Abstract
Initiation of the innate immune response requires agonist recognition by pathogen-recognition receptors such as the Toll-like receptors (TLRs). Toll/interleukin-1 receptor (TIR) domain-containing adaptors are critical in orchestrating the signal transduction pathways after TLR and interleukin-1 receptor activation. Myeloid differentiation primary response gene 88 (MyD88) adaptor-like (MAL)/TIR domain-containing adaptor protein (TIRAP) is involved in bridging MyD88 to TLR2 and TLR4 in response to bacterial infection. Genetic studies have associated a number of unique single-nucleotide polymorphisms in MAL with protection against invasive microbial infection, but a molecular understanding has been hampered by a lack of structural information. The present study describes the crystal structure of MAL TIR domain. Significant structural differences exist in the overall fold of MAL compared with other TIR domain structures: A sequence motif comprising a β-strand in other TIR domains instead corresponds to a long loop, placing the functionally important “BB loop” proline motif in a unique surface position in MAL. The structure suggests possible dimerization and MyD88-interacting interfaces, and we confirm the key interface residues by coimmunoprecipitation using site-directed mutants. Jointly, our results provide a molecular and structural basis for the role of MAL in TLR signaling and disease protection.
Keywords: innate immunity, protein--protein interactions, X-ray crystallography
Toll-like receptors (TLRs) are a family of single-pass transmembrane proteins that mediate innate immune responses to microbial stimuli such as bacterial lipids and nonself nucleic acids (1, 2). These molecules bind directly to the extracellular domains of the receptors and induce oligomerization, bringing the juxtamembrane sequences into close proximity and leading to the dimerization of the cytosolic Toll/interleukin-1 receptor (TIR) domains (3, 4). This activated conformation of the receptor provides a scaffold for the recruitment of downstream signal transducers into a postreceptor complex that causes activation of transcription factors such as NF-κB, p38 MAP kinase, and the IFN response factors.
The 10 human TLRs use five different signaling adaptor proteins that, like the receptors, contain TIR domains (5). A TIR domain has a globular fold with α/β secondary structure elements linked together by variable loops. The homo- and heterotypic TIR–TIR domain interactions play a pivotal role in the assembly of the TLR signalosomes and in initiation of the signaling pathway, with the loops providing the specificity of interactions (6). Adaptor TIR domains are proposed to bind at two symmetry-related sites formed upon receptor activation (7). The myeloid differentiation primary response gene 88 (MyD88) adaptor is used by all TLRs, except TLR3, and by the interleukin-1 family of inflammatory cytokine receptors (8). It is a bipartite molecule with a death domain in addition to a TIR domain. MyD88 death domains assemble into a highly oligomeric structure called the Myddosome (9). The other adaptor proteins fulfill more specialized roles. TIR domain-containing adaptor-inducing IFN-β (TRIF) couples directly to TLR3, leading to antiviral responses. The most complex adaptor use is displayed by TLR4. Activation of TLR4 by lipopolysaccharide, a complex glycolipid from the outer membrane of Gram-negative bacteria, leads to initial recruitment of either TRIF-related adaptor molecule (TRAM) or MAL/TIR domain-containing adaptor protein (TIRAP). These proteins are thought to function as “bridging” adaptors for MyD88 and TRIF, which transduce TLR4 signals originating from the cell surface or from the endosomal compartment, respectively (10).
Crystal structures for the TIR domains from TLR1, TLR2, TLR10, interleukin-1 receptor accessory protein-like (IL-1RAPL), a protein from Paracoccus denitrificans (PdTIR), a protein from Arabidopsis (AtTIR), and the flax resistance protein L6 (11–16), as well as the solution NMR structure of the TIR domain from MyD88 (17), have been determined. Although there is a lack of structural and biophysical information on the heterotypic TIR–TIR domain interactions, the lattice interfaces present in crystal structures, together with mutagenesis data, have yielded insights into homotypic TIR–TIR domain interactions, which have been extended to the structural modeling of TLR signalosome complexes. These studies have identified a number of conserved loop regions, including the BB loop, that are important in mediating complex assembly (7, 18–22).
