Abstract
A cryoelectron microscopy 8.5 Å resolution map of the 1,900 Å diameter, icosahedral, internally enveloped Paramecium bursaria chlorella virus was used to interpret structures of the virus at initial stages of cell infection. A fivefold averaged map demonstrated that two minor capsid proteins involved in stabilizing the capsid are missing in the vicinity of the unique vertex. Reconstruction of the virus in the presence of host chlorella cell walls established that the spike at the unique vertex initiates binding to the cell wall, which results in the enveloped nucleocapsid moving closer to the cell. This process is concurrent with the release of the internal viral membrane that was linked to the capsid by many copies of a viral membrane protein in the mature infectous virus. Simultaneously, part of the trisymmetrons around the unique vertex disassemble, probably in part because two minor capsid proteins are absent, causing Paramecium bursaria chlorella virus and the cellular contents to merge, possibly as a result of enzyme(s) within the spike assembly. This may be one of only a few recordings of successive stages of a virus while infecting a eukaryotic host in pseudoatomic detail in three dimensions.
Keywords: 3D structure, cell entry, conformation changes, minor proteins, PBCV-1
Paramecium bursaria chlorella virus (PBCV-1), a member of the Phycodnaviridae family (genus Chlorovirus), is a large, dsDNA virus (1, 2). Chlorella viruses are present in freshwater environments throughout the world, with titers as high as 100,000 infectious particles per mL of indigenous water. Phycodnaviruses, together with mimivirus, iridoviruses, asfarviruses, ascoviruses, and poxviruses have a common evolutionary ancestor and they are among the largest and most complex viruses known. Collectively these viruses are referred to as nucleocytoplasmic large DNA viruses (NCLDV) (3–8). With the exception of the poxviruses and ascoviruses, all of these viruses are roughly icosahedral in shape and have surfaces consisting of hexagonally closed packed arrays of capsomers organized into tri- and pentasymmetrons (SI Text). The NCLDV member with the most detailed structural information is probably Chilo iridescent virus (CIV), which has a diameter of about 1,850 Å (9). Analysis of CIV showed not only the organization of the major capsid proteins in the capsomers, but also recognized some minor capsid proteins between the outer protein capsid and an inner membrane surrounding the nucleocapsid core. Similar stabilizing proteins were identified in the dsDNA bacteriophage PRD1 (10, 11).
PBCV-1 has a 330-kbp genome containing 365 nonoverlapping genes (2) that also code for 11 tRNAs. A total of 148 virus encoded gene products have been detected in mature PBCV-1 virions. A 24 Å resolution map determined by cryoelectron microscopy (cryoEM) image reconstruction confirmed that PBCV-1 has a lipid bilayer membrane (12, 13) surrounded by an outer glycoprotein capsid with approximate icosahedral symmetry (14). The diameter of PBCV-1 virions is 1,900 Å measured along fivefold symmetry axes and about 1,660 Å measured along threefold symmetry axes. The surface of the virions consists of close packed arrays of pseudohexagonal capsomers, roughly consistent with the prediction of icosahedral virus surfaces (15) (Fig. 1A). The tri- and pentasymmetrons are separated by cleavage planes created by differences in capsomer orientation on either side of the fault lines (3) as first observed for Sericesthis iridescent virus (16) and later in other NCLDVs (3, 14) (Fig. 1A). The atomic structure of the PBCV-1 major capsid protein, Vp54, determined by X-ray crystallography (17) consists of two consecutive eight-stranded, antiparallel, β-barrell, “jelly-roll” folds. The structure fits well into the cryoEM density map of PBCV-1. Using fivefold (as opposed to icosahedral) symmetry in calculating the cryoEM image reconstruction Cherrier et al. (18) showed that there exists a unique fivefold vertex with a spike structure extending outward. Underneath the spike there is a pocket between the capsid and the internal membrane presumably enveloping the nucleocapsid composed of the genome and other proteins. A set of “finger proteins” (named because of their shape) was identified in the fivefold cryoEM reconstruction (18) below the five trisymmetrons associated with the unique vertex.
Fig. 1.
