Abstract
During human CMV infection, there is a preferential expansion of natural killer (NK) cells expressing the activating CD94–NKG2C receptor complex, implicating this receptor in the recognition of CMV-infected cells. We hypothesized that NK cells expanded in response to pathogens will be marked by expression of CD57, a carbohydrate antigen expressed on highly mature cells within the CD56dimCD16+ NK cell compartment. Here we demonstrate the preferential expansion of a unique subset of NK cells coexpressing the activating CD94–NKG2C receptor and CD57 in CMV+ donors. These CD57+NKG2Chi NK cells degranulated in response to stimulation through their NKG2C receptor. Furthermore, CD57+NKG2Chi NK cells preferentially lack expression of the inhibitory NKG2A receptor and the inhibitory KIR3DL1 receptor in individuals expressing its HLA-Bw4 ligand. Moreover, in solid-organ transplant recipients with active CMV infection, the percentage of CD57+NKG2Chi NK cells in the total NK cell population preferentially increased. During acute CMV infection, the NKG2C+ NK cells proliferated, became NKG2Chi, and finally acquired CD57. Thus, we propose that CD57 might provide a marker of “memory” NK cells that have been expanded in response to infection.
Keywords: innate immunity, lymphocyte
Natural killer (NK) cells are a subset of lymphocytes comprising 5% to 20% of peripheral blood mononuclear cells (PBMCs) in humans (1). NK cells participate in the innate immune response and play an important role in the defense against viral infections and tumor surveillance, and are also involved in shaping adaptive immune responses through their production of cytokines (2). Humans lacking NK cells are susceptible to certain infections, principally to life-threatening CMV infections (3) and other herpesviruses (4, 5).
In humans, two NK cell subsets have been characterized: CD56dimCD16+ NK cells represent approximately 90% of circulating NK cells and are considered the mature NK subset; CD56brightCD16neg/dim NK cells constitute approximately 10% and are considered immature (1, 6). NK cell function is regulated by a diverse array of inhibitory and activating receptors, including killer cell Ig-like receptors (KIRs), NKG2A, NKG2D, and natural cytotoxic receptors, which define functionally distinct subsets of NK cells within the total population. When activated, NK cells produce IFN-γ and other cytokines and kill susceptible target cells (7). NK cells preferentially recognize pathogen-infected cells, tumor cells, and stressed cells that express ligands for the activating NK receptors or cells that have down-regulated MHC class I molecules, which are ligands for NK inhibitory receptors (7).
CMV is often acquired in childhood. The primary infection is usually subclinical or mild, but it may cause a severe embryopathy during pregnancy or a lethal infection in neonates (8). Most humans are infected with CMV (50–100% depending on geographical location), and remain infected for life (9). In adults, severe disease occurs in immunocompromised subjects. The importance of NK cells in immunosurveillance against CMV is revealed by the multiple evasion strategies evolved by the virus to escape T and NK cell recognition (2, 10). Indeed, several CMV genes specifically down-regulate the surface expression of ligands for the activating receptors NKG2D and DNAM-1 that are induced by infection (2, 10–13). NKG2D, DNAM-1, and NKp46 have been implicated in recognition of CMV-infected cells (12, 14, 15). CMV also encodes a MHC class I homologue, UL18, which is recognized by the inhibitory LIR-1 (ILT2, CD85j, or LILRB1) receptor (10, 16, 17). UL18 also activates some NK cells by using an unknown receptor (17, 18).
CD94 forms an inhibitory receptor by disulfide bonding to the ITIM-bearing NKG2A subunit, but also forms an activating receptor by disulfide bonding to NKG2C and associating with the ITAM-bearing DAP12 protein (19). Both CD94–NKG2A and CD94–NKG2C receptors bind HLA-E, but NKG2C binds with a lower affinity than NKG2A (20, 21). Epidemiological studies have shown that a subset of NKG2C+ NK cells is increased in CMV-seropositive healthy individuals and aviremic HIV-1+ patients (22–24). NKG2C+ NK cells expanded when cocultured with CMV-infected fibroblasts in vitro (25). These observations suggest that NKG2C+ NK cells respond to CMV infection. This is supported by a report showing expansion of NKG2C+ NK cells in a SCID child infected with CMV (26).
Recent studies have shown that NK cells share features with B and T cells of the adaptive immune system, and indicate that mouse NK cells can acquire immunological memory after viral infection (27–29). In a mouse model of CMV infection, a specific population of NK cells expands, contracts after control of the virus, and generates long-lived “memory” NK cells that are more protective during a second encounter with this pathogen (28), raising the question of whether humans also possess virus-specific memory NK cells. In humans, CD57 is expressed preferentially by a subset of NK cells with a mature phenotype (30–32), suggesting that CD57 might mark NK cells that have been clonally expanded by infections (33). CMV infection is the most common viral complication after solid-organ transplantation (SOT) (34, 35). Therefore, in this study, we examined how NK cells coexpressing NKG2C and CD57 respond to CMV infection in SOT recipients.
Results
Unique NK Cell Population Is Detected in CMV+ Healthy Subjects.