The structure and function of the MAL adaptor is of particular interest because it is subject to multiple direct and indirect regulatory inputs. MAL associates constitutively with the plasma membrane through a phosphatidylinositol-4,5-bisphosphate (PIP2)-binding motif at the N terminus of the protein, an interaction essential for its function (23). Phosphorylation by Bruton's tyrosine kinase (Btk) leads to recruitment of suppressor of cytokine signaling 1 (SOCS1) and MAL degradation (24, 25), and phosphorylation by interleukin-1 receptor-associated kinase (IRAK)-1 and IRAK-4 promotes MAL ubiquitination and degradation (26). MAL function is also regulated by the cysteine protease caspase-1, which cleaves the protein in a region of the molecule that interacts with MyD88 (27, 28). The motif comprising residues 183–193 binds to the E3 ubiquitin ligase TNF receptor-associated factor 6 (TRAF6), which is a downstream transducer of the TLR pathway and is required for activation of NF-κB (29). Binding of TRAF6 to MAL appears to fulfill a secondary role in transactivation by inducing phosphorylation of the p65 NF-κB.
MyD88 binding and signaling are also impaired in a rare MAL polymorphism, D96N. The importance of the MAL adaptor for host defense is further emphasized by the properties of a common S180L human polymorphism. Individuals heterozygous for the serine and leucine forms are protected in malaria, tuberculosis, and pneumococcal pneumonia infections (30), but individuals homozygous for the leucine form have an increased risk of sepsis (31) and fail to respond to the Haemophilus influenzae serotype b (Hib) vaccine (32). It is likely that vaccine failure and sepsis occur because of an inability to mount a response to the vaccine or to the invading bacteria, respectively. Thus, subtle changes in MAL structure can have large effects on the development of protective immune responses.
This article describes the crystal structure of human MAL adaptor, revealing unexpected structural features and functional interfaces involved in homodimerization and interaction with MyD88, which we validate through site-directed mutagenesis.
Results
Crystal Structure of MAL TIR Domain.
We used a high-throughput recombinant cloning and expression methodology to identify stable constructs of human MAL TIR domain suitable for large-scale production and crystallization. We tested one of these constructs (lacking 29 N-terminal residues; MALΔ29) functionally, demonstrating that it retains the functional properties of full-length MAL except, as expected, plasma membrane localization (SI Appendix). The construct comprising residues 79–221 (herein termed MAL-TIR) eventually yielded the most useful crystals that diffracted to 3-Å resolution and allowed the structure to be determined (SI Appendix). The structure reveals a single molecule in the asymmetric unit composed of a TIR domain fold containing a five-stranded parallel β-sheet (βA–βE) surrounded by four α-helices (αA and αC–αE) (Fig. 1; the naming convention follows that in ref. 12). The MAL-TIR fold differs from other TIR domain structures because it lacks a helical segment (αB) between the βB- and βC-strands and instead contains a long loop (AB) connecting the first helix (αA) and the βB-strand (SI Appendix). Most of the residues comprising this loop were not visible in the electron density, presumably because of its inherent flexibility. Remarkably, however, the AB loop sequence shows significant similarity with the functionally important BB loop segment in other TIR domains, suggesting that a structural rearrangement has occurred in MAL despite the sequence retaining BB loop features (SI Appendix).
Fig. 1.
Crystal structure of human MAL-TIR. This cartoon representation of the structure shows the key secondary structure elements. The cysteine residues are shown in ball-and-stick representation, with the DTT adduct omitted for clarity.
Unexpectedly, the fold is further stabilized by two disulfide bonds (residues C89–C134 and C142–C174) (Fig. 1). The other two cysteine residues, C91 and C157, are 4.8 Å apart but displayed continuous electron density between the two thiol groups that was modeled as a single molecule of DTT, which was present throughout protein purification and crystallization (SI Appendix).
MAL-TIR shows a low level of amino acid sequence similarity to the other human TIR domains, with the closest homolog being MyD88-TIR (24% identity), followed by TLR2-TIR (23%). Searches using DALI (33) confirmed that these are the closest human structural homologs with, respectively, RMS distance values of 2.6 Å (over 96 Cα atoms) and 3.0 Å (over 95 Cα atoms) and Z scores of 10.4 and 10.2 (Fig. 2A). Surprisingly, the closest structural homologs are the bacterial protein PdTIR (15) and the plant protein AtTIR (16), with, respectively, RMS distance values of 2.3 Å (over 102 Cα atoms) and 3.0 Å (over 105 Cα atoms) and Z scores of 12.2 and 11.8 (Fig. 2B).
Fig. 2.
Comparison of TIR domain structures. (A) Superposition of human TIR domain structures: MAL-TIR (light green), MyD88-TIR (dark red), and TLR2-TIR (light blue). (B) Superposition of MAL-TIR (light green), PdTIR (yellow), and AtTIR (light blue).
MAL Oligomerization Interfaces.