The organization of minor proteins associated with a trisymmetron. (A) Structure of trisymmetrons and pentasymmetrons on the PBCV-1 icosahedral surface. (B) The blue densities represent the membrane proteins (MP); the red densities represent the long proteins; the yellow sausage-like densities represent the missing 24 amino acids of Vp54 (shown only within one trisymmetron); and white represents uninterpreted densities. Three red arrows point to the fibered capsomers in icosahedral reconstruction. However, the fivefold averaged reconstruction showed that only one of these capsomers is fibered. (C) One section of the icosahedrally reconstructed map shows densities connecting the capsid with the inner membrane. These densities are located at the boundaries between neighboring trisymmetrons.
PBCV-1 infects the unicellular, eukaryotic green alga Chlorella variabilis NC64A (19). The initial recognition and attachment process is probably complete in less than 1 min after mixing virus and algae (20). PBCV-1 protein Vp130 (A140/145R) is required for virus attachment (21). Antibody recognition of Vp130 indicates this protein is located near the viral surface (22). The inability of anti-Vp130 antibodies to bind to more than one site on the virion suggests that Vp130 might be located at the unique vertex. The chemical nature of the host receptor for PBCV-1 attachment is unknown although circumstantial evidence suggests it is a carbohydrate because PBCV-1 can attach to isolated cell walls treated with proteases and/or extracted with phenol to the same extent as healthy cells (23). After attachment to the host cell wall, a viral packaged enzyme(s) digests the cell wall around the binding site of the virus (24). Subsequently, viral DNA as well as some proteins are ejected into the cell cytoplasm, leaving an empty virus capsid on the cell wall (24, 25).
Here we have extended the resolution of the PBCV-1 structure to 8.5 Å and have identified a number of minor structural proteins that constitute the capsid. Some of these minor proteins were systematically missing from the internal surfaces closest to the unique vertex. Furthermore, the virus was studied by cryoEM while attaching to the cell walls of its host, allowing visualization of the initial conformational changes that occur during infection. This is one of the most detailed three-dimensional studies of a virus during successive stages of a virus infecting a eukaryotic host.
Results and Discussion
The Missing 24 Amino Acids in the X-ray Crystal Structure of the Major Capsid Protein (MCP) Vp54.
The X-ray crystallographic atomic structure of the Vp54 trimer, including the carbohydrate entities observed crystallographically, was fit into the 8.5 Å resolution, 66-fold averaged, electron density map of the capsomer using the EMfit program (26). Some secondary structural elements, such as three short α-helices of Vp54, were readily identified. A “difference” map was calculated by setting all density values to zero that were within 4.0 Å from any Cα atom. There were three large contiguous volumes (Fig. S1A) of densities in the asymmetric unit of the difference map (one Vp54 molecule) in regions that were on the outside of the virus particle, extending the region occupied by the 20 carbohydrate residues found in the crystallographic determination (17) to approximately 27 residues (based on the volume).
In addition to the external difference density, there is a “sausage-like” density, about 33 Å long and 10 Å wide, per asymmetric unit on the internal side of the capsomer (Fig. S1B). This feature is visible not only in the 8.5 Å resolution map of a single averaged capsomer, but is also easily seen in most of the capsomers in the 9.7 Å resolution icosahedral averaged map of the entire virus. One end of this density is near the threefold axis close to the N terminus of Vp54 and the other end of the density extends to the edge of the averaged capsomer. The diameter of the sausage-like density is similar to the densities representing α-helices in the averaged map. If this density were an α-helix it would represent about six or seven turns amounting to 22–25 amino acids. The first 24 amino acids of Vp54 were either disordered or missing in the crystal structure (17). Thus, the sausage-like density probably corresponds to the 24 amino-terminal disordered residues in the crystal structure. The N terminus of the 24 missing amino acids is close to a minor capsid protein (see below) (Fig. 1B). Therefore, the amino-terminal 24 amino acids of Vp54 are apparently ordered in the virus because of their association with a minor capsid protein (10). The Vp54 double jelly-roll structure is similar to the structure of the capsid protein P3 in the lipid enveloped bacteriophage PRD1, with approximately 59% of the Vp54 Cα atoms being superimposable onto the structure of P3 (27, 28). It is therefore noteworthy that P3 also has a 30 amino acid N-terminal α-helix.
Minor Capsid Proteins.