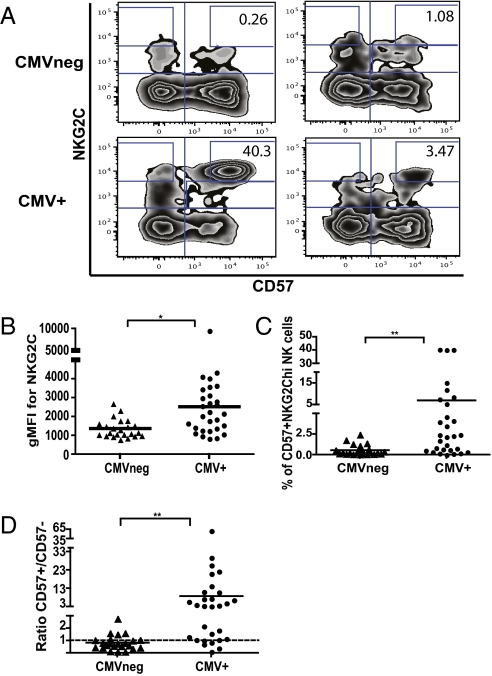
NK cells expressing NKG2C are increased in CMV+ blood donors, suggesting that this activating receptor is involved in the response against CMV (22). CD57 defines a subset of mature NK cells within the CD56dimCD16+ population (31, 32). We hypothesized that the subset of NK cells that has expanded in response to CMV will preferentially express CD57. We tested this hypothesis by examining the NK cell receptor repertoire within the CD57− and CD57+ NK cell subsets of CMV-seronegative and CMV+ donors, excluding immature CD56bright cells (Fig. S1A). No significant differences of expression between the CMV-seropositive and CMV-seronegative donors were detected for most of the NK receptors (e.g., NKG2D, natural cytotoxic receptors, and KIRs), activation, and maturation markers (e.g., CD69, CD62L, HLA-DR, and CD38) tested, even for receptors previously implicated in CMV recognition (NKG2D, DNAM-1, NKp46) (12, 14, 15, 24, 26). However, we observed a significant increase in NKG2C+ NK cells preferentially coexpressing CD57 in CMV+ donors (Fig. S1B). A unique population of CD57+NKG2Chi NK cells was detected in 23 of 28 CMV-seropositive donors, but was absent in CMV-seronegative donors (Fig. 1A) with the exception of two individuals. Of the 24 CMV-seronegative donors tested, in one donor, approximately 7.6% of CD56dimCD16+ NK cells were CD57+NKG2Chi; however, when stimulated with pp65 CMV peptides, we detected IFN-γ production by their T cells, demonstrating that, although seronegative, this individual had been exposed to CMV. The other individual within the CMV-seronegative cohort had 2.5% CD57+NKG2Chi NK cells without a detectable B- or T-cell response to CMV, possibly because of an NK cell, but not B- or T-cell, response to CMV infection or potentially in response to another pathogen. Of note, the cell surface density of NKG2C was increased specifically on the CD57+ NK cells from CMV+ (seropositive and/or CMV-specific T-cell response) compared with CMV− donors (Fig. 1B). The percentage of CD57+NKG2Chi cells varied between CMV+ donors (Fig. 1 A and C). The time of primary infection, as well as possible subclinical reactivation, is not known; therefore, the differences in frequencies might reflect the time since the NK cells last encountered virus. Finally, 22 of 29 CMV+ donors had a ratio of CD57+NKG2Chi/CD57−NKG2Chi greater than 1, whereas only four donors in the CMV− group had a ratio greater than 1 (Fig. 1D).
Fig. 1.
A unique population of CD57+NKG2Chi NK cells is detected in CMV+ donors. (A) CD3negCD56dimCD16+ NK cells from four CMV− (Upper) and CMV+ (Lower) donors were gated and analyzed for expression of CD57 and NKG2C. (B) The geometric mean fluorescence intensity (gMFI) for NKG2C in the CD57+ NK cell subset from CMV− and CMV+ donors is shown. (C) The percentage of CD57+NKG2Chi cells in the CD56dimCD16+ NK cell subset from CMV+ and CMV− donors is shown. (D) The ratio of CD57+NKG2Chi/CD57−NKG2Chi NK cells for each CMV− and CMV+ donors is shown. Triangles represent CMV− donors (n = 23), circles represent CMV+ donors (i.e., seropositive and/or CMV-specific T-cell responses; n = 29).
Activation of CD57+NKG2Chi NK Cells via NKG2C.
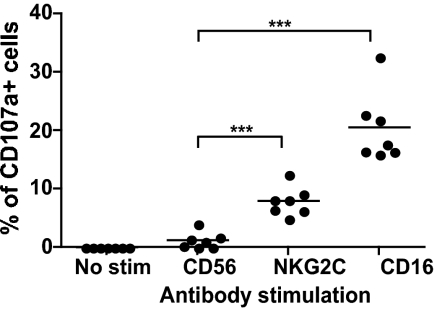
To determine whether the CD57+NKG2Chi NK cells are functional, PBMCs were stimulated with anti-CD56 (negative control), anti-NKG2C, or anti-CD16 (positive control) and then stained to identify the different NK subsets. CD16 crosslinking induced degranulation of approximately 20.5% of CD57+NKG2Chi NK cells (Fig. 2), and this was similar to the response detected in the other cells within the CD56dim NK subset. Moreover, NKG2C crosslinking specifically induced degranulation of approximately 7.9% of the CD57+NKG2Chi NK cells (Fig. 2).
Fig. 2.
CD57+NKG2Chi NK cells from CMV+ donors degranulated when activated through NKG2C. PBMCs from CMV+ donors where stimulated with plate-bound antibodies against CD56 (negative control), NKG2C, or CD16 (positive control). The percentage of CD107α+ cells was determined for CD57+NKG2Chi NK cells (n = 7).
Fewer Inhibitory NKG2A and KIR3DL1 Receptors on CD57+NKG2Chi NK Cells.