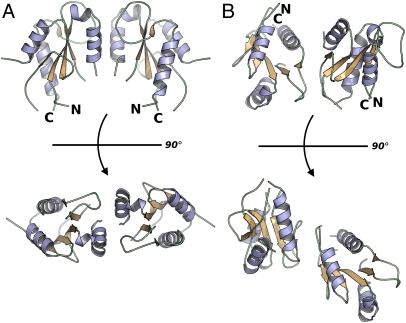
Two distinct dimers were observed in MAL-TIR crystals. We analyzed the possible physiological significance of each of these interactions by using the PISA server (34). The interface areas amounted to ∼750 Å2 and ∼640 Å2 for the two interactions, denoted hereinafter as interaction 1 and interaction 2. The interactions occur at distinct spatial locations (Fig. 3). The two monomers of the dimer in interaction 1 are related by a twofold symmetry axis (Fig. 3A). The most significant contributions, in terms of buried surface area, are from P155 and W156, both of which stack against a partially hydrophobic cavity formed by P189 and F193 of its symmetry partner. Other residues in the interface include Y159, E190, T166, and L162. The AB loops of both symmetry partners are not involved in interface formation and are exposed at the opposite sides of the assembly.
Fig. 3.
Potential dimerization interfaces in MAL-TIR. (A) Interaction 1 showing the association of the molecules related by a crystallographic twofold axis. (B) Interaction 2 demonstrating a putative asymmetric dimer.
In contrast to interaction 1, interaction 2 results in an asymmetric dimer (Fig. 3B). However, in this instance, it is the very N-terminal region and the AB loop of one protomer that participates in the interface. The indole ring of W82 is sequestered within a deep cleft formed by R184, Y187, P189, and R192. Notably, P125 of the AB loop is in close proximity to this pocket. Y196 is below the pocket, and its phenyl ring is oriented toward the sequestered tryptophan. Overall, there is a significantly higher density of hydrogen bonds and salt bridges across this interface than in is interaction 1.
Structural Basis for the Phenotype of MAL D96N and S180L Variants.
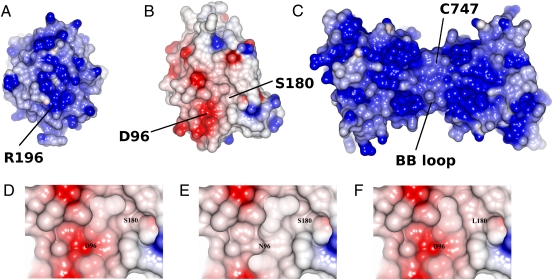
Previous modeling studies in our laboratories suggested that the recruitment of MyD88 to the TLR is mediated by the oppositely charged electrostatic surfaces of the two adaptors, contributing significantly toward their structural complementarity (7, 21, 35). Indeed, whereas the surface of MyD88-TIR and TLR4-TIR are largely basic, there are significant acidic patches on the MAL surface (Fig. 4 A–C). A prominent surface patch is centered on D96 of MAL. The D96N mutation affects the distribution of negative charge within this patch (Fig. 4 D–E), which could be sufficient for the failure of this variant of MAL to associate directly with MyD88. In both putative modes of MAL dimerization, the region surrounding D96 remains fully exposed and could participate in the recruitment of MyD88.
Fig. 4.
Structural environment of disease susceptibility polymorphisms S180L and D96N. (A) The surface of MyD88-TIR consists of significant patches of basic residues. The view of the interface predicted to associate with MAL is shown. (B) The surface of MAL-TIR is more hydrophobic with acidic patches in several locations, one of which is shown, comprising the D96 and S180 residues. (C) The surface of the homology model of the TLR4-TIR dimer from our previous study based on the crystal structure of TLR10-TIR (6). A very extensive basic surface can be observed throughout the dimer. (D) Close-up of the MAL-TIR surface containing the D96 and S180 residues. The surface cavity flanked by S180 can be seen in the center. (E) A model of the D96N variant showing the loss of the acidic patch in the region predicted to interact with MyD88-TIR. (F) A model of the S180L variant showing the physical occlusion of the hydrophobic surface cavity by the bulkier leucine side chain. The solvent-accessible surfaces are colored according to the electrostatic potential, ranging from deep blue (positive charge) to red (negative charge) in the range ±0.5 kT/e. The electrostatic potentials were calculated by solving the Poisson–Boltzmann equation with the CCP4MG program using the default settings.