After setting the density corresponding to all of the 1,680 trimeric capsomers in the virus to zero, a series of densities remain that has the same positional periodicity as the capsomers. These densities are presumably minor capsid proteins as occur in viruses CIV (9) and PRD1 (10, 11). However, the minor capsid proteins lack icosahedral symmetry. For instance, the finger protein (18) only occurs near the unique vertex, whereas other minor proteins occur at all icosahedrally related positions except around the unique vertex.
A row of five roughly parallel “long protein” densities can be recognized in the icosahedral difference map. These proteins form a hexagonal network over the internal face of the trisymmetrons (Fig. 1B) following the peripheries of the MCP capsomers. They have some similarity to the long glue proteins between capsomers of the PRD1 bacteriophage (10, 11) and adenovirus (29, 30) (Fig. S2A). The finger proteins can only be recognized on the fivefold averaged map and are seen only in the five trisymmetrons around the unique vertex. Furthermore, one row of five long proteins, closest to the unique pentasymmetron, is absent in these five trisymmetrons. No capsomers are associated with both a finger protein and a long protein (Fig. 2). Analysis of the volume of the averaged long proteins indicates that these proteins have a molecular weight of about 32 kDa when standardized against the observed volume of the Vp54 capsomer.
Fig. 2.
Diagrammatic organization of minor capsid proteins near the unique vertex viewed from outside of the virus. Each colored dot within each hexameric capsomer represents a double jelly-roll structure. Lighter colors are used in the neighboring trisymmetron. The spike at the unique fivefold vertex is shown in bright red. One of the other 11 local fivefold axes is indicated by a black pentagonal mark. There is only one fibered capsomer per trisymmetron positioned as shown. There is a second fibered capsomer in trisymmetrons not associated with the unique vertex. The occupancies of the various minor capsid proteins at the sites shown on this diagram is probably somewhat different from site-to-site and virion-to-virion. Black dotted lines shows the approximate limit of the dissociation of the virus when bound to isolated cell walls.
In addition to the hexagonal network of long proteins is a series of nine regularly repeated “membrane protein” (MP) densities located along the edge of the trisymmetrons and connected to the internal membrane (Fig. 1C). The distance between these MP densities is about the same as the distance between capsomers. Seven of these densities form a set of regular bridges across the trisymmetron boundary (Fig. 1B). The presence of the regularly spaced quasi-twofold axes relating the quasi-sixfold capsomers in one trisymmetron to the capsomers in the neighboring trisymmetron also relate the MPs along a trisymmetron edge to the MPs along the neighboring trisymmetron edge, suggesting that the MPs form a series of dimers between trisymmetrons. Thus, the dimeric minor MP binds to dimeric associated capsomers, which explains why the dimer cannot bind elsewhere to a trisymmetron in which the capsomers are all similarly oriented. The averaged MP dimer is about 120 Å in length and has lower density connecting it to the membrane component (Fig. S2B). Based on the averaged density volume, the molecular weight of the MP dimer external to the membrane is about 28 kDa.
A similar dimeric “Zip dimer” between trisymmetrons occurs in CIV, except that in CIV this is probably not a membrane protein (9). The fivefold averaged difference map, as opposed to the icosahedrally averaged difference map shows that the MP dimers are missing at the five trisymmetrons surrounding the unique vertex (Fig. 2). Indeed, the MP is too small to bridge the gap across the pocket between the viral membrane and the capsid at the unique vertex. The absence of interconnecting MP dimers linking trisymmetrons surrounding the unique vertex suggests that there might be a weakness in the capsid structure around this unique vertex, a feature that might be important during initial stages of viral entry. Similarly, the five icosahedral faces around the unique vertex of Mimivirus open up when the virus infects an amoeba (31).
Of the 148 virus encoded gene products in mature PBCV-1 that had been predicted to have membrane components, there were only three (A168R, A213L, A605L) that had a molecular weight in the range from 13 to 15 kDa (omitting the probable transmembrane component), consistent with the observed 14 kDa derived from the volume calculations. Of these proteins, A213L has the largest number of copies in the viron based on the exponentially modified protein abundance index in agreement with the large number of MP found in the PBCV-1 structure. Furthermore, A168R and A605L are predicted to be signal peptides (32, 33) and therefore would not be the MP identified structurally, leaving A213L as the most probable gene for MP.