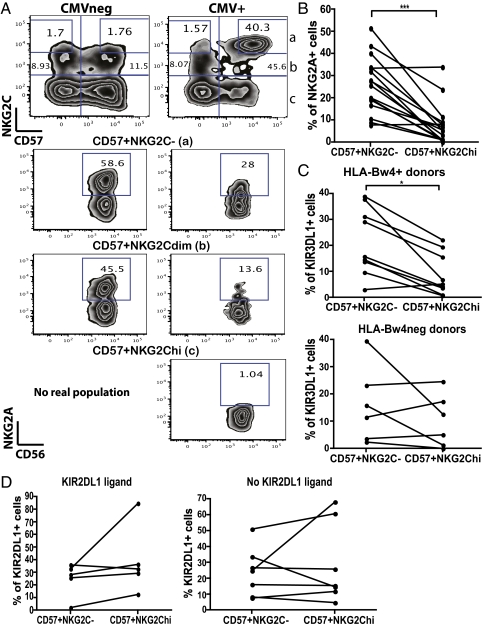
NK cells in mice that specifically respond to CMV preferentially lack inhibitory receptors for self-MHC class I ligands (36). Therefore, we compared expression of inhibitory receptors for HLA class I on the CD57+NKG2Chi and CD57+NKG2C− NK cells in the CMV+ individuals, which should be at a similar maturation stage. NKG2A, the inhibitory counterpart of NKG2C (20, 21), was preferentially decreased on the CD57+NKG2Chi NK cells from CMV+ donors (Fig. 3 A and B). The percentage of CD57+NKG2Chi NK cells expressing NKG2A was very low compared with CD57+NKG2C− and even CD57+NKG2Cdim NK cells in the same donor (Fig. 3A). By contrast, NKG2A was present on the CD57+NKG2Cdim NK cells present in CMV− donors (Fig. 3A). This preferential absence of NKG2A on the CD57+NKG2Chi NK cells was observed in most CMV+ donors examined (14 of 16; P < 0.0005; Fig. 3B). This suggests that most of the NKG2C+ NK cells that have acquired CD57 in the CMV+ donors were NKG2A− or had down-regulated NKG2A expression.
Fig. 3.
Certain inhibitory NK receptors are underrepresented on the CD57+NKG2Chi NK cell subset from CMV+ donors. (A) CD56dimCD16+ NK cells from CMV− (Left) and CMV+ (Right) donors were gated and analyzed for expression of CD57 and NKG2C. CD57+NKG2C− (a), CD57+NKG2Cdim (b), and CD57+NKG2Chi (c) NK cells were assessed for CD56 and NKG2A expression. (B) The percentage of NKG2A+ cells was determined on CD57+NKG2C− and CD57+NKG2Chi NK cells from CMV+ donors (n = 16). (C) The percentage of KIR3DL1+ cells was determined on CD57+NKG2C− and CD57+NKG2Chi NK cells from CMV+ donors. Graphs from HLA-Bw4+ (Upper; n = 9) and HLA-Bw4− donors (Lower; n = 6) are shown. (D) The percentage of KIR2DL1+ cells was determined on CD57+NKG2C− and CD57+NKG2Chi NK cells for CMV+ donors who have the KIR2DL1 ligand (Left; n = 5) or do not (Right; n = 7).
We also examined the expression of inhibitory KIRs that have well characterized HLA class I ligands. KIR3DL1 recognizes HLA-Bw4 (37, 38). We compared the frequency of KIR3DL1+ NK cells within the CD57+NKG2Chi and CD57+NKG2C− subsets in individuals who were HLA-Bw4+ (i.e., possess a self-ligand for KIR3DL1) or HLA-Bw4−. KIR3DL1 was selectively under-represented in the CD57+NKG2Chi NK cells in eight of nine individuals bearing HLA-Bw4 (P < 0.05; Fig. 3C). By contrast, the frequency of KIR3DL1+ cells in the CD57+NKG2Chi subset was not significantly skewed in four of six CMV+ donors lacking HLA-Bw4 (Fig. 3C). The frequency of NK cells expressing KIR2DL1, which recognizes HLA-C (39, 40), was not significantly skewed in CD57+NKG2Chi NK cells from CMV+ donors that had or lacked a KIR2DL1 ligand (Fig. 3D). Thus, the expression of some inhibitory receptors, NKG2A and KIR3DL1, was specifically underrepresented in CD57+NKG2Chi NK cells in CMV+ donors that expressed their respective ligands.
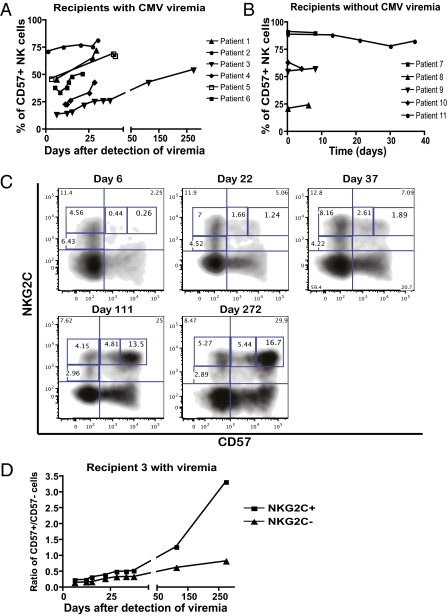
CD57+NKG2Chi NK Cells Selectively Expand During Acute CMV Infection.