One aspect in which the MAL crystal structure differs significantly from previous theoretical models is in the location of S180. Previously modeled as buried deep within the core of the protein, this residue is in fact solvent-exposed at the end of strand βD. This site is also in close proximity to the prominent acidic surface patch and ∼10 Å from D96. The side chain of the serine appears to border a prominent cavity within the structure, which is located at the edge of the acidic patch, and substitution with a bulkier leucine side chain would lead to steric occlusion of this cavity (Fig. 4F).
MyD88-Interacting Interface Includes D96 and S180 in MAL.
A number of surface residues in MyD88-TIR have been systematically investigated, through site-directed mutagenesis and reporter assays, for involvement in the interaction with MAL (17). It was found that mutation of two residues, R196 and R288, could abolish the TIR–TIR domain interaction and abrogate the signal response. Protein docking of the experimentally derived MyD88-TIR structure and a homology model of MAL-TIR was then used to propose a model in which a single MyD88-TIR binds two molecules of MAL-TIR at opposing interfaces. This model did not take into account more recent studies showing a direct role for MAL D96 in heterotypic MAL-MyD88 TIR–TIR domain association and the likelihood that homotypic TIR–TIR domain associations may have a role in the formation of the complex. We docked the experimental structures of both molecules, taking into account only the information provided by the structures themselves, such as surface and electrostatic complementarity. Five of the 10 best-docked complex models place the MyD88-TIR in very close proximity to the interface harboring the D96 and S180 residues in MAL (Fig. 5). Moreover, MyD88 R196, a residue known to be important for MAL binding (17), also consistently places in close proximity to MAL.
Fig. 5.
A structural model of the MAL-TIR:MyD88-TIR complex predicted through docking calculations. The 10 top-scoring solutions in ribbon representation. MAL-TIR is shown in light red, and MyD88-TIR is in light blue. The R196 residues in the MyD88-TIR molecules are shown as blue spheres, and the D96 and S180 residues in MAL-TIR are shown as red spheres. Five of the 10 best-scoring solutions place the MyD88-TIR structure within the structural vicinity of the interface comprising the D96 and S180 residues in MAL-TIR (and also R196 in three molecules).
Functional Significance of the Identified Interfaces.
To evaluate the functional consequences of disrupting the identified oligomerization and MyD88-binding interfaces, we generated a series of alanine substitutions of key interface residues. Coprecipitation experiments were carried out in HEK293T cells transiently transfected with FLAG-tagged MyD88 in conjunction with WT and mutant variants of HA-tagged MAL. As shown in Fig. 6A (lane 2), we were able to detect MAL- and MyD88-tagged protein complexes by coimmunoprecipitation. However, the substitution of residues in the region between residues 162 and 166 (Fig. 6A, lanes 6–8) consistently inhibited the ability of MAL to immunoprecipitate with MyD88, suggesting that these residues are critical for the MAL:MyD88 TIR:TIR domain heterotypic association. We also tested the interaction of MyD88 with WT MAL and its variants corresponding to polymorphisms D96N and S180L. Consistent with previous results, D96N MAL variant is unable to form a complex with MyD88, but S180L MAL variant is unaffected (Fig. 6B). Both variants are still able to form homodimers. This result is consistent with previous studies that suggest that S180L is hypomorphic, i.e., that it exhibits a partial loss of function (35). Finally, we show that the reducing agent DTT affects the interaction of MAL with MyD88 but not the interaction of MAL with TLR4 or MAL homodimer formation (SI Appendix).
Fig. 6.
Functional consequences of amino acid substitutions in MAL interfaces. (A) HEK293T cells (2 × 106) were transiently cotransfected with FLAG-tagged MyD88 and the indicated HA-tagged MAL constructs. Cells were coimmunoprecipitated with anti-FLAG M2-agarose beads, followed by immunoblot analysis with anti-HA antibody. Cell extracts were used to check expression of transfected HA-MAL constructs, and FLAG-MyD88 immunoprecipitation was checked by immunoblotting with anti-FLAG M2-horseradish peroxidase–conjugated antibody (n = 3). (B) HEK293T cells were cotransfected with either Myc-tagged MAL, MAL S180L, or MAL D96N constructs and FLAG-tagged MAL (lanes 1–3) or FLAG-tagged MyD88 (lanes 4–6). Interacting complexes were immunoprecipitated with anti-FLAG beads. Expression levels of each protein, along with detection of immunoprecipitated MAL and MAL mutants, were visualized by immunoblotting with anti-Myc antibody (n = 3).