Fivefold Unique Features.
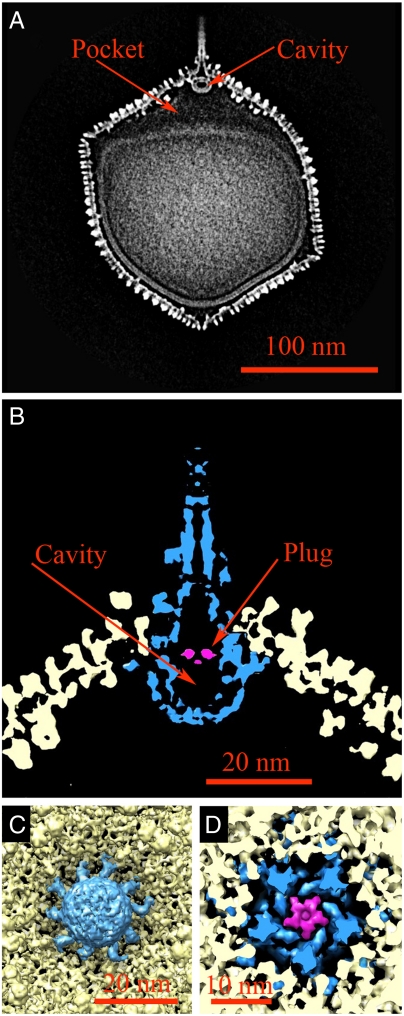
The PBCV-1 spike structure is a complex assembly inserted at the unique vertex (Fig. 3 A and B). Its total length is about 560 Å with about 340 Å protruding from the surface of the virus. The part of the spike structure that is outside the capsid has an external diameter of about 35 Å at the tip expanding to about 70 Å at the base. From the base on the viral surface, the spike structure widens to about 160 Å inside the capsid and forms a closed cavity inside the large pocket between the capsid and the membrane under the unique vertex. The spike assembly is held in place by the five peripentonal capsomers (18). The component of the spike assembly inside the viral capsid was previously described (18) as being about the same size and shape as the portal protein of tailed bacteriophages such as phi29 (34), SPP1 (35), and P22 (36, 37). However, the wide basal end of the PBCV-1 spike assembly encloses a roughly spherically closed cavity (Fig. 3A). This small spherical cavity has a plug on the side facing the spike whose diameter is about 62 Å (Fig. 3D). Apart from the fivefold symmetry imposed on the reconstruction, the additional small spherical cavity has pseudo-10-fold symmetry (Fig. 3C). If the spike assembly had sixfold symmetry as occurs in portal proteins of tailed phages, it would be unlikely that application of fivefold symmetry (used here) would yield any obvious structural features. Thus, the fivefold symmetric spike assembly differs from the portal proteins of tailed phages. There is a channel in the spike assembly that has an internal diameter of about 20 Å, starting at about 230 Å from the tip, leading to the plug covering the small internal spherical cavity (Fig. 3B). The narrowness of the internal diameter means that, unlike the tails of dsDNA phages, the spike is unlikely to serve as the vehicle for DNA delivery to the host cell. However, based on the ability of some small proteins to pass through apertures of this size (38–40), the PBCV-1 spike might deliver enzymes to digest the cell wall of the host to initiate infection. The primary function of the spherical cavity within the spike assembly could be to store the wall degrading enzyme(s) so that they are ready for ejection when the virus is in contact with the host. Other enzymes might be in the pocket between the membrane and spike assembly.
Fig. 3.
The PBCV-1 spike structure. (A) Central section of the fivefold symmetric reconstruction. (B) A magnified image of the spike structure. The spike assembly is colored in blue. (C) The cavity structure viewed from inside the virus shows it has pseudo-10-fold symmetry with 10 arms anchored to the capsid at the unique vertex on the inside of the viral capsid. (D) The plug of the cavity structure as seen from outside the virus is shown in pink.
