity.
A “Double Security” in apoE for Its Optimal Receptor-Binding Activity.
The unique ApoE3 structure reveals a “double security” that ensures the optimal apoE-receptor interaction. The first security is the shielding of the major LDLR-binding region by the CT domain that prevents lipid-free apoE from premature binding to apoE receptors. Interestingly, the observed interactions between the major LDLR-binding region and the CT domain are mainly salt-bridges and H-bonds, limiting the positively charged residues in the major LDLR-binding region from binding to receptors. The second security is an inactive conformation of the lipid-free NT helix-bundle (Table 2), which prevents both lipid-free and partially-lipidated apoE from receptor binding. This inactive conformation also facilitates the formation of the fully lipidated apoE which displays a completely opened conformation (5, Fig. 4), making the positively charged residues in the major LDLR-binding region completely exposed for an optimal receptor-binding activity (Table 2). Thus, this double security displayed in apoE structure ensures that only the fully lipidated apoE is able to bind to apoE receptors with an optimal activity.
Discussion
Human apoE carries out diverse biological functions in several major human diseases by binding to different partners. It remains unknown as to how apoE adjusts its conformation upon binding to these different partners (2), however, our results indicate that the unique tertiary structure of apoE3 precisely regulates its conformational adaptation upon binding, to only allow for one possible conformational change in a two-step manner. Indeed, the binding induced conformational transitions of apoE3 in vivo are hardly accessible using biophysical methods. However, useful information has been obtained using in vitro studies. When interpreted with caution, these studies shed light on physiological events of apoE upon binding to different partners.
In the lipid-binding case, this two-step binding process potentially generates partially lipidated and fully lipidated apoEs, permitting us to discuss the possible apoE3 structures on different lipoprotein particles. On discoidal HDLs, two molecules of apoE3 may adopt an antiparallel orientation (26). The distance between residues C112 and W264 in this lipid-bound apoE is > 80 Å (5, Fig. 3), whereas the same distance in the lipid-free apoE3 is ∼28 Å (1, Fig. 3), consistent with the previously reported FRET data (27). No intermolecular FRET was observed between residues C112 and W264 (27). Residue R172 of one monomer is located in a close proximity to the major LDLR-binding region of the other monomer in this antiparallel dimer orientation (5, Fig. 4). An R172A mutation of apoE3 significantly reduced its LDLR-binding activity (10).
Both lipid-bound apoE states are possible on spherical HDLs. The first state is a partially lipidated apoE (upper, Fig. S3A) and the second state is a fully lipidated apoE. Based on the crystal structure of apoE/DPPC particle at 10 Å resolution (28), we assume two molecules of apoE3 on a spherical HDL surface, which are staggered at a 42° angle to each other (lower, Fig. S3A). This relative orientation generates four possibilities. Using stereo hindrances as restraints and a distance constraint of > 80 Å between C112 and W264 (27) during modeling, we were able to eliminate three possibilities and only one configuration satisfies these restraints (lower, Fig. S3A). Similarly, both lipoprotein-associated apoEs may also be possible on VLDLs (Fig. S3B). VLDL has a much larger surface area than HDL, facilitating apoE3-binding. VLDL and VLDL remnant may contain more surface-located cholesterol because they are cholesterol-richer lipoprotein particles than HDLs (3). These differences cause a nearly 10-fold binding affinity difference of apoE3 between HDL and VLDL (24).
Both lipid-poor and lipid-bound apoEs are able to bind to the LRP (29) or HSPG (30). Saito and coworkers demonstrated two major heparin-binding sites in apoE: An NT-binding site that overlaps with the major LDLR-binding region and a CT-binding site involving basic residues around K233 (31). A two-step binding process was also suggested for apoE/heparin interaction. The basic residues around K233 in the apoE3 are completely exposed, potentially initiating HSPG-binding to trigger the same conformational change of the apoE-CT shown in Fig. 3. The experimental evidence indicated that lipid-free CT domain displayed a stronger affinity and faster kinetics in the first binding step to heparin, supporting this suggestion (Table S6) (30, 32). Such a conformational change interrupts domain interactions and makes the major LDLR-binding region available for the second-step of HSPG-binding, again consistent with the stronger affinity and faster kinetics of the NT domain binding to heparin in the second binding step (Table S6) (30). For lipid-bound apoE, both the major LDLR-binding region and the K233 site are exposed and available for binding to the cell surface located HSPG to initiate the LRP/HSPG pathway.
Because H-bonds are observed between the N terminus and the major LDLR-binding region, we suggest that these H-bonds are retained in the lipoprotein-associated structure, causing residue E3 to be in a close proximity to the major receptor-binding region. An E3K mutation enhances the positive potential of the major LDLR-binding region and increases apoE3 LDLR-binding activity (9). The spatial arrangement of buried H-bonds and salt-bridges between NT/CT domain interfaces further indicate they are critical in guiding recovery of the unique lipid-free structure of apoE3 once dissociation from the lipoprotein surface. Reestablishment of the observed buried H-bonds and salt-bridges serve as the specific interactions that secure apoE’s domain-domain interactions, ensuring apoE always adopts a unique tertiary structure for reversible lipoprotein binding and receptor-binding activities.
