Abstract
Npl3p, the major mRNA-binding protein of the yeast Saccharomyces cerevisiae shuttles between the nucleus and the cytoplasm. A single amino acid change in the carboxyl terminus of Npl3p (E409 → K) renders the mutant protein largely cytoplasmic because of a delay in its import into the nucleus. This import defect can be reversed by increasing the intracellular concentration of Mtr10p, the nuclear import receptor for Npl3p. Conversely, using this mutant, we show that Npl3p and mRNA export out of the nucleus is significantly slowed in cells bearing mutations in XPO1/CRM1, which encodes the export receptor for NES-containing proteins and in RAT7, which encodes an essential nucleoporin. Interestingly, following induction of stress by heat shock, high salt, or ethanol, conditions under which most mRNA export is blocked, Npl3p is still exported from the nucleus. The stress-induced export of Npl3p is independent of both the activity of Xpo1p and the continued selective export of heat-shock mRNAs that occurs following stress. UV-cross-linking experiments show that Npl3p is bound to mRNA under normal conditions, but is no longer RNA associated in stressed cells. Taken together, we suggest that the uncoupling of Npl3p and possibly other mRNA-binding proteins from mRNAs in the nucleus provides a general switch that regulates mRNA export. By this model, under normal conditions Npl3p is a major component of an export-competent RNP complex. However, under conditions of stress, Npl3p no longer associates with the export complex, rendering it export incompetent and thus nuclear.
Keywords: hnRNP, Npl3p, mRNA export, S. cerevisiae, stress
The partition created by the nuclear envelope requires that RNA molecules exit the nucleus in a specific and efficient manner. Most, if not all, RNA molecules exist within cells as ribonucleoprotein (RNP) complexes (Dreyfuss et al. 1993). For mRNAs, their association of various RNA-binding proteins begins as the mRNA precursors are being transcribed (Dreyfuss et al. 1993). The resulting RNP complexes must then move to and pass through the aqueous channels created by the nuclear pore complexes (NPCs). Some proteins appear to dissociate from the RNA before it enters the pore (Pinol-Roma and Dreyfuss 1992), whereas others, such as the cap-binding complex and some heterogeneous nuclear ribonucleoproteins (hnRNPs), remain associated during transit (Daneholt 1997). These proteins are then exchanged for cytoplasmic mRNA-binding proteins and rapidly re-enter the nucleus for new rounds of RNA packaging and transport.
The dynamics of the HIV Rev protein are perhaps the best understood example of the interplay between protein and RNA export. In HIV-infected cells, Rev enters the nucleus by association with the importin β nuclear import receptor (Henderson and Percipalle 1997). Once inside, it binds to RNAs containing the Rev Response Element. The resulting complex moves out of the nucleus and the viral RNA is released in the cytoplasm. Free Rev then re-enters the nucleus for another round of transport. Rev contains a short stretch of leucine-rich amino acids that is both necessary and sufficient for its nuclear export (Fischer et al. 1995). Similar nuclear export signals (NESs) have now been found in a number of proteins known to shuttle between the nucleus and the cytoplasm, such as the protein kinase inhibitor (PKI; Wen et al. 1995; for review, see Lee and Silver 1997; Izaurralde and Adam 1998). This phenomenon of NES-dependent export led to the identification of export receptors (e.g., mammalian exportin and yeast Xpo1p/Crm1p) that bind to NESs and guide their cargo to the cytoplasm (Fornerod et al. 1997; Fukuda et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997). The formation of the export complex is supported by the small GTPase, Ran, in its GTP-bound state (Fornerod et al. 1997; Izaurralde et al. 1997). The induction of Ran’s intrinsic GTPase activity by the cytoplasmically localized GAP then causes the dissociation of the cargo from exportin in the cytoplasm (Kutay et al. 1997).
Can the Rev example be generalized to the export of mRNAs? By such a mechanism, one or more of the mRNA-binding proteins that accompany the mRNA out of the nucleus would contain signals to confer interaction with an export receptor. There are a number of mRNA-binding proteins that shuttle between the nucleus and the cytoplasm, including the highly abundant hnRNPA1 (Nakielny and Dreyfuss 1996; Nakielny et al. 1997). However, none have been shown to rely on exportin for its export. In addition, a direct role for exportin in mRNA export in higher eukaryotes remains controversial (Fischer et al. 1995; Fornerod et al. 1997; Pasquinelli et al. 1997). The transported RNP complex, which can consist of transcripts up to 30 kb, covered with proteins and packed into a complex as large as 50 nm, must pass through the 25-nm-active channel formed by the NPC (Ohno et al. 1998). The large size of the transported RNP complex might mandate that it use a number of export proteins such as the recently described Mex67p, Mtr2p, and Gle1p.
The yeast Saccharomyces cerevisiae has proven to be particularly useful for studies of mRNA export. In particular, mutations in a number of genes have revealed roles for certain nucleoporins and RNA-binding proteins (for review, see Corbett and Silver 1997). Among the best characterized of the latter class is the abundant mRNA-binding protein Npl3p, also referred to as Nop3p, Mts1p, Nab1p, and Mtr13p (Bossie et al. 1992; Russell and Tollervey 1992; Ellis and Reid 1993; Kadowaki et al. 1994; Wilson et al. 1994). A number of observations have led to the idea that Npl3p is an important mediator of mRNA export. First, loss-of-function temperature-sensitive NPL3 alleles accumulate poly(A)+ RNA in the nucleus at the nonpermissive temperature (Lee et al. 1996). Second, Npl3p shuttles between the nucleus and the cytoplasm and mutations in Npl3p that prevent its nuclear export result in a concurrent block in mRNA export (Lee et al. 1996). Further, Npl3p can be purified with poly(A)+ RNA after UV-cross-linking ( Russell and Tollervey 1995; Segref et al. 1997). However, it remains an open question as to whether Npl3p is an active carrier of mRNAs or a passive cargo that is required for proper packaging of mRNAs for export. In the latter case, when Npl3p is mutated, mRNA might fail to assemble properly into a complex competent for export.
Under various conditions of stress, cells respond by selectively exporting mRNAs encoding heat shock proteins (Saavedra et al. 1996). There is a simultaneous cessation in the export of normal mRNAs (Saavedra et al. 1996; Tani et al. 1996). It remains unknown how this novel form of regulation occurs. Interestingly, in yeast, in which this phenomenon has been studied in the greatest detail, the export of heat shock mRNA does not require the functions of the Ran GTPase or Npl3p, which are required for export of normal mRNAs (Saavedra et al. 1996, 1997). In contrast, hs mRNA export requires the function of several different export factors, including Rip1p and Gle1p (Saavedra et al. 1997). This result has led to the proposal that there is a special pathway for the export of heat shock RNAs. However, the mechanism by which normal mRNAs fail to be exported under heat shock conditions has not been explored.
Here, we present data that indicate that the reason constitutively exported mRNAs fail to leave the nucleus in stressed cells is because an export incompetent RNP complex, lacking Npl3p, is produced. This hypothesis is based on the observation that Npl3p is exported from the nucleus following onset of stress, whereas mRNA remains nuclear. However, this is not due to exit of Npl3p from the nucleus in association with heat shock mRNAs. In UV-cross-linking experiments, we show that Npl3p is not bound to poly(A)+ RNA after the cells were stressed. Taken together, we propose that the uncoupling of Npl3p, and perhaps other hnRNPs, from mRNAs render the RNPs incompetent for export and that it is this uncoupling that provides part of the rapid switch in gene expression observed under stress conditions.