Discussion
In this article, we describe the crystal structure of the TIR domain from the adaptor protein MAL. The structure differs significantly from previously published homology models (Cα RMS distance of 4.3 Å) (6) (SI Appendix). This observation reflects the low level of amino acid sequence identity (17–24%) with TIR domains with known structures and highlights the difficulty of computational modeling and the importance of determining experimental structures at this level of sequence identity. Of particular note, although the TIR domain BB loop signature motif is conserved in MAL, it is found in an entirely different structural arrangement in MAL, preceding strand βB (and therefore instead corresponding to the AB loop in MAL) (SI Appendix). The resulting configuration may be a key structural feature distinguishing the function and specificity of MAL and bridging adaptors from other TIR domains. Furthermore, the polymorphic residue S180 is surface-exposed, rather than being buried as predicted in the homology model (6), and is within ∼10 Å of D96, a residue required for MyD88 recruitment.
The crystal structure reveals two potential dimerization interfaces. Although purified MAL behaves as a monomer in solution, it forms a homodimer in vivo (21). It appears that weak dimerization is a general feature of TIR domain function and that stabilization of this interaction by membrane attachment or within the context of a larger complex represents a key regulatory mechanism. This concept is consistent with the observation that all TIR domain-only protein constructs that have been biochemically characterized to date are monomeric in solution, but several crystal structures reveal substantial dimeric interfaces that have been presented as reflective of natural associations (11, 14–16). Intriguingly, the dimer interfaces differ substantially in every structure reported (SI Appendix). For example, TLR10-TIR contains a symmetric dimer within the asymmetric unit of the crystal, with the most significant contacts made by residues of the BB loops, the αC-helices, and a part of the DD loop (14). In the TLR2-TIR structure, a different spatial arrangement is observed, with the proposed asymmetric dimeric interface mediated by the residues of the αB-, αC-, and αD-helices and CD and DD loops of one symmetry partner, with the αB-helix and BB loop of another (12). In the case of MAL, interaction 1 (Fig. 2A) is likely to represent the physiologically relevant interface. First, this configuration has the most extensive interface and, crucially, has twofold symmetry, which means that the N termini of the two subunits are oriented in the same direction, allowing the phosphatidylinositol-4,5-bisphosphate (PIP2)-binding motifs of both protomers to associate with the membrane. By contrast, interaction 2 (Fig. 2B) is asymmetric, and it is unlikely that such a dimer would be able to dock stably with the membrane. Cyclic symmetry, such as that imposed by a twofold crystallographic axis, is prevalent in proteins that interact with membranes because it imposes directionality, which in turn leads to thermodynamically favorable protein–lipid associations (36). In a more general sense, what is clearly emerging is that there may not be a universal dimerization interface in TIR domains but that specific features of the dimer interface may correlate with the specific function of the TIR domain and its interacting partners. This concept is consistent with the observation that the conservation of quaternary structure decreases significantly when the protein sequences are less than 30% identical (37).
This work also provides a molecular explanation for the properties of two polymorphisms in MAL in humans: S180L and D96N. Individuals who carry the S180L allele are susceptible to a variety of diseases (30–32), and the rare D96N allele has been shown in a Chinese population to predispose individuals to the development of tuberculosis (38). The structure reveals that these residues are within ∼10 Å of each other on the surface of MAL in a probable heterotypic TIR domain-binding site. Docking of the experimental structures of MAL and MyD88 identifies the acidic surface that includes S180 and D96 as the likely binding interface for MyD88, a conclusion further supported by the location of MyD88 residue R196 in the interface in the docked models. Humans homozygous for the R196C variant in MyD88 are highly susceptible to infection by Gram-positive bacteria, although they are able to resist infection by other pathogens (39). The importance of this acidic patch for MyD88 recruitment is supported by binding experiments (Fig. 6B). MAL D96N variant is unable to bind MyD88, although the interaction is observed in the case of the S180L variant. The structure shows that S180 is in a surface-exposed cavity and that substitution by leucine is likely to cause steric occlusion of the cavity. MyD88 may bind in a different configuration to MAL S180L variant, which might interfere with the formation of the receptor ternary complexes required to initiate signaling. This result is also consistent with the weak dominant-negative effects of MAL D96N, preventing MAL from recruiting MyD88 to the plasma membrane and interfering with MAL phosphorylation (35). Thus, our model supports the hypothesis that loss or alteration of the direct interaction between MAL and MyD88 normally mediated through R196, S180, and D96 may be responsible for the clinical symptoms associated with the increased risk of infection. It is also likely that heterotypic interactions between the receptor and MAL adaptor TIR domains is mediated by the AB loop, in agreement with previous studies (7, 35). In addition, a preliminary docking analysis suggests that this arrangement places the MyD88 TIR domain in position where it can form a second interface with the receptor TIR homodimer (SI Appendix), which is consistent with MAL acting to sensitize MyD88-directed signaling, as proposed previously (35).