The buried nature of the major LDLR-binding region may have functional significance for apoE3 during folding, maturation, and intracellular trafficking of apoE receptors. Indeed, receptor-associated protein (RAP), apoE and apoE receptors are all folded within the Endoplasmic Reticulum (ER) (33). ApoE and RAP fold quickly, whereas apoE receptors fold slowly and require chaperones including RAP. ApoE and RAP share a similar binding affinity (nM) to its receptors (34, 35) and can compete for binding to apoE receptors during receptor folding and maturation. These two unique structural features of apoE eliminate the possible premature binding, thus RAP can perform its chaperone and escort functions (35). Any mutations in apoE3 that interrupt apoE domain interaction may expose the major LDLR-binding region. The “double security” feature displayed in apoE3 structure also suggests that VLDLR, apoER2, and LPR may have different binding modes to apoE, because these receptors can bind to apoE in their lipid-free state (29, 36). This new aspect directly ties apoE3 domain interaction with receptor folding, maturation, and intracellular trafficking and may have important clinical implications, because misfolding of the apoE receptors causes several major human diseases (37, 38).
Many human naturally occurring mutants of apoE carry mutations of the critical LDLR-binding residues, potentially interrupting apoE domain interactions (Fig. S4) and facilitating premature lipid-free apoE-binding to receptors and formation of the partially-lipidated apoE. A premature binding between lipid-free/partially-lipidated apoE and the newly synthesized apoE receptors may interrupt folding and trafficking of apoE receptors inside the cells. Similarly, a premature binding between partially-lipidated apoE and mature apoE receptors on the cell surfaces during lipoprotein transport and metabolism in circulation may also affect optimal apoE-receptor interaction. However, the double security in apoE structure eliminates this possible premature receptor binding, enabling apoE to accurately regulate its structure for an optimal apoE-receptor binding. This structural feature ensures proper folding and intracellular trafficking of apoE receptors in the cells and warrants that only the fully lipidated apoE binds to mature apoE receptors on the cell surface for an optimal lipoprotein transport and metabolism in circulation.
We recognize the speculative nature of our model for HDL and VLDL, as many important questions remain unanswered and require experimental investigations. Nonetheless, the apoE3 structure reported here demonstrates that the unique topology of human apoE regulates its biological functions, providing a structural framework for further studies of the diverse biological functions of apoE in several major human diseases including atherosclerosis, diabetes, and Alzheimer’s diseases.
Materials and Methods
NOE distance restraints were generated using 3D/4D Nuclear Overhauser Effect Spectroscopy (NOESY) experiments, including 3D 15N-edited NOESY and 4D-13C, 15N-edited NOESY using both uniformly and segmentally labeled apoE3 samples. Unique segmental labeling strategy was developed using a 2H, 15N-apoE-NT-13C, 15N-apoE-CT sample. The 3D 15N-edited NOESY spectrum of this unique NMR sample allowed us to unambiguously identify 78 interdomain NOEs between apoE3-NT amide protons and apoE3-CT aliphatic protons (39).
A total of 3,459 NOE derived distance restraints, 408 dihedral angle restraints, and 318 hydrogen bond restraints were used for structure calculation using the software package CYANA (40). Structure calculations were carried out in an iterative manner and each iteration generated and energy minimized 200 NMR structures in CYANA, including 10,000 steps of simulated annealing. The generated structures were analyzed for distance and dihedral angle restraint violations. A new restraint set was generated after these analyses for the next calculation. Final twenty best-fit NMR structures were used for further analysis. Further details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors would like to express sincere thank to Professor Robert Ryan for his valuable insights and suggestions during preparation of this manuscript. We also thank Ms Victoria Murray for critical reading of the manuscript. This work is supported by NIH R01 grants (HL074365 to J.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank www.pdb.org and BMRB- BioMagResBank, www.bmrb.wisc.edu (PDB code: 2L7B) (BMRB accession no. 15744).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106420108/-/DCSupplemental.