A previous study indicated that each of the 20 trisymmetrons have one instead of three capsomers with a central fiber (18). This improved fivefold symmetric reconstruction indicates that the fiber has a 60 Å-long radial component followed by a 90° turn and at least another 130 Å component (Fig. S3A). Based on some single images, the fiber is probably longer than what is visible in the fivefold averaged map. These fibers have also been seen in an atomic force microscopy study (41) and in a quick freeze and deep etched microscopy study (25). The correlation coefficient between the averaged Vp54 trimer, from the icosahedrally averaged reconstruction, and any of the general capsomers from the fivefold averaged map was approximately 0.71, but only approximately 0.44 when correlated with the fibered capsomer (excluding the volume known to be carbohydrate moieties). Furthermore, the difference map in which Vp54 was subtracted indicates that the capsomers with fibers do not have the same sugar density at the surface glycosylation sites, nor do the fibered capsomers have a density for the first 24 amino acids that are ordered in the other capsomers but not in the crystal structure (see above) (Fig. 1B and Fig. S3B). Thus, the fibered capsomers are presumably constructed of a different, but homologous, protein than the Vp54 capsomers, probably corresponding to one of the gene products that have sequences similar to Vp54.
An estimate of the number of minor capsid proteins is given in Table S1. These numbers should be compared with the number of 1,680 hexameric capsomers in the virus, which would imply that there are 5,040 copies of the Vp54 monomer. However, this number is an overestimation because of the special capsomers such as the fibered and peripentonal capsomers.
Initial Events Associated with Infection.
To explore the function of the PBCV-1 structures in the initial stages of cell infection, CCD cryoEM images were recorded of PBCV-1 attached to isolated C. variabilis NC64A cell walls. For most virus–host interactions it is difficult to have confidence in electron micrographs because most of the particles are noninfectious. However, for PBCV-1 the ratio of infectious virus to total virus is approximately 30% (42), which is higher than for most viruses and therefore validates electron micrographic observations for attachment and wall digestion.
The virus was seen in various stages of gaining entry into the cell. Some particles were excluded from the analysis because either they were far away from the cell wall or on top of the cell wall (Fig. 4A). Other particles were less than 450 Å from the cell wall (measured to the position of highest contrast), but their unique vertex could still be identified by the pocket under the spike (red arrows in Fig. 4A). Of these particles, 93 out of 94 had their unique vertex oriented toward the cell wall. Of these 93 particles, 51 did not overlap with other particles. These 51 particles were used to calculate a three-dimensional fivefold symmetric reconstruction. The orientation of these particles was determined by comparison with the three-dimensional PBCV-1 model. This reconstruction was notable in that the tip of the unique spike was significantly larger (90 Å diameter) and denser (Fig. 4 B and C) than the tip of the spike (35 Å diameter) observed in the reconstruction using the in vitro prepared virions (Fig. 3A and Fig. S3A). To ascertain that the unmistakable larger spike tip did not result from the small number of particles used in the reconstruction, 51 randomly selected particles from the 17,016 in vitro particle database were selected and used in a reconstruction. The result gave a poorly defined spike that did not show any enlargement of its tip. These results suggest that a conformational change occurs in PBCV-1’s structure when it initially comes in contact and recognizes the receptor on the host cell wall.
Fig. 4.
Initial attachment to C. variabilis NC64A cell walls by PBCV-1. (A) The cryoEM CCD image shows some virus particles attached to the cell wall. The virus particles marked with white arrows are either too far away from the cell wall or overlap with the cell wall in the projection to be considered as definitely having recognized the cell wall. The orientation of the unique vertex (recognizable by the pocket under the vertex) can be seen for the virus particles marked with red arrows. These virus particles and the majority of similar particles in other micrographs have their unique vertex facing the cell wall indicating that these particles recognized the cell surface. All such particles that were not overlapped with neighboring particles were included in the reconstruction shown in panels B and C. The virus particles marked by black arrows have approached the cell wall such that the unique vertex is no longer recognized and the virion has presumably partially disassembled. Some of the cell wall appears to be digested in the vicinity of the contact area. Furthermore, about 135 capsomers around the unique vertex (see dotted line in Fig. 3) were not recognizable in the contact region. In addition, the viral membrane changes from having a roughly icosahedral to a spherical shape. Such particles were included in the reconstruction shown in panels D and E. (B) and (D) The external surface of the reconstructions. (C) and (E) A central cross-section through the reconstructions.