Results
Npl3p has been shown to shuttle rapidly between the nucleus and the cytoplasm (Flach et al. 1994). Its export is thought to be coupled in some manner to the export of poly(A)+ RNA. Under steady-state conditions, Npl3p appears to localize exclusively to the nucleus because its rate of reimport into the nucleus following export is quite rapid. We now further analyze the dynamics of Npl3p by taking advantage of a novel mutant allele that allows us to dissect effects on both import and export in more detail.
A single amino acid change in Npl3p slows its nuclear reimport
npl3-27 was originally identified in a screen for plasmid-borne temperature-sensitive alleles of NPL3 (Lee et al. 1996). Sequencing of the mutated gene revealed a single-point mutation resulting in the change of glutamate 409 to lysine (E409K). A haploid yeast strain was constructed in which genomic NPL3 was replaced by the npl3-27 allele. Interestingly, this strain, termed npl3-27, was viable at all temperatures; no temperature-sensitive growth defects were observed. This might be because once integrated into the yeast genome, only a single copy of the gene is present. The original mutant was isolated on a multicopy plasmid and may, therefore, have produced a slightly dominant effect on cell growth.
Npl3-27p is localized mainly to the cytoplasm. Yeast cells were analyzed by indirect immunofluorescence microscopy with an anti-Npl3p antibody (Fig. 1A). Wild-type Npl3p is concentrated in the nucleus at all temperatures (Fig. 1A, top). Npl3-27p on the other hand, localizes to the cytoplasm (Fig. 1A, middle). Note that Npl3-27p is not completely excluded from the nucleus but appears to equilibrate between the two compartments at 37°C.
Figure 1.
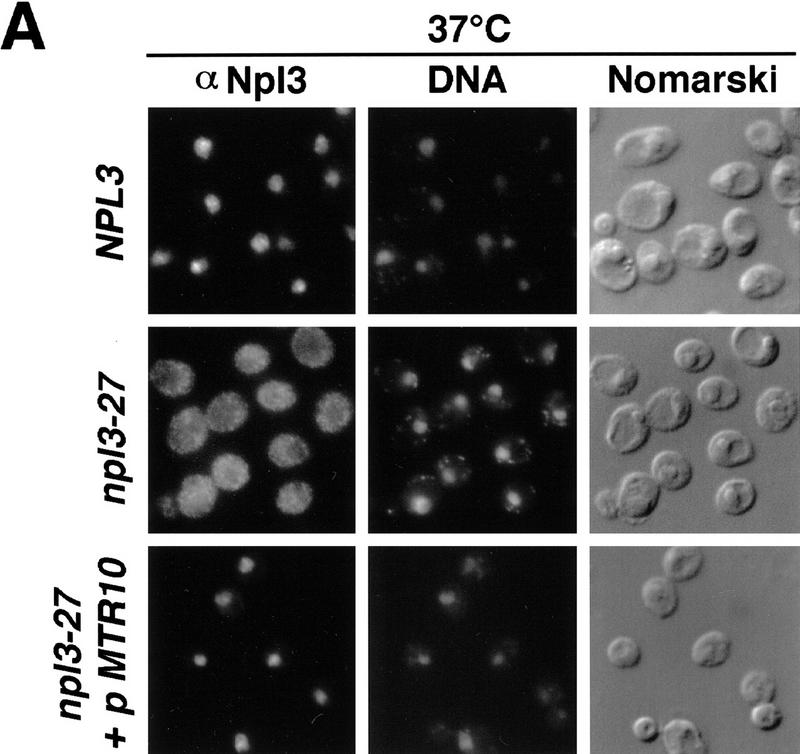
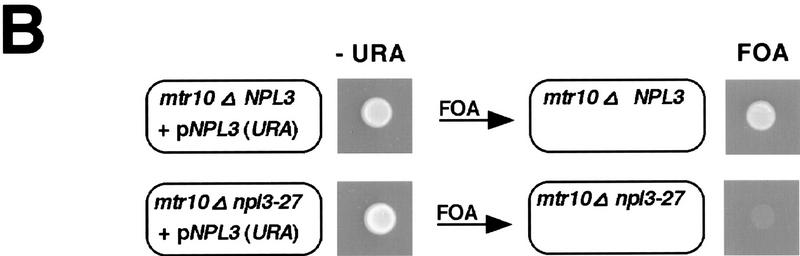
(A) Npl3-27p is slow in nuclear import. Wild-type (top), npl3-27 (middle), and npl3-27 cells overexpressing MTR10 on a 2μ plasmid (bottom) were grown to log phase and shifted to 37°C for 30 min. Npl3p localization was analyzed with an α-Npl3p antibody by immunofluorescence, the DNA was stained with DAPI, and the cells were photographed by Nomarski optics. (B) mtr10Δ and npl3-27 are synthetically lethal. The mtr10Δ npl3-27 double-mutant strain is not able to grow on a plate containing FOA, which allows only those cells to grow that are able to lose the URA3 plasmid (bottom, right). In the presence of wild-type NPL3 on a URA3 plasmid, cells are able to grow (bottom, left). Growth on FOA-containing medium is not impaired in the mutant mtr10Δ that contains genomic wild-type NPL3 (top).
One possible explanation for the behavior of Npl3-27p is that its rate of import is diminished. This is consistent with reports that the carboxyl terminus of Npl3p contains the information for nuclear targeting (Flach et al. 1994; Senger et al. 1998) because the mutation in npl3-27 lies in this carboxy-terminal encoding region. To test whether the import of Npl3-27p was altered, we investigated the effect of overproducing its import receptor, Mtr10p. A multicopy plasmid encoding Mtr10p was introduced into npl3-27 cells and the localization of Npl3-27p was analyzed by immunofluorescence after shifting the cells to 37°C. As shown in Figure 1A (bottom), Npl3-27p is completely relocalized to the nucleus when Mtr10p is overproduced. This is consistent with the interpretation that the E409K mutation reduces the efficiency of the Npl3p NLS and that the presence of additional Mtr10p is able to accelerate the slowed import. Together, these results demonstrate that Npl3-27p is still capable of re-entering the nucleus.
Cells deleted for MTR10 (mtr10Δ) are viable at 20°C but mislocalize Npl3p to the cytoplasm (Pemberton et al. 1997; Senger et al. 1998). However, cells harboring a combination of both mutant alleles, mtr10Δ and npl3-27, are inviable supporting their genetic interaction. This was demonstrated by showing that mtr10Δ cells can lose a NPL3 URA3-marked plasmid and are thus able to grow on 5-fluoro-orotic acid (FOA)-containing medium (Fig. 1B; top), whereas mtr10Δ npl3-27 cells cannot (Fig. 1B; bottom). These data suggest that mtr10Δ cells can live because some Npl3p can still enter the nucleus. However, when the NLS of Npl3p is mutated as in Npl3-27p, cells die because the intranuclear level of the protein is insufficient to support proper growth. Taken together, these data show that import of Npl3p is crucial for the cell. Furthermore, Npl3-27p offers a novel tool for studying the export of Npl3p.