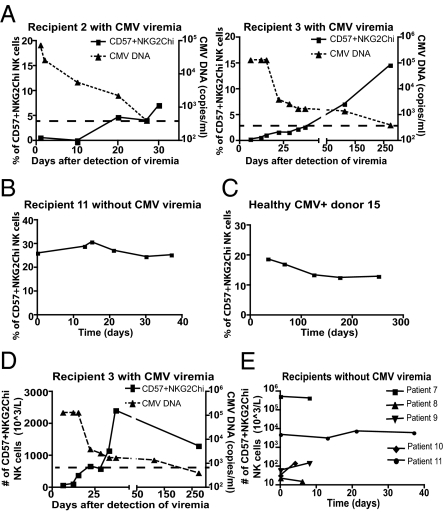
To determine if CD57+NKG2Chi NK cells preferentially respond to CMV, we longitudinally followed patients with acute CMV infection. Immunosuppressive protocols used in SOT recipients target the activation of T cells and impair specific antiviral immunity (41). NK cell function appears to be less affected by such therapeutic regimens (42–44), allowing us to study NK cell response to CMV. We monitored the NKG2C+ NK cells from SOT recipients with CMV viremia during their hospital stay (Table S1). In all these patients, a significant number of CD57+NKG2Chi NK cells appeared within 1 to 2 wk after detection of CMV viremia (Fig. 4A). The frequency of CD57+NKG2ChiNK cells increased within the total CD56dimCD16+ NK population as the viral load decreased after initiation of antiviral drug therapy (Fig. 4A). We compared these patients with SOT recipients with previous CMV infection without CMV viremia during the study (Table S1). In patients without viremia, the percentage of CD57+NKG2Chi NK cells remained stable (Fig. 4B), suggesting that the increase in CD57+NKG2Chi NK cells observed in the viremic patients was specific to acute CMV infection. This difference of behavior was statistically significant: the percentage of CD57+NKG2Chi NK cells increased in SOT recipients with viremia (delta mean ± SEM, 6.04 ± 2.13; n = 6) and not in patients without viremia (delta mean ± SEM, −1.848 ± 0.928; n = 5; P = 0.0116). CD57+NKG2Chi NK cells were not detected in a CMV− SOT recipient (Table S1), and did not increase in a healthy CMV+ blood donor who was monitored for 9 mo (Fig. 4C).
Fig. 4.
CD57+NKG2Chi NK cells increase during acute CMV infection in SOT recipients. (A) NKG2C and CD57 expression on CD56dimCD16+ NK cells from SOT recipients with acute CMV infection was analyzed. Antiviral treatment was administrated immediately after detection of viremia (day 0). The percentage of CD57+NKG2Chi NK cells detected over time and the CMV viral load detected in plasma (value > 400 copies/mL) from representative patients are shown (n = 6). Solid line and square represent CD57+NKG2Chi NK cells, dashed line and triangle represent CMV DNA. (B and C) The percentages of CD57+NKG2Chi NK cells detected over time from one representative SOT recipient without CMV viremia (B; n = 5) and from a healthy CMV+ donor (C) are shown. (D and E) Approximate absolute numbers (in 103 cells/L) of CD57+NKG2Chi NK cells detected over time in one representative SOT recipient with CMV viremia (D; CMV viral load shown; n = 6) and in SOT recipients without CMV viremia (E; n = 5).
In mice, Ly49H+ NK cells specifically expand during CMV infection, contract after control of the virus, and generate memory NK cells that can be detected months later (28). We addressed if we could detect expansion and contraction of a specific subset of human NK cells responding to CMV infection. In SOT recipients, CD57+NKG2Chi NK cells increased in numbers as CMV viremia decreased (Fig. 4D), and they began to contract after control of the infection. Thus, both the absolute number and the percentage of CD57+NKG2Chi NK cells increased after viremia, and after resolution of the infection the CD57+NKG2Chi NK cells persisted at a higher frequency, suggesting their preferential survival or slower contraction than other NK cells. We could not determine when the CD57+NKG2Chi NK cell subset stabilized because blood samples were not available after resolution of the infection. In contrast, in SOT recipients without CMV viremia, the numbers of CD57+NKG2Chi NK cells did not vary significantly over time (Fig. 4E).
NKG2C+ NK Cells Acquire CD57 After Acute CMV Infection.
The percentage of CD57+ cells within the CD56dimCD16+ subset increased in SOT recipients during CMV viremia (Fig. 5A); however, it did not change over time in patients without CMV viremia (Fig. 5B) or in healthy CMV+ donors who were monitored for approxiately 1 y. The frequency of CD57+ NK cells increased within both NKG2C+ and NKG2C− subsets after viremia (Fig. 5C), but there was preferential expansion of the NKG2C+ NK cells within the total NK cell population. Of note, NKG2C+ NK cells were predominantly CD57− just after the detection of viremia, but over time, they increased the amount of NKG2C on their cell surface and acquired CD57 expression, becoming CD57dimNKG2Chi, and then CD57+NKG2Chi (Fig. 5C). This suggests that NKG2C+ NK cells that were responding to CMV acquired CD57 after clonal expansion. If NKG2C+ NK cells were the NK cells mainly responding to CMV, they should preferentially acquire CD57. Indeed, when we analyzed changes in the ratio of CD57+/CD57− cells on NKG2C+ versus NKG2C− NK cells, the ratio increased faster and higher in the NKG2C+ NK cell subset (Fig. 5D). This indicates that NKG2C+ NK cells preferentially acquired CD57 during the course of infection in SOT recipients with viremia.
Fig. 5.
During CMV acute infection, NKG2C+ NK cells acquire CD57 expression and become CD57+NKG2Chi NK cells. (A and B) The percentages of CD57+ NK cells detected over time in SOT recipients with acute CMV infection (A; n = 6) or without CMV viremia (B; n = 5) are shown. (C) CD56dimCD16+ NK cells were analyzed for expression of CD57 and NKG2C. Plots for different time points from a representative SOT recipient with CMV viremia are shown (n = 6). Population gates: CD57−NKG2Cdim, CD57−NKG2Chi, CD57dimNKG2Chi, and CD57+NKG2Chi NK cells. (D) CD56dimCD16+ NK cells were gated on NKG2C+ and NKG2C− cells and the percentage of CD57+ cells for each subset was assessed over time. The ratio of CD57+/CD57− cells on each subset from a representative SOT recipient with CMV viremia is shown (n = 6). Solid square is from NKG2C+, solid triangle is from NKG2C− NK cells.
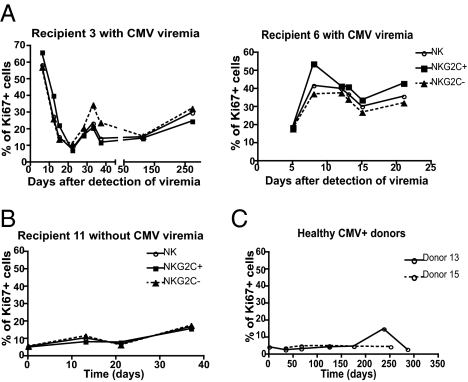
NK Cells Proliferate in Response to Acute CMV Infection.