Other particles had progressed further in the infection process by attaching to the cell wall. These particles appeared to have fused with the cell and, hence, the pocket marking the unique vertex could not be recognized (black arrows in Fig. 4A). A three-dimensional reconstruction of particles actually attached to the cell wall was computed by selecting 42 particles that were fused with the cell wall. The spike and the associated capsid proteins were no longer recognized in this reconstruction. In addition, the membrane that formed the base of the pocket shifted to replace the space previously occupied by the viral pocket, whereas the rest of the membrane envelope became more spherical (Fig. 4 D and E).
Apparently the cell wall and the viral capsid close to the site of attachment had been digested and disassembled, respectively. Considering the size of the approximately 800 Å diameter hole created in the cell wall, it is likely that an enzyme(s), stored in the spherical cavity of the spike assembly or less likely in the pocket around the unique vertex, is ejected through the central channel of the spike to digest the cell wall. Approximately half of each of the five trisymmtrons surrounding the attachment site were not seen in the reconstruction. Considering the absence of MPs and the missing long minor capsid proteins in the vicinity of the unique vertex (see above), the surrounding trisymmetrons may have reduced stability, allowing them to partially disassemble, presenting the viral membrane to the membrane of the cell.
As mentioned above, PBCV-1 has one fiber attached to a special capsomer in each trisymmetron. The reconstruction indicates that this fiber is bent and at least 190 Å long, although it is likely to be flexible and, hence, even longer, but not visible on an averaged reconstruction. Furthermore, longer fibers attached to the surface of the virus can be seen occasionally on raw images of PBCV-1 (25, 41). Indeed, these images show that some of these special fibers might be anchoring the virus to the cell wall. If the fibers are responsible for PBCV-1 initially recognizing the host cell, then the orientation of the unique spike relative to the cell surface would be random. Because this does not occur, it is more likely that the spike provides the first contact with the cell and that the fibers then aid in holding the virus to the cell wall once the spike has been jettisoned. Similarly, tail fibers of some tailed bacteriophages are required for binding of phage to a host bacterium (43, 44).
The MP that attaches the viral membrane with the capsid causes the membrane to have an icosahedral shape. However, in particles that have attached to host walls, the virus membrane has a sphere-like shape in which the membrane is separated from the capsid at all fivefold vertices (Fig. 4E). Consequently, the MP has either been cleaved during release of the cell wall digesting enzyme(s) after attachment, been extracted from the membrane or become disattached from the capsid. Thus, on initial recognition of the host cell, the viral membrane extends toward the site of attachment, displacing the contents of the pocket region while moving away from the other fivefold vertices, presumably leaving the volume enclosed by the membrane approximately unchanged. The digestion of the cell wall, the release and extension of the virus membrane to the site of attachment and the partial disassembly of the capsid preceed infection by fusion of the viral membrane with the host cell membrane.
Materials and Methods
PBCV-1 was grown and purified as described previously (42). Chlorella variabilis NC64A cell walls were isolated by disrupting cells with sonication followed by vortexing in a Genie Disruptor with 0.3 mm glass beads for 15 min at 4 °C. Following centrifugation, the wall pellet was extracted with Tris-buffered phenol to remove proteins followed by a wash with toluene. The wall preparation was then washed several times with 50 mM sodium acetate, pH 6.5 buffer by resuspending and centrifuging. The pellet was suspended in the sodium acetate buffer and incubated overnight at 37 °C with 8 units per mL of α-amylase (Sigma, Aspergillus oryaae) to digest residual starch. The sample was then heated to 100 °C for 90 s, cooled, centrifuged and the pellet was incubated with 5% SDS at 40 °C for 30 min. The walls were subjected to five washes in sterile distilled water and centrifugations and the final pellet was suspended in sterile distilled water.
PBCV-1 particles or isolated cell walls incubated with PBCV-1 in Bold’s basal medium (45) for 10 min at room temperature were frozen using the Gatan Cryoplunge3 device. Grids were blotted for 5 s before plunging into ethane at 100 K while the humidity was greater then 85%. CryoEM images were collected on Kodak films (Kodak Electron Image Film SO-163) using a CM300 FEI electron microscope with approximately 20 electrons/Å2 dose. Images were recorded using a magnification of 33,000 with a defocus range from 1.5 to 4 μm. Films were digitized with a Nikon scanner (Super Coolscan9000) at 6.35 μm/pixel, corresponding to 1.92 Å/pixel at the sample.