Factors affecting export of Npl3p
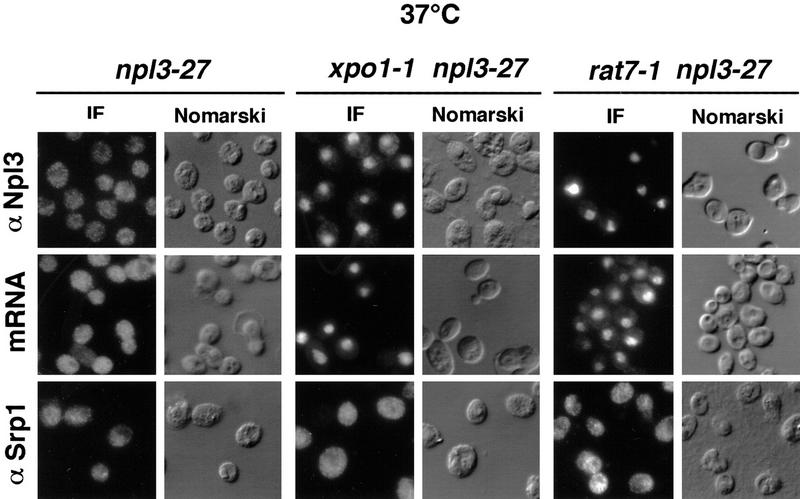
In principle, mutations that slow the rate of Npl3p export will result in accumulation of Npl3-27p in the nucleus instead of the cytoplasm. To test this idea, double mutants were constructed containing the npl3-27 allele and either the NES-receptor mutant xpo1-1 or the nucleoporin mutant, rat7-1. Following a shift of the single and double mutants to 37°C for 30 min, cells were fixed and the localization of Npl3-27p was determined by immunofluorescence. The distribution of poly(A)+ RNA in the same cells was visualized by in situ hybridization with an oligo[d(T50)] probe. Under these conditions, Npl3-27p and mRNA were both cytoplasmic in the npl3-27 single mutant, indicating no defect in export (Fig. 2, left). However, Npl3-27p and poly(A)+ RNA both accumulated in the nuclei of xpo1-1 and rat7-1 cells following the shift to the nonpermissive temperature (Fig. 2, center and right).
Figure 2.
xpo1-1 and rat7-1 exhibit Npl3-27p localization and mRNA export defects. npl3-27 (left), xpo1-1 npl3-27 (middle), and rat7-1 npl3-27 (right) strains were grown to log phase then shifted to 37°C for 30 min before they were prepared for immunofluorescence (IF) with α-Npl3p (top), oligo[d(T50)] probe (middle), or α-Srp1p (bottom).
To investigate whether the export of other proteins is altered at the nonpermissive temperature in xpo1-1 and rat7-1 cells, the localization of Srp1p/Kap65p, the NLS receptor was also analyzed by immunofluorescence (Fig. 2, bottom). Srp1p shuttles between the cytoplasm and the nucleus. Its export from the nucleus depends on the importin-β homolog Cse1p; in cse1-1 cells, Srp1p accumulates inside the nucleus (Hood et al. 1998; Kunzler and Hurt 1998; Solsbacher et al. 1998). However, in npl3-27, npl3-27 xpo1-1, and npl3-27 rat7-1 cells, Srp1p was partly cytoplasmic as it is in wild-type cells, demonstrating that export can occur through the NPCs at the restrictive temperature. Together these data demonstrate that the mRNA-export factors Xpo1p and Rat7p are important for Npl3p export as well.
Nuclear export of Npl3-27p during conditions of heat shock
Cells undergoing various forms of stress such as heat shock retain a substantial amount of their mRNA in the nucleus. Heat shock mRNAs, however, exit rapidly to allow the cell to selectively cope with the stress situation (Saavedra et al. 1996). Given this rapid switch in mRNA export, it was of interest to analyze the behavior of Npl3p under conditions in which mRNA export is perturbed.
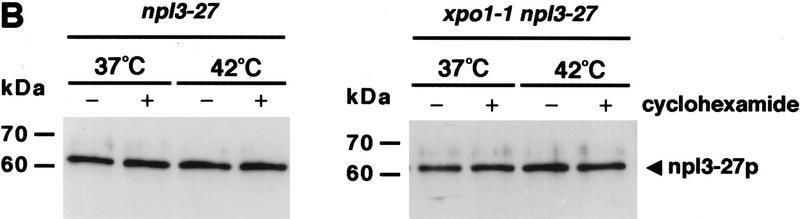
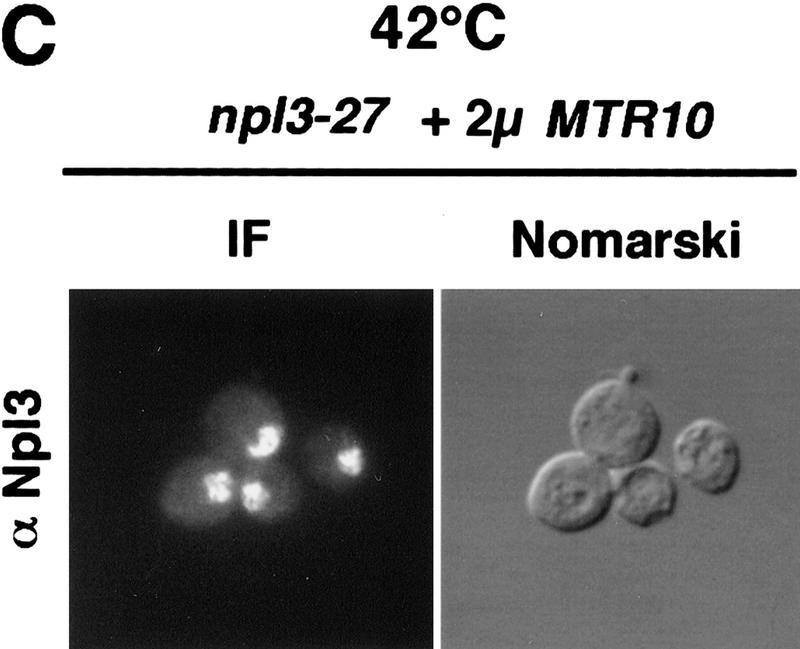
Surprisingly, Npl3-27p was still exported rapidly under heat shock conditions. npl3-27 cells were shifted for 15 min to 42°C and analyzed for the intracellular location of Npl3-27p and poly(A)+ RNA, as well as for the SSA4 mRNA, which encodes Hsp70p. Following heat shock, an accumulation of poly(A)+ RNA in the nucleus was observed (Fig. 3A) as has been described previously (Saavedra et al. 1996). Similarly, under conditions of heat shock, the SSA4 mRNA moved to the cytoplasm as has been reported previously (Saavedra et al. 1996; Fig. 3A). However, Npl3-27p remained in the cytoplasm in the heat-shocked cells, indicating that it was still being exported from the nucleus (Fig. 3A; top). Even under conditions in which Npl3-27p is normally trapped inside the nucleus, such as in the xpo1-1 mutant, Npl3-27p instead exited the nucleus after a 15 min shift to 42°C. The poly(A)+ RNA, on the other hand, accumulated in the nucleus following heat shock of the xpo1-1 cells. Interestingly, although bulk mRNA export is blocked in xpo1-1 cells, heat shock mRNA (SSA4) export is not impaired (Fig. 3A, bottom). The same results were obtained following heat shock treatments of 5 to 30 min and in the presence of the protein synthesis inhibitor cycloheximide (data not shown). Moreover, Npl3-27p remains intact under all conditions as determined by immunoblot analysis (Fig. 3B). These data show that under heat shock conditions, Npl3-27p can dissociate from the mRNA and exit the nucleus. As is the case for cells grown at 37°C, overexpression of Mtr10p at 42°C drives Npl3-27p into the nucleus (Fig. 3C). Thus, Npl3-27p is still capable of nuclear entry even under heat shock conditions.