References
- 1.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 2.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res. 2009;50(Suppl):S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahley RW, Ji ZS. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- 4.Luc G, et al. Impact of apolipoprotein E polymorphism on lipoproteins and risk of myocardial infarction. The ECTIM Study. Arterioscler Thromb. 1994;14:1412–1419. doi: 10.1161/01.atv.14.9.1412. [DOI] [PubMed] [Google Scholar]
- 5.Mahley RW, Rall SC Jr, editors. The Metabolic and Molecular Base of Inherited Disease. 8 Ed. New York: (McGraw-Hill); 2001. pp. 2835–2862. [Google Scholar]
- 6.Burt TD, et al. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression. Proc Natl Acad Sci USA. 2008;105:8718–8723. doi: 10.1073/pnas.0803526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma SW, Benzie IF, Yeung VT. Type 2 diabetes mellitus and its renal complications in relation to apolipoprotein E gene polymorphism. Transl Res. 2008;152:134–142. doi: 10.1016/j.trsl.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 9.Wardell MR, Rall SC, Jr, Schaefer EJ, Kane JP, Weisgraber KH. Two apolipoprotein E5 variants illustrate the importance of the position of additional positive charge on receptor-binding activity. J Lipid Res. 1991;32:521–528. [PubMed] [Google Scholar]
- 10.Morrow JA, et al. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J Biol Chem. 2000;275:2576–2580. doi: 10.1074/jbc.275.4.2576. [DOI] [PubMed] [Google Scholar]
- 11.Wilson C, et al. Salt bridge relay triggers defective LDL receptor binding by a mutant apolipoprotein. Structure. 1994;2:713–718. doi: 10.1016/s0969-2126(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 12.Dong LM, et al. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol. 1996;3:718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 13.Dong LM, et al. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 14.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Chen J, Wang J. A complete backbone spectral assignment of lipid-free human apolipoprotein E (apoE) Biomol NMR Assigm. 2008;2:207–210. doi: 10.1007/s12104-008-9122-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Zhang Y, Cui C, Li Q, Wang J. An efficient on-column expressed protein ligation strategy: application to segmental triple labeling of human apolipoprotein E3. Protein Sci. 2008;17:736–747. doi: 10.1110/ps.073383708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J Biol Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 19.Sivashanmugam A, Wang J. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity. J Biol Chem. 2009;284:14657–14666. doi: 10.1074/jbc.M901012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wetterau JR, Aggerbeck LP, Rall SC, Jr, Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 21.Willard L, et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrow JA, et al. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J Biol Chem. 2002;277:50380–50385. doi: 10.1074/jbc.M204898200. [DOI] [PubMed] [Google Scholar]
- 23.Chroni A, et al. Biophysical analysis of progressive C-terminal truncations of human apolipoprotein E4: insights into secondary structure and unfolding properties. Biochemistry. 2008;47:9071–9080. doi: 10.1021/bi800469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry. 2009;48:3025–3032. doi: 10.1021/bi9000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Kypreos K, Zanni EE, Zannis V. Domains of apoE required for binding to apoE receptor 2 and to phospholipids: implications for the functions of apoE in the brain. Biochemistry. 2003;42:10406–10417. doi: 10.1021/bi027093c. [DOI] [PubMed] [Google Scholar]
- 26.Narayanaswami V, et al. Helix orientation of the functional domains in apolipoprotein E in discoidal high density lipoprotein particles. J Biol Chem. 2004;279:14273–14279. doi: 10.1074/jbc.M313318200. [DOI] [PubMed] [Google Scholar]
- 27.Narayanaswami V, Szeto SS, Ryan RO. Lipid association-induced N- and C-terminal domain reorganization in human apolipoprotein E3. J Biol Chem. 2001;276:37853–37860. doi: 10.1074/jbc.M102953200. [DOI] [PubMed] [Google Scholar]
- 28.Peters-Libeu CA, Newhouse Y, Hatters, DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J Biol Chem. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
- 29.Narita M, et al. Cellular catabolism of lipid poor apolipoprotein E via cell surface LDL receptor-related protein. J Biochem. 2002;132:743–749. doi: 10.1093/oxfordjournals.jbchem.a003282. [DOI] [PubMed] [Google Scholar]
- 30.Futamura M, et al. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J Biol Chem. 2005;280:5414–5422. doi: 10.1074/jbc.M411719200. [DOI] [PubMed] [Google Scholar]
- 31.Saito H, et al. Characterization of the heparin binding sites in human apolipoprotein E. J Biol Chem. 2003;278:14782–14787. doi: 10.1074/jbc.M213207200. [DOI] [PubMed] [Google Scholar]
- 32.Libeu CP, et al. New insights into the heparan sulfate proteoglycan-binding activity of apolipoprotein E. J Biol Chem. 2001;276:39138–39144. doi: 10.1074/jbc.M104746200. [DOI] [PubMed] [Google Scholar]
- 33.Herz J, Marschang P. Coaxing the LDL receptor family into the fold. Cell. 2003;112:289–292. doi: 10.1016/s0092-8674(03)00073-4. [DOI] [PubMed] [Google Scholar]
- 34.Innerarity TL, Friedlander EJ, Rall SC, Jr, Weisgraber KH, Mahley RW. The receptor-binding domain of human apolipoprotein E. Binding of apolipoprotein E fragments. J Biol Chem. 1983;258:12341–12347. [PubMed] [Google Scholar]
- 35.Bu G, Schwartz AL. RAP, a novel type of ER chaperone. Trends Cell Biol. 1998;8:272–276. doi: 10.1016/s0962-8924(98)01283-5. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz J, et al. The apoE isoform binding properties of the VLDL receptor reveal marked differences from LRP and the LDL receptor. J Lipid Res. 2005;46:1721–1731. doi: 10.1194/jlr.M500114-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 38.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Wang J. A segmental labeling strategy for unambiguous determination of domain-domain interactions of large multi-domain proteins. J Biomol NMR. 2011;50:403–410. doi: 10.1007/s10858-011-9526-0. [DOI] [PubMed] [Google Scholar]
- 40.Guntert P. Automated NMR structure calculation with CYANA. Method Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.