A total of 17,016 particles were selected and boxed using the e2boxer software from the EMAN2 system (46). These particles were selected from about 800 micrographs that had Thon rings extending beyond 9 Å resolution. The initial model was created by program starticos from the EMAN system (47) and refined to 9.7 Å based on the point where the Fourier shell correlation fell below 0.5 between maps computed from the odd numbered and even numbered particles (48).
The atomic structure of Vp54 was fitted into the cryoEM density of the icosahedrally reconstructed map using the program EMfit (26). An averaged map of the 22 independent capsomers within the asymmetric unit of a trisymmetron was calculated assuming the initial translation and orientation matrix from EMfit using the program AVE (49). The 22 rotational and translational matrices were refined by aligning each individual capsomer in the cryoEM map with the averaged map using the program IMP (49). These refined matrices were used to calculate a new averaged map. This iterative process was repeated five times. The orientation and position of the threefold axis in the refined average capsomer was similarly determined by again using the program IMP to generate a final 66-fold averaged map. The resolution of this map was 8.5 Å based on applying the refined matrices separately onto the odd numbered and even numbered particles and then calculating the Fourier shell correlation between the two maps using the same criterion as given above.
To obtain a fivefold average reconstruction that included the entire spike at the unique vertex, the particles were reboxed with a larger box size with 700 instead of 600 pixels where the pixel separation was 3.84 Å. The center of the particle was retained at the center of the box. To obtain a unique orientation of one vertex, a model of PBCV-1 was constructed that had a pocket in the vicinity of one fivefold vertex, corresponding to the observation of numerous single cryoEM particles. This model was then compared with approximately 200 particle images that showed a recognizable pocket. This model was projected using the 60 icosahedrally related angular orientations found for each of these images in the previous icosahedral reconstruction. Of these 60 projections there were five identical results that were seen to bring the orientation of the pocket in the projected model into the same orientation as the pocket in the image. The resultant reconstruction based on these 200 particles was then used to identify the unique fivefold orientation of all the other 16,816 particles. The resultant reconstruction was then used for selecting orientations in the next iteration. The procedure converged after only two cycles.
Supplementary Material
Acknowledgments.
We thank Jim Gurnon for growing PBCV-1 and for isolating the Chlorella variabilis NC64A cell walls. We are grateful to Siyang Sun and Pavel Plevka for many insightful discussions. We thank Sheryl Kelly for help in the preparation of the manuscript. The work was supported by National Institutes of Health (NIH) Grant R01 AI11219 (M.G.R.), National Science Foundation-Experimental Program to Stimulate Competitive Research Grant EPS-1004094 (J.L.V.E.) and NIH/National Center for Research Resources Grant P30 RR031151-01 (J.L.V.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107847108/-/DCSupplemental.
References
- 1.Van Etten JL. Unusual life style of giant chlorella viruses. Annu Rev Genet. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 2.Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Adv Virus Res. 2006;66:293–336. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr D Biol Crystallogr. 2003;59:2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 4.Hawes PC, Netherton CL, Wileman TE, Monaghan P. The envelope of intracellular African swine fever virus is composed of a single lipid bilayer. J Virol. 2008;82:7905–7912. doi: 10.1128/JVI.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao C, et al. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Dunigan DD, Fitzgerald LA, Van Etten JL. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 9.Yan X, et al. The capsid proteins of a large, icosahedral dsDNA virus. J Mol Biol. 2009;385:1287–1299. doi: 10.1016/j.jmb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuma R, Bamford JKH, Bamford DH, Russell MP, Thomas GJ. Structure, interactions and dynamics of PRD1 virus. I. Coupling of subunit folding and capsid assembly. J Mol Biol. 1996;257:87–101. doi: 10.1006/jmbi.1996.0149. [DOI] [PubMed] [Google Scholar]
- 11.San Martín C, et al. Minor proteins, mobile arms, and membrane–capsid interactions in the bacteriophage PRD1 capsid. Nat Struct Biol. 2002;9:756–763. doi: 10.1038/nsb837. [DOI] [PubMed] [Google Scholar]