Figure 3.
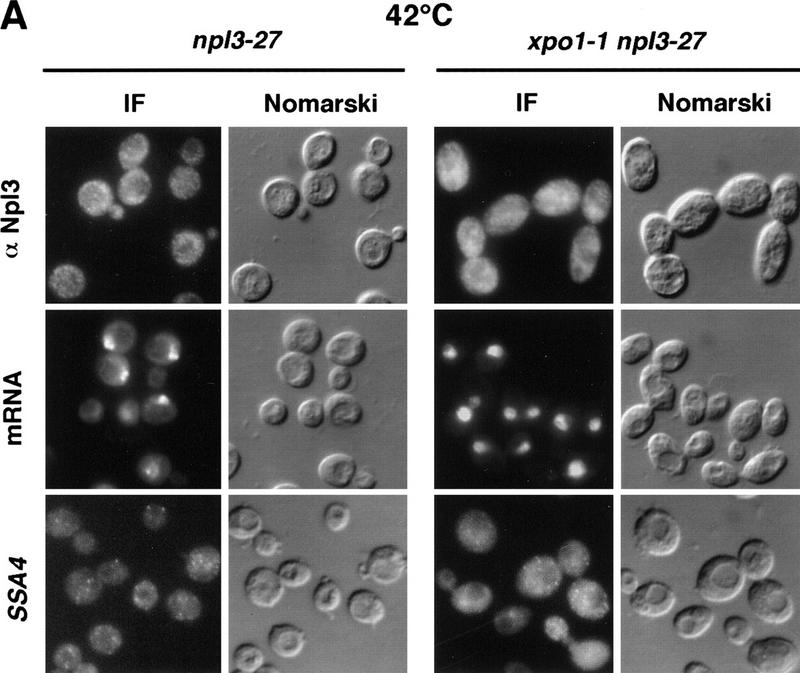
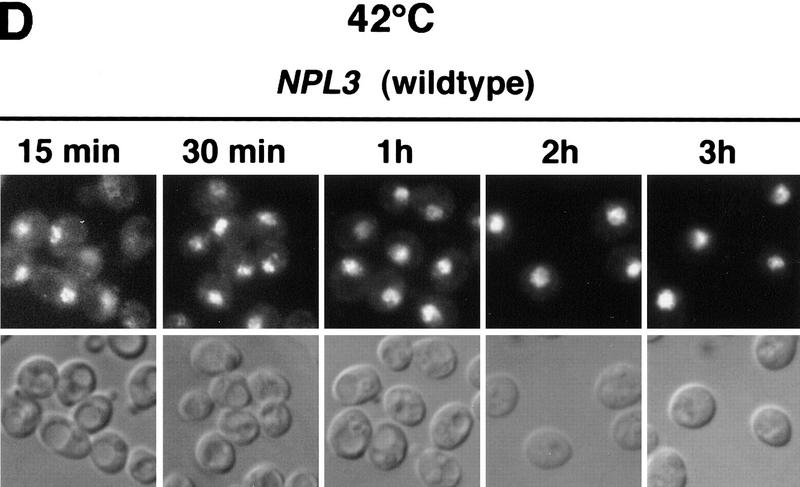
(A) At 42°C, Npl3-27p, but not mRNA exits the nucleus. Both strains, npl3-27 (left) and xpo1-1 npl3-27 (right), were grown to log phase, then shifted to 42°C for 15 min, before they were prepared for IF with α-Npl3p (top), oligo[d(T50)] probe (middle), and a SSA4 probe (bottom). (B) Yeast lysates of the same strains were prepared after they were grown to log phase and either incubated at 37°C for 3 hr (lanes 1,2) or to 42°C for 15 min (lanes 3,4) without [− (lanes 1,3)] or with cycloheximide addition [ + (lanes 2,4)]. The lysates were separated on a 12% SDS-polyacrylamide gel and analyzed on a Western blot with antibodies against Npl3p. (C) npl3-27 cells overexpressing MTR10 on a 2μ plasmid were grown to log phase and shifted to 42°C for 15 min. (D) Wild-type strains were grown to log phase before they were shifted to 42°C for different times. The localization of Npl3p was monitored by immunofluorescence.
Wild-type Npl3p also accumulates in the cytoplasm to some extent under conditions of stress. Wild-type cells were shifted to 42°C for 15 and 30 min and 1, 2, and 3 hr. The intracellular localization of Npl3p was determined by immunofluorescence with anti-Npl3p antibodies (Fig. 3D). At 15 and 30 min, some Npl3p is observed in the cytoplasm. However, the effect is not as complete as that observed for Npl3-27p, presumably because the reimport of Npl3p is faster than that of Npl3-27p. As cells adapt to the heat shock, after ∼1 hr, cytoplasmic Npl3p disappears.
Npl3-27p does not exit the nucleus with heat shock mRNAs
The observation that both Npl3-27p and heat shock mRNA exit the nucleus, whereas bulk mRNA remains nuclear after heat shock, indicates that the export of Npl3p and mRNA have become uncoupled. There are at least three possible explanations for this apparent separation of Npl3p from most of the mRNA. By one mechanism, Npl3p might exit the nucleus alone and unbound to mRNA. By a second mechanism, Npl3p could move out of the nucleus with a specific pool of mRNA that is not detectable in the in situ hybridization experiments. A third possibility is that Npl3p exits the nucleus together with heat shock mRNA under conditions of stress.
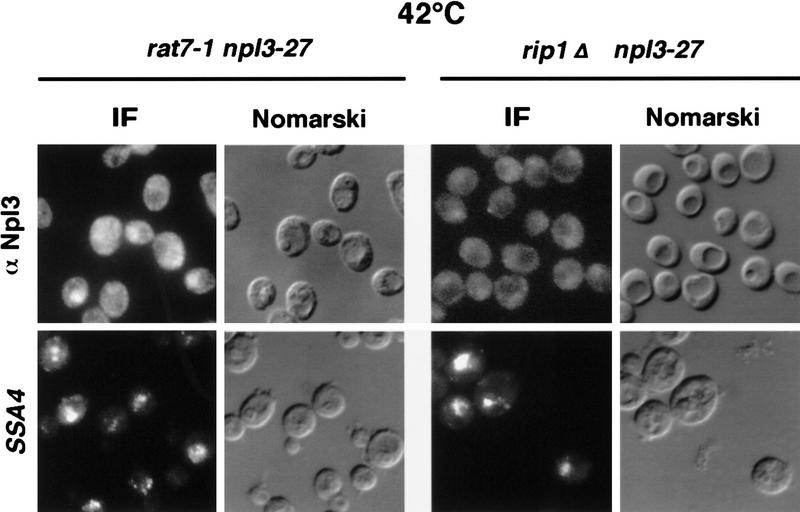
It has been reported previously that the nuclear pore-associated proteins Rat7p and Rip1p are essential for the nuclear export of heat shock mRNAs (Saavedra et al. 1997). To test whether or not Npl3-27p is exported together with SSA4 heat shock mRNA, the localization of Npl3-27p was investigated in rat7-1 and rip1Δ mutants. rat7-1 npl3-27 and rip1Δ npl3-27 double mutants were constructed and shifted for 15 min to 42°C to induce the heat shock stress response. The cells were then analyzed by immunofluorescence with anti-Npl3p antibodies and by in situ hybridization with a probe specific for SSA4 mRNA. As expected, a strong accumulation of the SSA4 mRNA in the nucleus was observed in both strains (Fig. 4, bottom). However, Npl3-27p was largely localized in the cytoplasm (Fig. 4, top). These results indicate that under heat shock conditions, Npl3p exits the nucleus without heat shock mRNAs.