A high percentage of NK cells in all SOT recipients were Ki-67+ (between 30% and 70% depending on the donor; Fig. 6A) just after detection of CMV viremia, indicating that NK cells were highly dividing at that time. After control of CMV, a lower percentage of NK cells continued to proliferate (Fig. 6A). There was no significant difference in Ki-67 detected between NKG2C+ and NKG2C− NK cells (Fig. 6A) or between CD57+ and CD57− NK cells (Fig. S2). To confirm that the NK cells divided in response to the acute CMV infection, we stained NK cells for Ki-67 expression in SOT recipients without CMV viremia and in healthy CMV+ donors. In both cases, few NK cells divided [∼5–15% (Fig. 6B) and <5% (Fig. 6C), respectively], and this remained stable over time.
Fig. 6.
All NK cells divide in response to acute CMV infection. (A and B) CD56dimCD16+ NK cells were gated on NKG2C+ and NKG2C− cells, and Ki-67 expression on all CD56dimCD16+ NK cells and in each subset was assessed. Graphs from representative SOT recipients with CMV viremia (A; n = 4) and without CMV viremia (B) are shown. Solid line and empty circle represent CD56dimCD16+ NK cells, solid line and square represent NKG2C+ NK cells, and dashed line and triangle represent NKG2C− NK cells. (C) The percentage of Ki-67+ cells within the CD56dimCD16+ NK cells detected over time in healthy CMV+ donors is shown.
Discussion
Here, we demonstrate that CD57+NKG2Chi NK cells preferentially respond to acute CMV infection and can be detected in CMV+ healthy adults years after the primary infection. Our findings confirm and extend previous observations, in particular demonstrating formally that these NKG2C+ NK cells preferentially expand during acute CMV infection and up-regulate cell surface density of NKG2C and acquire CD57. Similarly, we previously documented an increase in the cell surface density of the CMV-specific Ly49H receptor on memory NK cells that were expanded after infection with mouse CMV (28).
The ligand on CMV-infected cells recognized by NKG2C+ NK cells has not been identified. CD94–NKG2C receptor binds HLA-E, but CD94–NKG2C binds with a lower affinity than CD94–NKG2A, its inhibitory counterpart (20, 21). The difference in the affinity of the two receptors depends largely on the peptide bound to HLA-E (45, 46). NKG2C+ NK cells can proliferate in the presence of IL-15 and HLA-E–expressing targets in vitro (47). NKG2C recognition of CMV-infected cells might depend on HLA-E associated with peptides induced during the infection. Whereas classical MHC class I molecules are down-regulated, HLA-E expression is up-regulated by CMV (48–50). Induction of HLA-E surface expression by CMV protein gpUL40 inhibits NK cell-mediated lysis by interacting with NKG2A (51, 52), but might allow recognition by NKG2C+ NK cells. Future studies are needed to determine if NKG2C directly recognizes CMV-infected cells and if this recognition is HLA-E–dependent. Nonetheless, our studies documenting preferential expansion of NKG2C+ NK cells during acute CMV infection and in CMV+ healthy individuals (22–24), as well as the finding that CMV-infected fibroblasts induced proliferation of NKG2C+ NK cells in vitro (25), strongly suggest that NKG2C is involved in the response to CMV. Moreover, a higher percentage of NKG2C+ NK cells was associated with a lower rate of acute CMV infection in SOT recipients (53). This hypothesis is also supported by the following observations: (i) CD57+NKG2Chi NK cells were observed in CMV+ and not in CMV− healthy individuals, independently of infection by other common pathogens; (ii) NKG2C+ NK cells, specifically CD57+NKG2Chi NK cells, increased as the CMV viremia decreased in SOT recipients; (iii) CD57+NKG2Chi NK cells were not detected in a CMV− SOT recipient; and (iv) CD57+NKG2Chi NK cells were detected and remained stable over time in CMV+ SOT recipients with no viremia.
In mice, all NK cells proliferate early after CMV infection in response to the cytokines induced by the infection; thereafter, CMV-specific Ly49H+ NK cells preferentially continue to proliferate and increase in frequency (28, 54). When measuring Ki-67 in NK cells from SOT recipients with CMV acute infection, we observed that all NK cells, including CD57+NKG2Chi NK cells, proliferated. The fact that a high percentage of CD56dimCD16+ NK cells, independently of NKG2C expression, were proliferating, suggests that it was probably mainly in response to cytokines induced by the infection. However, Ki-67 expression does not reflect how many rounds of division a cell has undergone; therefore, we cannot directly measure the extent of proliferation in the NKG2C− versus NKG2C+ NK cell subsets. Further studies are needed to determine if NKG2C+ NK cells not only proliferate in responce to cytokines but also specifically to CMV-infected cells, as suggested by their specific expansion during CMV acute infection in SOT recipients.
CD57+ T cells are considered to be senescent (55–57), but this is controversial (58, 59). We and others reported that CD57+ NK cells have a proliferation defect in vitro (31, 32), and after human stem cell transplantation (31). Even if, in some patients, we could observe a gradually less frequent Ki-67 expression on subsets in the order CD57−NKG2C+ > CD57−NKG2C− > CD57+NKG2C+ > CD57+NKG2C− after detection of CMV viremia, there was no statistically significant difference in the percentage of Ki-67+ cells between NK cells from the CD56dimCD16+ subset depending on their NKG2C or CD57 expression. This suggests that CD57+ NK cells likely proliferate in vivo in response to viral infection and that CD57 may not reflect senescence in NK cells, in agreement with another recent study (59). However, it has been reported in rhesus macaques that Ki-67 staining indicates that a T cell has undergone DNA synthesis in the previous 3 to 4 d (60). It is not known how long Ki-67 can be detected after DNA synthesis in human NK cells, but these previous observations suggest that it marks cells that have divided in the past days, and thus it is possible that CD57 expression might be acquired on NK cells after division. Further studies are needed to determine if CD57+ NK cells have a proliferation defect in vivo, and if this defect is abolished or less pronounced during acute viral infection.