Intriguingly, functional investigation of alanine-substitution mutants revealed that the region observed structurally to be important for self-association in MAL (residues 162–166) is also crucial for the ability of MAL to associate with MyD88. This finding suggests that, for the TLR adaptor proteins, the homo- and heterotypic associations within their TIR regions are likely to be closely intertwined events with substantial functional crosstalk.
An unusual feature of the MAL-TIR structure is the stabilization of the TIR fold by two internal disulfide bonds, a unique phenomenon among the TIR domain structures reported so far and an unusual feature for a cytosolic protein (40). Another two thiol groups, from C91 and C157, are within close structural proximity (Fig. 1 and SI Appendix) and are exposed on the protein surface. S-nitrosylation of a single cysteine residue by nitric oxide (NO) within the TIR domain of MyD88 was found to lead to the attenuation of interaction with MAL and downstream signaling (41). It is interesting that one of the unpaired cysteines observed in the MAL structure, C157, is highly conserved in other TIR domains (SI Appendix). Recently, C747 residue in the TIR domain of TLR4 (corresponding to C157 in MAL) was demonstrated to be a critical residue for binding the small-molecule inhibitor of TLR4-mediated signaling, TAK-242 (resatorvid) (42). C747 and C706 are also critical for NF-κB activation (7). TAK-242 acts by interfering with protein–protein interactions between the TIR domains of TLR4 and MAL or TRAM (43). These studies underscore the potential role of reactive cysteines in the formation of heterotypic TIR–TIR domain associations.
MAL is known to be regulated by several different posttranslational modification events, and the structure sheds light on the molecular basis of some of these. The cleavage by caspase-1 would remove helix αE and modify the predicted MyD88-interacting surface. The Btk phosphorylation sites Y86 and Y106 are solvent-exposed, whereas Y187 is less so and phosphorylation could induce some local conformation changes. None of these residues are near the predicted dimerization interface, but Y106 and Y187 are near the predicted MyD88-interacting interface. The IRAK-1 and IRAK-4 phosphorylation site T28 is outside of the TIR domain. The TRAF6-interacting region corresponds to the surface segment connecting β-strands βD and βE.
TLR signaling pathways are increasingly being targeted for therapeutic intervention in an effort to develop new treatments for pathologies of deregulated immune response, such as acute sepsis and chronic inflammatory diseases (44, 45). Recently, a single mutation within the MyD88 TIR domain, L265P, was found to be present in almost one-third of all diffuse large B-cell lymphomas, an aggressive form of cancer with poor treatment prognosis (46). L265P is a gain-of-function mutation that promotes tumor survival through stabilization of complexes with downstream kinases. Components of the TLR signaling pathways may therefore be integral to oncogenesis and could also be targeted in the development of novel cancer treatments. Future efforts toward determining the 3D structures of adaptor:receptor complexes will prove a critical step toward realizing the potential of effective, rationally designed drugs targeting the regulation of TLR signaling pathways.
Materials and Methods
Protein Production and Crystallization.
Human MAL construct comprising residues 79–221 was expressed in Escherichia coli. After metal-affinity purification and size-exclusion chromatography, crystals were obtained in 10–12% PEG 10,000, 5% PEG 3,350, 0.2 M NaCl, 0.1 M Tris⋅HCl (pH 7.3), and 20 mM DTT. For data collection, the stabilizing solution was supplemented with 20% (vol/vol) glycerol. Full methods are described in SI Appendix.
Data Collection and Structure Determination.
Data were collected on the MX2 beamline of the Australian Synchrotron, Melbourne, Australia, with Blu-Ice software (47). Diffraction data were processed and reduced with XDS and Scala (48, 49). The structure was determined by using a procedure (50) that combined molecular replacement with energy- and electron density-guided model building and refinement protocols with the program ROSETTA as implemented in the PHENIX suite (51, 52) and by using a search model based on the coordinates of the MyD88-TIR NMR structure (PDB ID code 2Z5V). Model improvement was monitored through Rfree during rounds of density modification and reciprocal-space refinement. After extensive manual rebuilding in COOT (53), combined with maximum likelihood-based restrained refinement in BUSTER-TNT (54), the final model had Rwork/Rfree values of 0.23/0.25 (SI Appendix). The stereochemistry of the structure was assessed and validated with MolProbity (55). Figures were generated with PyMOL (http://www.pymol.org) and CCP4MG (56).