Figure 4.
Npl3p does not exit the nucleus coupled to heat-shock mRNA. Both double mutants rat7-1 npl3-27 (left columns) and rip1Δ npl3-27 (right columns) were grown to log phase before they were shifted to 42°C for 15 min. Then they were analyzed by immunofluorescence with α-Npl3p (top) and by in situ hybridization with a SSA4 probe (bottom).
Uncoupling of Npl3p from mRNA export is a general response to different stress conditions
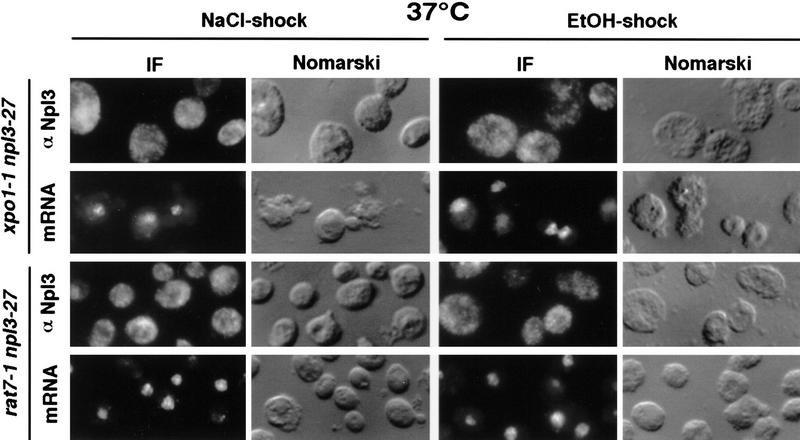
As we have shown that Npl3p exits the nucleus under heat shock conditions independently of poly(A)+ RNA, the question arose as to whether this represented a general stress response of the cell. To determine this, the double mutant strains xpo1-1 npl3-27 and rat7-1 npl3-27 were shifted to 37°C for 30 min, conditions that trap Npl3-27p and mRNA inside the nucleus (Fig. 2). The cells were then exposed to either high salt (0.4 m NaCl) for 10 min or 10% ethanol for 30 min. As shown in Figure 5, although poly(A)+ RNA accumulated inside the nucleus, Npl3-27p was separated from the bulk mRNA and transported into the cytoplasm. These data suggest a rapid dissociation of Npl3p from mRNA as a response to stress.
Figure 5.
Salt- and ethanol shocks result in the export of Npl3-27p. Both double-mutant strains xpo1-1 npl3-27 (top two rows) and rat7-1 npl3-27 (bottom two rows) were grown to log phase and shifted to 37°C for 30 min before they were stressed with 0.4 m NaCl for 10 min (left) or with 10% ethanol for 30 min (right). Cells were then prepared for immunofluorescence (IF) with α-Npl3p (first and third row) and a dT probe (second and fourth row).
Cross-linking of Npl3p to poly(A)+ RNA
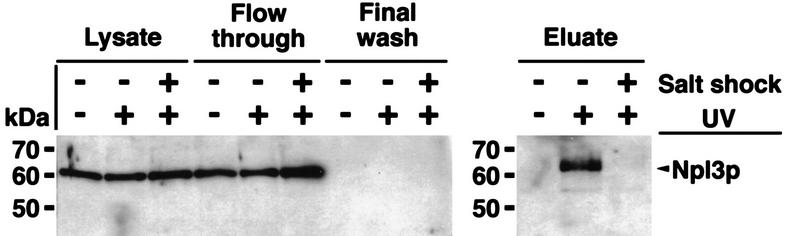
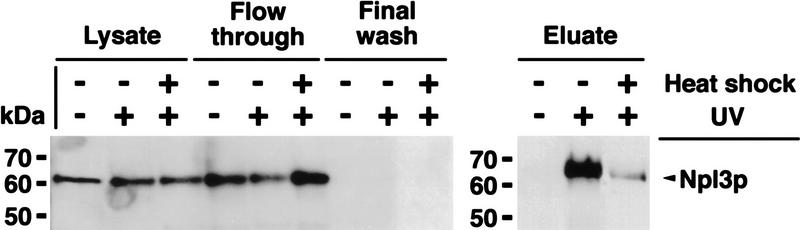
From the data shown above, we suggest a model in which Npl3p is released from poly(A)+ RNA prior to stress. To further test this proposal, we performed UV cross-linking experiments, which have shown previously that Npl3p can be photocross-linked to poly(A)+ RNA (Russell and Tollervey 1995; Segref et al. 1997). A wild-type strain was grown to log phase and then separated into three portions. One was not UV-irradiated, one was UV-irradiated, and the third portion was exposed to high salt (0.4 m NaCl) for 10 min before UV-irradiation. Approximately the same amounts of poly(A)+ RNA were isolated by oligo(dT)-cellulose chromatography (as determined by OD260) and tested for the presence of Npl3p by Western blot analysis, loading 10 OD260 units per lane. Npl3p was UV-cross-linked to poly(A)+ RNA under normal growth conditions (Fig. 6, top, eluate; −salt shock +UV). However, Npl3p was completely absent from purified poly(A)+ RNA when isolated under the identical conditions from cells that had been salt shocked before the UV-irradiation (Fig. 6, eluate; +salt shock +UV). The same experiment was performed on cells incubated at 42°C for 15 min (heat shock conditions). Following the heat treatment, cells were transferred to ice and subjected to UV irradiation just as with the salt-shocked cells. As predicted and as in the case of salt-shocked cells, very little Npl3p was found associated with the poly(A)+ RNA under these conditions (Fig. 6, bottom, cf. eluate −heat shock +UV with +heat shock +UV). It could be that this small amount of Npl3p reassociates with the mRNA during the incubation on ice.
Figure 6.
UV cross-linking of Npl3p to poly(A)+ RNA. (Top) Cells were either treated with or without high salt (+ or − Salt shock) and exposed to UV for cross-linking (+ or −UV-irradiation). The lysates were taken before application to the oligo(dT)–cellulose-column. (Flow through) The material that did not bind to the column. The final wash is derived from the last wash before elution (see Materials and Methods). The eluate contains the cross-linked poly(A)+ RNA-associated proteins. (Bottom) Cells were either shifted to 42°C for 15 min and exposed to UV (+heat shock +UV) or UV cross-linked without a heat shock (− heat shock +UV). The immunoblot was carried out with a polyclonal antibody against Npl3p.
Discussion
In all cells, mRNAs are bound by a number of hnRNPs (Nakielny and Dreyfuss 1996; Nakielny et al. 1997). The precise role(s) of the numerous hnRNP proteins in mRNA processing and transport remains one of the outstanding questions in RNA biology. In the yeast S. cerevisiae, NPL3 encodes an essential and abundant mRNA-binding protein that shuttles between the nucleus and the cytoplasm (Bossie et al. 1992; Flach et al. 1994; Wilson et al. 1994). Moreover, loss-of-function mutations in NPL3 result in a block in export of mRNA out of the nucleus (Kadowaki et al. 1994; Lee et al. 1996). Thus, Npl3p has been implicated in the proper export of mRNAs out of the nucleus.