CD57+NKG2Chi NK cells increased in percentage and in numbers in the blood of SOT recipients shortly after detection of CMV viremia, and when the viremia had been controlled, CD57+NKG2Chi NK cells contracted. The number of CD57+NKG2Chi NK cells decreased, but the percentage of this subset remained stable or continued to increase, suggesting that the other NK cells contracted more rapidly or that CD57+NKG2Chi NK cells might preferentially survive and be less susceptible to death. Importantly, we could detect CD57+NKG2Chi NK cells years after primary CMV infection in CMV+ healthy adults. This NK cell subset might be maintained by periodic subclinical reactivation of CMV replication in the host. In these SOT recipients, the kinetics of the response of these CD57+NKG2Chi NK cells might be affected by the immunosuppressive drug treatment of these patients, delaying the response compared with the response in healthy individuals.
NKG2C+ NK cells expanded when cocultured with CMV-infected cells, and this expansion was correlated with a decrease in NKG2A+ NK cells (25). In mice, “unlicensed” NK cells expressing the CMV-specific Ly49H activating receptor proliferated better and more efficiently controlled mouse CMV replication in vivo than NK cells expressing inhibitory receptors for self-MHC class I (36). In humans, a minor subset of NK cells coexpress NKG2A and NKG2C (61). NKG2A can be transiently induced on NKG2C+ NK cells by stimulation with IL-12 alone or by coculture with CMV-infected cells (61). During acute CMV infection in SOT recipients, most of the NKG2C+ NK cells that expanded lacked NKG2A, although transient expression of NKG2A was detected in some patients. Most of the CD57+NKG2Chi NK cells in CMV+ blood donors did not express NKG2A or KIR3DL1 in HLA−Bw4+ individuals. In contrast, KIR2DL1 was not underrepresented on the CD57+NKG2Chi NK cells from CMV+ donors expressing the HLA-C ligands of KIR2DL1. These findings suggest that the NKG2C+ NK cells less restrained by inhibitory receptors for certain HLA class I ligands preferentially respond during CMV infection and become CD57+NKG2Chi NK cells. The absence of HLA ligands for inhibitory KIR has been associated with a lower rate of CMV infection in SOT recipients (53), suggesting that mounting an NK cell response to CMV that cannot be inhibited might be more protective. However, a preferential expression of self-specific KIRs (KIR2DL1 or KIR2DL2/3, depending on the donors) on NKG2C+ NK cells was observed after expansion of these cells in response to hantavirus (47), suggesting that NKG2C+ NK cells might be licensed in response to HLA-C. We observed an increased percentage of LIR-1+ cells in the CD57+NKG2Chi NK subset. LIR-1 recognizes UL18 in CMV-infected cells, and this interaction has been described as inhibitory (10, 16, 17). However, the functional properties of LIR-1 are uncertain, as we have observed lower inhibitory function of LIR-1 compared with KIRs and NKG2A, and it has been reported that LIR-1 interaction with UL18 can be activating (62). It is possible that the selective lack of expression of KIR3DL1 on CD57+NKG2Chi NK cells in HLA-Bw4-bearing individuals may reflect a selective expression of other inhibitory receptors. Further studies are required to examine the role of the different inhibitory receptors and their specific HLA class I ligands during NK cell responses to CMV and to determine why only certain inhibitory receptors to self-MHC class I were underrepresented on the CD57+NKG2Chi NK cells.
A recent study reported that NKG2C+ NK cells also expanded during acute hantavirus infection; of note, this expansion was observed only in CMV+ donors (47). NKG2C+ NK cells remained elevated in these individuals for more than 60 d (47), longer than what was previously considered typical for an innate immune response. The authors speculated that some NKG2C+ NK cells in CMV+ individuals might possess the memory-like features described for mouse NK cells (27, 28, 33, 63), which allowed them to expand during a subsequent hantavirus infection. In mice, Ly49H+ memory NK cells specific to CMV are more responsive to stimulation through other activating receptors and to cytokines (28). Future experiments are required to determine if CD57+NKG2Chi NK cells have the memory features described for mouse NK cells (33, 64).
Although speculative, we propose that CD57+NKG2Chi NK cells might be memory NK cells specific for CMV. It will be of interest to further characterize these cells in individuals highly susceptible to CMV infection, for example, SOT and hematopoietic stem cell transplant recipents, as well as in HIV-infected individuals.
Materials and Methods
PBMCs.
PBMCs were isolated from leukocyte reduction Pall filters (Blood Centers of the Pacific) from healthy adults and from healthy blood donors by Ficoll–Paque (GE Healthcare) density gradient centrifugation. All volunteers gave informed consent (approved by University of California, San Francisco, Committee on Human Research, institutional review board approval no. 10–00265). PBMCs were HLA and KIR genotyped, tested for CMV-specific T-cell responses, and stained as described in SI Materials and Methods. Discarded blood was obtained from SOT recipients with acute CMV infection and CMV+ SOT recipients without CMV viremia, as well as CMV− SOT recipients (Table S1). All patients with active CMV infection were treated with antiviral therapy for at least 21 d from the time of diagnosis. Patients were followed longitudinally during hospitalization (approved by University of California, San Francisco, Committee on Human Research, institutional review board approval no. 10–03608).
CMV DNA Detection.
DNA was extracted from patient plasma samples using a QIAsymphony SP automated instrument with a Virus/Bacteria Midi kit (Qiagen). Quantitative PCR for CMV DNA was performed as previously described (65).