Immunoprecipitation and Immunoblot Analysis.
Site-directed mutants of MAL were generated by GenScript. All mutations are unlikely to have destabilizing effects on the protein (SI Appendix). For immunoprecipitation experiments, HEK293T cells (2 × 106) were transiently transfected with FuGENE 6 (Roche Applied Science) (29) and lysed in KalB buffer [50 mM Tris (pH 7.4), 1.0% Triton X-100, 150 mM NaCl, 1 mM EDTA, 2 mM Na3VO4, 10 mM NaF, 1 mM PMSF, and protease cocktail inhibitor mixture], and MyD88:MAL complexes were immunoprecipitated with anti-FLAG Sepharose beads. Beads were washed, eluted by the addition of sample buffer followed by SDS/PAGE, and immunoblotted. Protein samples were separated by SDS/PAGE, transferred to nitrocellulose, and immunoblotted. Complexes were visualized by using SuperSignal West Pico Chemiluminescent Substrate (Pierce) followed by exposure to X-ray film (Hyperfilm ECL; Amersham Biosciences) to detect chemiluminescence.
Supplementary Material
Acknowledgments
We thank Susan Lea for access to the computing facilities; Gerard Bricogne and the Global Phasing Consortium for access to the beta version of autoBUSTER software; Tom Caradoc-Davies for assistance with microfocus synchrotron data collection; and Rafael Counago, Daniel Ericsson, Simon Williams, Mary Marfori, and Thomas Ve for discussions. We acknowledge the use of the University of Queensland Remote Operation Crystallization and X-ray (UQ ROCX) Diffraction Facility. P.R. was funded by a Wellcome Trust Project Grant to Susan Lea. J.L.M. is an Australian Research Council Laureate Fellow and Honorary National Health and Medical Research Council (NHMRC) Fellow. B.K. is an NHMRC Research Fellow. This work was supported by the Australian Research Council (J.L.M. and B.K.); the NHMRC (B.K., A.M., and N.J.G.); the Medical Research Council, Biotechnology and Biological Sciences Research Council; the Victorian Government Operational Infrastructure Support Program; and the Wellcome Trust (N.J.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2Y92).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104780108/-/DCSupplemental.
References
- 1.Gay NJ, Gangloff M, O'Neill LA. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32:104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 3.Jin MS, Lee JO. Structures of the Toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Bryant CE, Spring DR, Gangloff M, Gay NJ. The molecular basis of the host response to lipopolysaccharide. Nat Rev Microbiol. 2010;8:8–14. doi: 10.1038/nrmicro2266. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins KA, Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine. 2010;49:237–244. doi: 10.1016/j.cyto.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Toshchakov VU, Basu S, Fenton MJ, Vogel SN. Differential involvement of BB loops of Toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4- versus TLR2-mediated signal transduction. J Immunol. 2005;175:494–500. doi: 10.4049/jimmunol.175.1.494. [DOI] [PubMed] [Google Scholar]
- 7.Núñez Miguel R, et al. A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signalling adaptor proteins. PLoS ONE. 2007;2:e788. doi: 10.1371/journal.pone.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 9.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan JC, et al. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernoux M, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9:200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 13.Khan JA, Brint EK, O'Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279:31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- 14.Nyman T, et al. The crystal structure of the human Toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 15.Chan SL, et al. Molecular mimicry in innate immunity: Crystal structure of a bacterial TIR domain. J Biol Chem. 2009;284:21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci. 2010;19:155–161. doi: 10.1002/pro.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohnishi H, et al. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc Natl Acad Sci USA. 2009;106:10260–10265. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loiarro M, et al. Pivotal Advance: Inhibition of MyD88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J Leukoc Biol. 2007;82:801–810. doi: 10.1189/jlb.1206746. [DOI] [PubMed] [Google Scholar]
- 19.Gangloff M, Gay NJ. Baseless assumptions: Activation of TLR9 by DNA. Immunity. 2008;28:293–294. doi: 10.1016/j.immuni.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Loiarro M, et al. Peptide-mediated interference of TIR domain dimerization in MyD88 inhibits interleukin-1-dependent activation of NF-κB. J Biol Chem. 2005;280:15809–15814. doi: 10.1074/jbc.C400613200. [DOI] [PubMed] [Google Scholar]