We now present data defining further the role of Npl3p in mRNA export. By using a novel mutant form, Npl3-27p, that is slowed in nuclear reuptake, we are able to monitor the requirements for its nuclear export. We show that under conditions of stress, Npl3p is not associated with mRNA and is thus capable of exiting the nucleus while the mRNA remains nuclear. We propose a model in which the uncoupling of Npl3p from mRNAs results in the failure of the majority of mRNA to exit the nucleus under conditions of stress. This model is supported by UV-cross-linking experiments in which the Npl3p binding to poly(A)+ RNA is greately reduced under two different stress conditions. We propose that this uncoupling, in turn, allows for the rapid export of heat shock mRNAs in an Npl3p-independent manner. This switch in mRNA export is thus necessary for the rapid onset of the ability of cells to cope with stress conditions.
A novel mutant to study hnRNP nuclear protein export
The experiments presented here were carried out with a novel form of Npl3p that contains a single amino acid change in the carboxyl terminus. Cells bearing the npl3-27 allele are still viable at all temperatures, but the amount of Npl3-27p in the cytoplasm under steady-state conditions is greatly increased as compared with wild-type Npl3p. Previous studies have indicated that the carboxyl terminus of Npl3p is important for its nuclear import (Flach et al. 1994; Senger et al. 1998). Removal of amino acids 397–414 results in the failure of Npl3p to enter the nucleus (Senger et al. 1998). However, the amino acid sequence important for Npl3p nuclear targeting bears no similarity to the classical NLS.
MTR10 encodes one of several nuclear import receptors and, as such, promotes the nuclear import of Npl3p and other potential RNA-binding proteins (Pemberton et al. 1997; Senger et al. 1998; E.P. Lei and P.A. Silver, unpubl.). In fact, MTR10 was originally identified as a mutant defective in mRNA export (Kadowaki et al. 1994). Cells missing MTR10 accumlate Npl3p in the cytoplasm and show a simultaneous accumulation of poly(A)+ RNA in the nucleus. In contrast, npl3-27 cells, which also have cytoplasmic Npl3p, do not have a pronounced mRNA export defect. Possible explanations for this apparent paradox are as follows: (1) there still might be sufficient Npl3p moving in and out of the nucleus to sustain mRNA export in npl3-27; and/or (2) additional mRNA-binding proteins important for mRNA export may not be imported into the nucleus in mtr10 mutants.
Npl3p has been shown to interact directly with Mtr10p and amino acids 397–414 are important for this interaction, further confirming their function as an NLS (Senger et al. 1998). To reflect the decreased rate of Npl3-27p import because of a mutation in its NLS, we show that overexpression of Mtr10p can drive Npl3-27p into the nucleus. Taken together, these data indicate that Npl3-27p is still capable of being exported from the nucleus but is slowed in its reimport, thus resulting in a net accumulation of protein in the cytoplasm but not a block in mRNA export.
Why can cells still survive normally with so much Npl3-27p in the cytoplasm? One interpretation is that at any given time there is still enough Npl3p in the nucleus to package mRNAs and escort them into the cytoplasm. Similarly, in mtr10Δ cells, Npl3p is localized mainly to the cytoplasm, but the cells are still viable (Pemberton et al. 1997; Senger et al. 1998). It is possible that there is a second importer for Npl3p that has reduced affinity for its cargo, thus resulting in reduced nuclear uptake. However, when MTR10 is deleted from npl3-27 cells, they can no longer survive, suggesting that any backup import system is not sufficient when the NLS is mutated.
The mechanism by which most hnRNPs are exported from the nucleus to the cytoplasm remains an open question. Our data suggest that in yeast there may be an interplay between the export of Npl3p and the function of the NES nuclear export receptor encoded by XPO1. However, our failure to demonstrate a direct interaction between Npl3p and Xpo1p (data not shown) leaves open the possibility that these effects may be indirect.
In higher eukaryotes, the role of the Xpo1p homolog exportin in mRNA export remains ambiguous. On one hand, treatment with the exportin inhibitor leptomycin B results in a block in NES-dependent export but not in mRNA export in Xenopus oocyctes (Fischer et al. 1995; Fornerod et al. 1997). Moreover, overexpression of the Nup214 carboxyl terminus in COS cells causes a coincident relocalization of exportin to the nucleoplasm and a block in NES-dependent export, but no block in mRNA export (Bogerd et al. 1998). In contrast, others report inhibition of mRNA export when certain NESs are injected at high concentration into the Xenopus oocyte nucleus (Pasquinelli et al. 1997).
Npl3p is uncoupled from mRNA when cells are exposed to stress
Eukaryotic cells respond to stress in a number of ways, including the retention of mRNA. To make the response to stress as efficient as possible, mRNAs encoding heat shock proteins are rapidly transcribed and exported from the nucleus to the cytoplasm for translation (Saavedra et al. 1996). At the same time, there is a shutdown in the export of bulk mRNAs, presumably to allow for the selective and rapid translation of the heat shock mRNAs (Saavedra et al. 1996; Tani et al. 1996). One outstanding question is how the cell executes this rapid switch in RNA export.
We now present the novel finding that the major yeast hnRNP protein, Npl3p, is no longer mRNA associated under conditions of stress that result in a general block in poly(A)+ RNA nuclear export. The first indication for the dissociation of Npl3p from mRNA came from our observations of the behavior of Npl3-27p in xpo1-1 and rat7-1 mutants, in which at the restrictive temperature both Npl3-27p and mRNA remain in the nucleus. However, when salt or ethanol is then added to these shifted mutants, Npl3-27p is exported, whereas the bulk of the mRNA remains nuclear. These findings regarding the behavior of Npl3p can be interpreted in at least three different ways: Under conditions of stress, Npl3p could (1) leave the nucleus together with heat shock mRNAs, (2) fail to re-enter the nucleus, and/or (3) dissociate from the majority of the mRNA and redistribute independently.
Whether Npl3p leaves the nucleus together with heat shock mRNAs was tested by use of mutants that are defective in heat shock mRNA export, rat7-1 and rip1Δ. At the nonpermissive temperature, heat shock mRNA export was blocked, whereas Npl3-27p export was not affected, indicating movement of Npl3p independent of heat shock mRNAs. This finding is further supported by the fact that heat shock mRNA export is not blocked in npl3 mutants in which poly(A)+ RNA export is severely slowed (Saavedra et al. 1997).
Blockage of Npl3p nuclear import following stress is contraindicated by the mostly nuclear localization of wild-type Npl3p at all times. Could the accumulation of mRNA during stress be due to a reduced rate of Npl3p import? If cytoplasmic Npl3p is sufficient to block mRNA export, we would also expect to see trapped mRNA in the npl3-27 strain, in which much of the protein is cytoplasmic. The cytoplasmic localization of mRNA in npl3-27 strains at 25°C and 37°C argues that the cytoplasmic appearance of Npl3p is not sufficent for the accumulation of mRNA during stress.
Failure of Npl3p to maintain its mRNA association following stress is perhaps the simplest explanation. In this way, any protein no longer bound to mRNA would appear where it is localized at steady state, that is, wild-type protein mostly in the nucleus and much of Npl3-27p in the cytoplasm. Support for this view comes from the results of our UV cross-linking experiments. Although wild-type Npl3p was detected bound to mRNA in unstressed cells, association was greatly reduced in heat-shocked and salt-stressed cells. Thus, stress may lead to the dissociation of Npl3p and mRNA.