Immunofluorescent Staining.
PBMCs from SOT recipients and healthy donors were stained with the following reagents: FITC–anti-CD57, PE–anti-NKG2C, PerCPCy5.5–anti-CD3, pacific blue (PB)–anti-CD16, Alexa 700–anti-CD56, biotin–anti-NKG2A and Qdot605–streptavidin, and Near-IR live/dead fixable dead cell dye. After surface staining, cells were fixed 10 min at room temperature with BD Cytofix and then permeabilized 5 min at room temperature with 0.1% Triton in PBS solution, and were finally stained with Alexa 647–anti–Ki-67. Cells were fixed and analyzed with a BD three-laser LSR-II instrument and FlowJo software (TreeStar).
Antibody Stimulation.
Plates were coated with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (Sigma) at 1 mg/mL and then with anti-CD56 (My31.13), anti-NKG2C (134572), or anti-CD16 (Leu11a) at 10 μg/mL. PBMCs from healthy CMV+ donors were incubated on the mAb-coated plates for 1 h with FITC-anti-CD107α, brefeldin A was added, and they were then cultured for 5 h. Cells were then stained with PE–anti-NKG2C, ECD–anti-CD3, PE–Cy5–anti-CD57, PE–Cy7–anti-CD56, PB–anti-CD16, and Amine Aqua live/dead fixable dead cell dye. Cells were fixed and analyzed with a four-laser LSR-II instrument and FlowJo software (TreeStar).
Statistics.
The two-tailed paired t test was used in the phenotypic studies to compare CD57+ and CD57− NK cells. An unpaired t test with Welch correction was used to compare the phenotype of cells of CMV+ and CMV− donors. The nonparametric Wilcoxon matched-pairs t test was used to compare CD57+NKG2Chi NK cells with other NK subsets and in the functional analysis. The statistical significance threshold was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank Janice Arakawa-Hyot for assistance, Suchitra Pandey and Kien Nyugen for providing blood samples, and the L.L.L. laboratory for helpful discussion. This study was supported by National Institutes of Health Grants AI068129 and HL095470. S.L.V. is a Cancer Research Institute/Irvington Institute Postdoctoral Fellow. L.L.L. is an American Cancer Society Professor.
Footnotes
Conflict of interest statement: J.P.H. is an employee of R&D Systems. The other authors have no conflicts.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110900108/-/DCSupplemental.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 4.Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002;4:1545–1558. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 5.Etzioni A, et al. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr. 2005;146:423–425. doi: 10.1016/j.jpeds.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL, Le AM, Civin CI, Loken MR, Phillips JH. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J Immunol. 1986;136:4480–4486. [PubMed] [Google Scholar]
- 7.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pass R. Cytomegalovirus. In: Knipe D, Howley PM, Griffin DE, Lamb RA, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 2675–2705. [Google Scholar]
- 9.Ho M. Epidemiology of cytomegalovirus infections. Rev Infect Dis. 1990;12(suppl 7):S701–S710. doi: 10.1093/clinids/12.supplement_7.s701. [DOI] [PubMed] [Google Scholar]
- 10.López-Botet M, Angulo A, Gumá M. Natural killer cell receptors for major histocompatibility complex class I and related molecules in cytomegalovirus infection. Tissue Antigens. 2004;63:195–203. doi: 10.1111/j.1399-0039.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 11.Bennett NJ, et al. Intracellular sequestration of the NKG2D ligand ULBP3 by human cytomegalovirus. J Immunol. 2010;185:1093–1102. doi: 10.4049/jimmunol.1000789. [DOI] [PubMed] [Google Scholar]
- 12.Magri G, et al. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood. 2011;117:848–856. doi: 10.1182/blood-2010-08-301374. [DOI] [PubMed] [Google Scholar]
- 13.Welte SA, et al. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol. 2003;33:194–203. doi: 10.1002/immu.200390022. [DOI] [PubMed] [Google Scholar]
- 14.Cosman D, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland CL, et al. UL16-binding proteins, novel MHC class I-related proteins, bind to NKG2D and activate multiple signaling pathways in primary NK cells. J Immunol. 2002;168:671–679. doi: 10.4049/jimmunol.168.2.671. [DOI] [PubMed] [Google Scholar]
- 16.Beck S, Barrell BG. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988;331:269–272. doi: 10.1038/331269a0. [DOI] [PubMed] [Google Scholar]
- 17.Prod’homme V, et al. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1- NK cells. J Immunol. 2007;178:4473–4481. doi: 10.4049/jimmunol.178.7.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leong CC, et al. Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: The role of endogenous class I major histocompatibility complex and a viral class I homolog. J Exp Med. 1998;187:1681–1687. doi: 10.1084/jem.187.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 20.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 21.Valés-Gómez M, Reyburn HT, Erskine RA, López-Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999;18:4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gumá M, et al. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 23.Gumá M, et al. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis. 2006;194:38–41. doi: 10.1086/504719. [DOI] [PubMed] [Google Scholar]
- 24.Monsiváis-Urenda A, et al. Influence of human cytomegalovirus infection on the NK cell receptor repertoire in children. Eur J Immunol. 2010;40:1418–1427. doi: 10.1002/eji.200939898. [DOI] [PubMed] [Google Scholar]
- 25.Gumá M, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 26.Kuijpers TW, et al. Human NK cells can control CMV infection in the absence of T cells. Blood. 2008;112:914–915. doi: 10.1182/blood-2008-05-157354. [DOI] [PubMed] [Google Scholar]