- 21.Dunne A, Ejdeback M, Ludidi PL, O'Neill LA, Gay NJ. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J Biol Chem. 2003;278:41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Z, et al. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc Natl Acad Sci USA. 2006;103:10961–10966. doi: 10.1073/pnas.0603804103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 24.Gray P, et al. MyD88 adapter-like (Mal) is phosphorylated by Bruton's tyrosine kinase during TLR2 and TLR4 signal transduction. J Biol Chem. 2006;281:10489–10495. doi: 10.1074/jbc.M508892200. [DOI] [PubMed] [Google Scholar]
- 25.Mansell A, et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 26.Dunne A, et al. IRAK1 and IRAK4 promote phosphorylation, ubiquitination, and degradation of MyD88 adaptor-like (Mal) J Biol Chem. 2010;285:18276–18282. doi: 10.1074/jbc.M109.098137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miggin SM, et al. NF-κB activation by the Toll-IL-1 receptor domain protein MyD88 adapter-like is regulated by caspase-1. Proc Natl Acad Sci USA. 2007;104:3372–3377. doi: 10.1073/pnas.0608100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulrichts P, et al. Caspase-1 targets the TLR adaptor Mal at a crucial TIR-domain interaction site. J Cell Sci. 2010;123:256–265. doi: 10.1242/jcs.056002. [DOI] [PubMed] [Google Scholar]
- 29.Verstak B, et al. MyD88 adapter-like (Mal)/TIRAP interaction with TRAF6 is critical for TLR2- and TLR4-mediated NF-κB proinflammatory responses. J Biol Chem. 2009;284:24192–24203. doi: 10.1074/jbc.M109.023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khor CC, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferwerda B, et al. Functional and genetic evidence that the Mal/TIRAP allele variant 180L has been selected by providing protection against septic shock. Proc Natl Acad Sci USA. 2009;106:10272–10277. doi: 10.1073/pnas.0811273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladhani SN, et al. Association between single-nucleotide polymorphisms in Mal/TIRAP and interleukin-10 genes and susceptibility to invasive Haemophilus influenzae serotype b infection in immunized children. Clin Infect Dis. 2010;51:761–767. doi: 10.1086/656236. [DOI] [PubMed] [Google Scholar]
- 33.Holm L, Kääriäinen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006;14:5.5.1–5.5.24. doi: 10.1002/0471250953.bi0505s14. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Nagpal K, et al. A TIR domain variant of MyD88 adapter-like (Mal)/TIRAP results in loss of MyD88 binding and reduced TLR2/TLR4 signaling. J Biol Chem. 2009;284:25742–25748. doi: 10.1074/jbc.M109.014886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 37.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YX, et al. Association of TIRAP (MAL) gene polymorhisms with susceptibility to tuberculosis in a Chinese population. Genet Mol Res. 2011;10:7–15. doi: 10.4238/vol10-1gmr980. [DOI] [PubMed] [Google Scholar]
- 39.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Østergaard H, Tachibana C, Winther JR. Monitoring disulfide bond formation in the eukaryotic cytosol. J Cell Biol. 2004;166:337–345. doi: 10.1083/jcb.200402120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Into T, et al. Regulation of MyD88-dependent signaling events by S nitrosylation retards toll-like receptor signal transduction and initiation of acute-phase immune responses. Mol Cell Biol. 2008;28:1338–1347. doi: 10.1128/MCB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawamoto T, Ii M, Kitazaki T, Iizawa Y, Kimura H. TAK-242 selectively suppresses Toll-like receptor 4-signaling mediated by the intracellular domain. Eur J Pharmacol. 2008;584:40–48. doi: 10.1016/j.ejphar.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 43.Matsunaga N, Tsuchimori N, Matsumoto T, Ii M. TAK-242 (resatorvid), a small-molecule inhibitor of Toll-like receptor (TLR) 4 signaling, binds selectively to TLR4 and interferes with interactions between TLR4 and its adaptor molecules. Mol Pharmacol. 2011;79:34–41. doi: 10.1124/mol.110.068064. [DOI] [PubMed] [Google Scholar]
- 44.Ulevitch RJ. Therapeutics targeting the innate immune system. Nat Rev Immunol. 2004;4:512–520. doi: 10.1038/nri1396. [DOI] [PubMed] [Google Scholar]
- 45.Wittebole X, Castanares-Zapatero D, Laterre PF. Toll-like receptor 4 modulation as a strategy to treat sepsis. Mediators Inflamm. 2010;2010:568396. doi: 10.1155/2010/568396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McPhillips TM, et al. Blu-Ice and the Distributed Control System: Software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 50.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afonine PV, et al. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 55.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.