- 12.Skrdla MP, Burbank DE, Xia Y, Meints RH, Van Etten JL. Structural proteins and lipids in a virus, PBCV-1, which replicates in a Chlorella-like alga. Virology. 1984;135:308–315. doi: 10.1016/0042-6822(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 13.Meints RH, Lee K, Van Etten JL. Assembly site of the virus PBCV-1 in a Chlorella-like green alga: Ultrastructural studies. Virology. 1986;154:240–245. doi: 10.1016/0042-6822(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 14.Yan X, et al. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Mol Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 16.Wrigley NG. An electron microscope study of the structure of Sericesthis iridescent virus. J Gen Virol. 1969;5:123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- 17.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherrier MV, et al. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc Natl Acad Sci USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L. The systematics of Zoochlorella revisited employing an integrative approach. Environ Microbiol. 2011;13:350–364. doi: 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- 20.Thiel G, Moroni A, Dunigan D, Van Etten JL. Initial events associated with virus PBCV-1 infection of Chlorella NC64A. Prog Bot. 2010;71:169–183. doi: 10.1007/978-3-642-02167-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onimatsu H, Sugimoto I, Fujie M, Usami S, Yamada T. Vp130, a chloroviral surface protein that interacts with the host Chlorella cell wall. Virology. 2004;319:71–80. doi: 10.1016/j.virol.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Onimatsu H, Suganuma K, Uenoyama S, Yamada T. C-terminal repetitive motifs in Vp130 present at the unique vertex of the chlorovirus capsid are essential for binding to the host chlorella cell wall. Virology. 2006;353:433–442. doi: 10.1016/j.virol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Meints RH, Burbank DE, Van Etten JL, Lamport DTA. Properties of the Chlorella receptor for the virus PBCV-1. Virology. 1988;164:15–21. doi: 10.1016/0042-6822(88)90614-9. [DOI] [PubMed] [Google Scholar]
- 24.Meints RH, Lee K, Burbank DE, Van Etten JL. Infection of a Chlorella-like alga with the virus, PBCV-1: Ultrastructural studies. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- 25.Van Etten JL, Lane LC, Meints RH. Viruses and virus-like particles of eukaryotic algae. Microbiol Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossmann MG, Bernal R, Pletnev SV. Combining electron microscopic with X-ray crystallographic structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 27.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 28.San Martín C, et al. Combined EM/X-Ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9:917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 29.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 Å resolution. Science. 2010;329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zauberman N, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 33.Songsri P, Hiramatsu S, Fujie M, Yamada T. Proteolytic processing of Chlorella virus CVK2 capsid proteins. Virology. 1997;227:252–254. doi: 10.1006/viro.1996.8298. [DOI] [PubMed] [Google Scholar]
- 34.Simpson AA, et al. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebedev AA, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 38.Menetret JF, et al. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 39.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 40.Yip CK, Strynadka NC. New structural insights into the bacterial type III secretion system. Trends Biochem Sci. 2006;31:223–230. doi: 10.1016/j.tibs.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Kuznetsov YG, Gurnon JR, Van Etten JL, McPherson A. Atomic force microscopy investigation of a chlorella virus, PBCV-1. J Struct Biol. 2005;149:256–263. doi: 10.1016/j.jsb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Van Etten JL, Burbank DE, Xia Y, Meints RH. Growth cycle of a virus, PBCV-1, that infects Chlorella-like algae. Virology. 1983;126:117–125. doi: 10.1016/0042-6822(83)90466-x. [DOI] [PubMed] [Google Scholar]
- 43.Simon LD, Anderson TF. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology. 1967;32:279–297. doi: 10.1016/0042-6822(67)90277-2. [DOI] [PubMed] [Google Scholar]
- 44.Xiang Y, et al. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell. 2009;34:375–386. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nichols HW, Bold HC. Trichosarcina polymorpha Gen. et sp. Nov. J Phycol. 1965;1:34–38. [Google Scholar]
- 46.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 48.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 49.Jones TA. A, yaap, asap, @#*? A set of averaging programs. In: Dodson E, Gover S, Wolf W, editors. Molecular Replacement Proceedings of the CCP4 Study Weekend, 31 January–1 February 1992; Daresbury Laboratory, Warrington, United Kingdom: Science and Engineering Research Council; 1992. pp. 91–105. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.