- 27.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 28.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE. 2010;5:e11966. doi: 10.1371/journal.pone.0011966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Björkström NK, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Vergès S, et al. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116:3865–3874. doi: 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun JC, Lopez-Verges S, Kim CC, DeRisi JL, Lanier LL. NK cells and immune “memory”. J Immunol. 2011;186:1891–1897. doi: 10.4049/jimmunol.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med. 2007;357:2601–2614. doi: 10.1056/NEJMra064928. [DOI] [PubMed] [Google Scholar]
- 35.Fishman JA, et al. Cytomegalovirus in transplantation - challenging the status quo. Clin Transplant. 2007;21:149–158. doi: 10.1111/j.1399-0012.2006.00618.x. [DOI] [PubMed] [Google Scholar]
- 36.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11:321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanier LL, et al. The NKB1 and HP-3E4 NK cells receptors are structurally distinct glycoproteins and independently recognize polymorphic HLA-B and HLA-C molecules. J Immunol. 1995;154:3320–3327. [PubMed] [Google Scholar]
- 39.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 40.Valés-Gómez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villard J. Immunity after organ transplantation. Swiss Med Wkly. 2006;136:71–77. doi: 10.4414/smw.2006.11010. [DOI] [PubMed] [Google Scholar]
- 42.Chiossone L, et al. Molecular analysis of the methylprednisolone-mediated inhibition of NK-cell function: Evidence for different susceptibility of IL-2- versus IL-15-activated NK cells. Blood. 2007;109:3767–3775. doi: 10.1182/blood-2006-07-037846. [DOI] [PubMed] [Google Scholar]
- 43.Wai LE, Fujiki M, Takeda S, Martinez OM, Krams SM. Rapamycin, but not cyclosporine or FK506, alters natural killer cell function. Transplantation. 2008;85:145–149. doi: 10.1097/01.tp.0000296817.28053.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, et al. The unexpected effect of cyclosporin A on CD56+CD16- and CD56+CD16+ natural killer cell subpopulations. Blood. 2007;110:1530–1539. doi: 10.1182/blood-2006-10-048173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaiser BK, et al. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174:2878–2884. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- 46.Pietra G, Romagnani C, Moretta L, Mingari MC. HLA-E and HLA-E-bound peptides: recognition by subsets of NK and T cells. Curr Pharm Des. 2009;15:3336–3344. doi: 10.2174/138161209789105207. [DOI] [PubMed] [Google Scholar]
- 47.Björkström NK, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011;208:13–21. doi: 10.1084/jem.20100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomasec P, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 49.Llano M, Gumá M, Ortega M, Angulo A, López-Botet M. Differential effects of US2, US6 and US11 human cytomegalovirus proteins on HLA class Ia and HLA-E expression: impact on target susceptibility to NK cell subsets. Eur J Immunol. 2003;33:2744–2754. doi: 10.1002/eji.200324182. [DOI] [PubMed] [Google Scholar]
- 50.Ulbrecht M, et al. HCMV glycoprotein US6 mediated inhibition of TAP does not affect HLA-E dependent protection of K-562 cells from NK cell lysis. Hum Immunol. 2003;64:231–237. doi: 10.1016/s0198-8859(02)00788-7. [DOI] [PubMed] [Google Scholar]
- 51.Cerboni C, et al. Synergistic effect of IFN-gamma and human cytomegalovirus protein UL40 in the HLA-E-dependent protection from NK cell-mediated cytotoxicity. Eur J Immunol. 2001;31:2926–2935. doi: 10.1002/1521-4141(2001010)31:10<2926::aid-immu2926>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 52.Wang EC, et al. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc Natl Acad Sci USA. 2002;99:7570–7575. doi: 10.1073/pnas.112680099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hadaya K, et al. Natural killer cell receptor repertoire and their ligands, and the risk of CMV infection after kidney transplantation. Am J Transplant. 2008;8:2674–2683. doi: 10.1111/j.1600-6143.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- 54.Dokun AO, et al. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 55.Brenchley JM, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 56.Focosi D, Bestagno M, Burrone O, Petrini M. CD57+ T lymphocytes and functional immune deficiency. J Leukoc Biol. 2010;87:107–116. doi: 10.1189/jlb.0809566. [DOI] [PubMed] [Google Scholar]
- 57.Le Priol Y, et al. High cytotoxic and specific migratory potencies of senescent CD8+ CD57+ cells in HIV-infected and uninfected individuals. J Immunol. 2006;177:5145–5154. doi: 10.4049/jimmunol.177.8.5145. [DOI] [PubMed] [Google Scholar]
- 58.Chong LK, et al. Proliferation and interleukin 5 production by CD8hi CD57+ T cells. Eur J Immunol. 2008;38:995–1000. doi: 10.1002/eji.200737687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lutz CT, et al. Human NK cells proliferate and die in vivo more rapidly than T cells in healthy young and elderly adults. J Immunol. 2011;186:4590–4598. doi: 10.4049/jimmunol.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pitcher CJ, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 61.Sáez-Borderías A, et al. IL-12-dependent inducible expression of the CD94/NKG2A inhibitory receptor regulates CD94/NKG2C+ NK cell function. J Immunol. 2009;182:829–836. doi: 10.4049/jimmunol.182.2.829. [DOI] [PubMed] [Google Scholar]
- 62.Saverino D, et al. Specific recognition of the viral protein UL18 by CD85j/LIR-1/ILT2 on CD8+ T cells mediates the non-MHC-restricted lysis of human cytomegalovirus-infected cells. J Immunol. 2004;172:5629–5637. doi: 10.4049/jimmunol.172.9.5629. [DOI] [PubMed] [Google Scholar]
- 63.Sun JC, Lanier LL. Natural killer cells remember: an evolutionary bridge between innate and adaptive immunity? Eur J Immunol. 2009;39:2059–2064. doi: 10.1002/eji.200939435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller S, Seet H, Khan Y, Wright C, Nadarajah R. Comparison of QIAGEN automated nucleic acid extraction methods for CMV quantitative PCR testing. Am J Clin Pathol. 2010;133:558–563. doi: 10.1309/AJCPE5VZL1ONZHFJ. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.