Whereas mRNA is most likely always protein bound, the composition of the RNA–protein complex is probably dynamic. For example, proteins important for holding the mRNA in the correct conformation for splicing and transport might come on and off in a temporal manner. Once bound to the mRNA, proteins such as Npl3p might further promote its interaction with the export machinery. From our findings of the dynamic behavior of Npl3p during stress conditions, we suggest a model in which Npl3p (and perhaps other proteins) dissociates from the RNP, leaving an export-incompetent complex behind (Fig. 7). The biological consequences of this dissociation and the resulting block in mRNA export could then be to allow the cell to cope with stress, for example by reducing the general traffic at the nuclear pore. The transport machinery and nuclear pores would then be free for the necessary rapid export of heat shock mRNAs.
Figure 7.
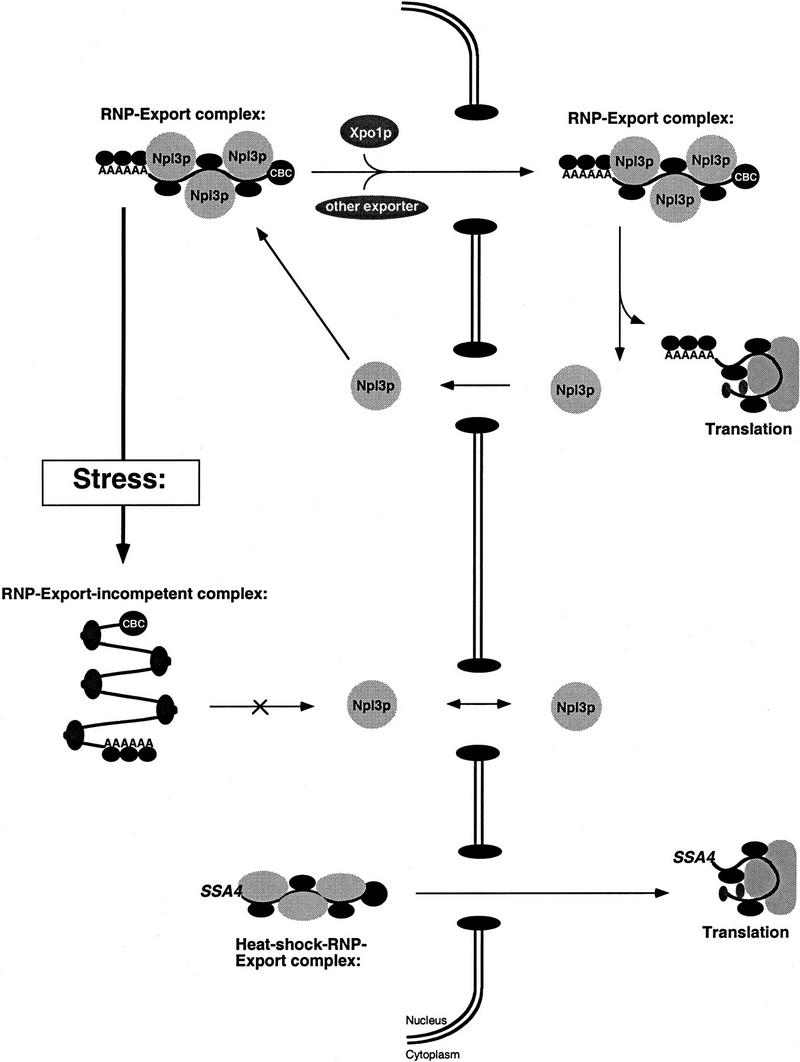
Model for RNP export under normal conditions and during stress. According to this model, mRNAs are packaged into export complexes by proteins such as Npl3p, which may interact with the as yet to be defined mRNA export machinery. Under conditions of stress, Npl3p dissociates from the RNP leaving it incompetent for export.
Materials and methods
Plasmid constructs
All plasmid constructs used in this study are listed in Table 1. To integrate npl3-27 into the genome, a 4.5-kb ClaI fragment from pPS873 containing npl3-27 was ligated into ClaI-linearized integration vector pPS750 (Sikorski and Hieter 1989).
Table 1.
Plasmids used in this study
| Plasmid
|
Features
|
Source
|
|---|---|---|
| pPS750 | CEN URA3 yeast integration vector | Sikorski and Hieter (1989) |
| pPS873 | 4.2-kb ClaI fragment containing npl3-27 in YCp50 LEU2 | this study |
| pPS878 | 4.2-kb ClaI fragment from pPS873 into pPS750 URA3 | this study |
| pPS1164 | 3.5-kb ScaI fragment of NPL3 in YEp352 URA3 (pMH7) | Lee et al. (1996) |
| pPS1370 | chromosomal sequence containing MTR10 in YEp24 URA3 | A.M. Tartakoff (Case Western Reserve University School of Medicine, Cleveland, OH) |
| pPS1743 | chromosomal sequence containing SSA4 in 2μ LEU2 | C. Cole (Dartmouth Medical School, Hanover, NH) |
| pPS1745 | chromosomal sequence containing SSA4 in 2μ URA3 | C. Cole |
Yeast strains
Growth and maintenance of yeast strains, as well as all genetic manipulations were performed as described (Rose and Broach 1990). The strains used in this study are listed in Table 2. To integrate npl3-27 into the genome, the diploid strain PSY1121 was transformed with the integration vector pPS878 linearized within npl3-27 with SexAI. After selection on plates lacking uracil, single colonies were streaked to FOA to loop out one copy of NPL3. The diploid strain was sporulated, genomic DNA was prepared from the viable spores, and a part of the NPL3 gene was amplified with the following primers: forward, 5′-GACAATCCTCCACCAATCAGAAGATCAAA-3′; reverse, 5′-TTATTCTATCTACTGGAGTGAAGTCG-3′, under standard PCR conditions. The resulting 705-bp fragment was analyzed for the loss of a PmlI site because the mutation (E409K) is based on a single nucleotide exchange (GAA to AAA) within this restriction site. In each tetrad, two spores had lost the PmlI site, indicating retention of the npl3-27 allele, whereas two spores retained the PmlI site, indicating retention of the wild-type NPL3 allele. In this way, the haploid strains PSY1031, 1032, 1033, and 1034 containing npl3-27 were obtained. Crosses of these strains with PSY653, 1105, 1169, sporulation of the resulting diploid stains, and subsequent tetrad analysis resulted in the double-mutant strains PSY1637, PSY1638, and PSY1639.
Table 2.
Strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| PSY580 | MATa ura3 leu2 trp1 | Winston et al. (1995) |
| PSY635 | MATα rat7-1 ura3 leu2 his3 | C. Cole |
| PSY814 | MATα npl3∷HIS3 ura3 leu2 his3 lys1 trp1 ade2 ade8 can1 + YCp50 NPL3 URA3 | Henry and Silver (1996) |
| PSY1031 | MATα npl3-27 ura3 leu2 trp1 lys1 his3 ade2 ade8 can1 | this study |
| PSY1032 | MATa npl3-27 ura3 leu2 lys1 his3 ade2 ade8 can1 | this study |
| PSY1033 | MATα npl3-27 ura3 leu2 lys1 his3 ade2 ade8 can1 | this study |
| PSY1034 | MATa npl3-27 ura3 leu2 trp1 lys1 his3 ade2 ade8 can1 | this study |
| PSY1105 | MATα xpo1∷LEU2 ura3 leu2 trp1 his3 ade2 can1 + pKW457 xpo1-1 HIS3 | Stade et al. (1997) |
| PSY1121 | MATa/α npl3∷HIS3/NPL3 ura3/ura3 leu2/leu2 trp1/TRP1 lys1/lys1 his3/his3 ade2/ade2 ade8/ade8 can1/can1 | Shen et al. (1998) |
| PSY1169 | MATα rip1∷HIS3 ura3 leu2 his3 | C. Cole |
| PSY1637 | MATα npl3-27 rat7-1 ura3 leu2 his3 ade− | this study |
| PSY1638 | MATα npl3-27 xpo1∷LEU2 ura3 lys1 his3 ade2 + pKW457 xpo1-1 HIS3 | this study |
| PSY1639 | MATα npl3-27 rip1∷HIS3 ura3 leu2 his3 | this study |
| PSY1670 | MATα mtr10∷HIS3 ura3 leu2 lys1 his3 ade− + pMTR10 URA3 CEN | this study |
| PSY1671 | MATa mtr10∷HIS3 npl3-27 ura3 leu2 trp1 lys1 his3 ade− + pMTR10 URA3 CEN | this study |
Indirect immunofluorescence
Cells were grown to 1 × 107 to 5 × 107 cells/ml at 25°C. Cells (5 ml) were then shifted to 37°C or 42°C in a shaking water bath for different time points, before 350 μl of 37% formaldehyde was added for 30 min to fix the cells. Cells were washed once in 0.1 m K2HPO4–KH2PO4 at pH 6.5 and once in P solution (0.1 m K2HPO4–KH2PO4 at pH 6.5, 1.2 m Sorbitol), spheroplasted with Zymolase (100T, ICN) at a concentration of 500 μg/ml, placed on slides coated with 0.3% polylysine, and permeabilized by addition of 0.5% Triton X-100 for 5 min. Antibodies against Npl3p were used at a 1:1000 dilution, those against Srp1p at a 1:2000 dilution, and a fluorescein isothiocyanate-conjugated anti-rabbit secondary antibody was used at a 1:1000 dilution (Lee et al. 1996). Nuclei were stained with 10 μg/ml DAPI as described in Lee et al. (1996).
In situ poly(A)+ RNA hybridization
Localization of poly(A)+ RNA by in situ hybridization was performed essentially as described (Amberg et al. 1992), with some modifications. Cells were fixed, washed, spheroplasted, and permeabilized as described for immunofluorescence. After incubation with a digoxigenin-labeled oligo[d(T50)] probe, cells were then processed for immunofluorescence as described in Lee et al. (1996), beginning with the antibody-blocking buffer step. A sheep anti-digoxigenin Fab-FITC antibody (Boehringer) was used at a 1:200 dilution.
In situ SSA4 RNA hybridization
Localization of SSA4 by in situ hybridization was performed as described for localization of poly(A)+ RNA, with the PCR-labeled probe described below at a final concentration of 1 ng/μl. To generate the SSA4 probe, PCR with the following primers (forward, 5′-ATGTCAAAAGCTGTTGGTATTGATTTAGG-3′; reverse, 5′-GGCCCTTTCAGCGGCGGTCC-3′) resulted in a 800-bp fragment, incorporating digoxigenin-11-dUTP (Boehringer Mannheim) into the product when used at 40 μm concentration. A signal was only obtained when SSA4 was overexpressed (pPS1743 or pPS1745). No signal was observed in stressed cells not carrying the plasmid (data not shown).
Stress conditions
Heat shock conditions used in this study were as follows: Cells were grown to 1 × 107 to 5 × 107 cells/ml and then incubated in a shaking water bath for 5, 10, 15, and 30 min at 42°C, before fixation. Both salt and ethanol shock were performed at the same cell density. However, cells were shifted to 37°C for 30 min before 0.4 m NaCl or 10% ethanol was added to a final concentration. Salt-shocked cells were incubated for 5, 10, and 15 min at 37°C before fixation. Ethanol-shocked cells were further incubated for 30 and 60 min at 37°C, before fixation. Cultures (10 ml) were grown under the same conditions to 2 × 107 cells/ml. Cyclohexamide was added before the temperature shift at a final concentration of 10 μg/ml. Cells were then lysed in 50 μl of SDS-PAGE sample buffer. Five microliters of these extracts were separated on a 12% SDS-polyacrylamide gel before Western blotting. α-Npl3p was used at a 1:1000 dilution.
UV cross-linking of Npl3p to poly(A)+ RNA
Isolation of UV-cross-linked poly(A)+ RNA–RNP complexes and analysis of cross-linked Npl3p by SDS-PAGE and Western blotting were done essentially as described (Anderson et al. 1993). A total of 1.5 liters of the wild-type strain PSY580 was grown to OD600 of 1.0. Cells were harvested, washed twice in PBS, and split in three equal portions. The first portion was not UV-irradiated, the second portion was UV-irradiated for 2 × 2.5 min on ice in the Stratalinker (Stratagene). The third portion was treated with 0.4 m NaCl for 10 min before it was UV-irradiated and treated as portion two. For the heat shock, cells were incubated at 42°C for 15 min. All samples were washed in PBS and lysed in lysis buffer. The poly(A)+ RNA was purified and the proteins analysed as described previously (Anderson et al. 1993; Russell and Tollervey 1995). All eluate samples were loaded normalized to the purified RNA and 10 OD260 units were loaded per lane.
Acknowledgments
We thank the following people for generously providing plasmids and yeast strains: C. Cole for the plasmids encoding SSA4 and the mutant strains of RAT7 and RIP1, A.M. Tartakoff for the plasmid encoding Mtr10p, and K. Weis for the mutant strain of XPO1. We also thank H. Bastians, A. McBride, and E. Shen for helpful discussion and P. Ferrigno, J. Hood, E. Lei, and T. Zimmerman for reading the manuscript. H.K. was supported by a fellowship from the Deutsche Forschungsgemeinschaft, and T.T. was supported by fellowships from the Japanese Society for the Promotion of Science and The Uehara Memorial Foundation. M.S.L. was supported by a fellowship of the Medical Foundation. This work was funded by grants from the National Institutes of Health to P.A.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pamela_silver@dfci.harvard.edu; FAX (617) 632-5103.
References
- Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: A yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Echarri A, Ross TM, Cullen BR. Inhibition of human immunodeficiency virus rev and human T-cell leukemia virus rex function, but not mason-pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie MA, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Ellis EM, Reid GA. The Saccharomyces cerevisiae MTS1 gene encodes a putative RNA-binding protein involved in mitochondrial protein targeting. Gene. 1993;132:175–183. doi: 10.1016/0378-1119(93)90193-7. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins DA, Silver PA. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: The Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+ RNA-binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JK, Silver PA. Cse1p is required for export of Srp1p/importin-{alpha} from the nucleus in Saccharomyces cerevisiae. J Biol Chem. 1998;273:35142–35147. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzler M, Hurt EC. Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS LETT. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lee MS, Silver PA. RNA movement between the nucleus and the cytoplasm. Curr Opin Genet Dev. 1997;7:212–219. doi: 10.1016/s0959-437x(97)80131-1. [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Fischer U, Michael WM, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: The last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Powers MA, Lund E, Forbes D, Dahlberg JE. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc Natl Acad Sci. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- Russell ID, Tollervey D. Yeast Nop3p has structural and functional similarities to mammalian pre-mRNA binding proteins. Eur J Cell Biol. 1995;66:293–301. [PubMed] [Google Scholar]
- Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes & Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat-shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes & Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Lührmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes & Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Bischoff FR, Schlenstedt G. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Tani T, Derby RJ, Hiraoka Y, Spector DL. Nucleolar accumulation of poly (A)+ RNA in heat-shocked yeast cells: Implication of nucleolar involvement in mRNA transport. Mol Biol Cell. 1996;7:173–192. doi: 10.1091/mbc.7.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson MS. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupera-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]