Abstract
Environmental stress leads to dramatic transcriptional reprogramming, which is central to plant survival. Although substantial knowledge has accumulated on how a few plant cis-regulatory elements (CREs) function in stress regulation, many more CREs remain to be discovered. In addition, the plant stress cis-regulatory code, i.e., how CREs work independently and/or in concert to specify stress-responsive transcription, is mostly unknown. On the basis of gene expression patterns under multiple stresses, we identified a large number of putative CREs (pCREs) in Arabidopsis thaliana with characteristics of authentic cis-elements. Surprisingly, biotic and abiotic responses are mostly mediated by two distinct pCRE superfamilies. In addition, we uncovered cis-regulatory codes specifying how pCRE presence and absence, combinatorial relationships, location, and copy number can be used to predict stress-responsive expression. Expression prediction models based on pCRE combinations perform significantly better than those based on simply pCRE presence and absence, location, and copy number. Furthermore, instead of a few master combinatorial rules for each stress condition, many rules were discovered, and each appears to control only a small subset of stress-responsive genes. Given there are very few documented interactions between plant CREs, the combinatorial rules we have uncovered significantly contribute to a better understanding of the cis-regulatory logic underlying plant stress response and provide prioritized targets for experimentation.
Keywords: machine learning, motif discovery, transcription factor binding site
Environmental stress, both abiotic and biotic, is the key constraint to plant productivity (1). Under stressful environments, plants undergo significant physiological and/or morphological alterations (2). Such plastic responses are particularly relevant for plants, which need to respond to ambient conditions due to their sessile nature (2). At the molecular level, one of the most immediate responses to stress is the extensive reprogramming of temporal and spatial transcription. In the past two decades, substantial progress has been made in understanding how this reprogramming occurs via the interaction of transcription factors with a handful of cis-regulatory elements (CREs). Examples of these well-characterized CREs include abscisic acid-responsive element (ABRE) (3, 4), dehydration-responsive element (DRE) (5), C-repeat (6), and W-box (7). In Arabidopsis thaliana, there are an estimated 1,346–2,290 putative transcription factor genes (8, 9), and many are likely involved in regulating stress-responsive transcription. However, the corresponding CREs of most of these transcription factors are not known. In addition, it is not clear to what degree the known CREs can explain expression changes in response to stress.
On the basis of knowledge of transcription factors and CREs, transcriptional regulatory models in yeast and mammals have been established that can explain the transcription profiles of different developmental stages and environmental conditions (10–12). One approach to building transcriptional regulatory models is to consider the presence or absence of CREs and their combinatorial relationships, copy number, and/or location relative to their regulatory targets. Such models, or cis-regulatory codes, have been applied to explain gene expression in yeast (13, 14), humans (15), and fruit flies (16). In plants, a number of CREs are known to be essential for stress-responsive transcription (e.g., refs. 3 and 4). The importance of a few CRE combinations has also been demonstrated (17–19), indicating that stress-responsive genes are regulated by multiple transcription factors. In addition, the roles of plant CRE copy number and location in transcriptional regulation of plant stress have also been studied for a few CREs (5, 20–22). Although these pioneering studies have clearly demonstrated the existence of stress cis-regulatory codes in plants, there are few examples and a global description of CRE-based stress regulatory rules is not available.
To globally decipher cis-regulatory code, detailed knowledge of CREs is required. However, many experimentally verified (referred to as “known”) plant CREs in public depositories, such as the Arabidopsis Gene Regulatory Information Server (23) and Plant Cis-Acting Regulatory DNA Elements database (24), are derived from multiple plant species and some are highly similar or identical. Furthermore, known plant CREs are mostly available in the form of consensus sequences with little information on binding site degeneracy. Thus, to complement our current knowledge of plant CREs, we first identified putative CREs (pCREs) involved in stress response through analysis of A. thaliana stress expression data. We then demonstrated that these pCREs exhibit characteristics of authentic cis-elements. Finally, the presence, combination, copy number, and location of these pCREs were used to establish the cis-regulatory code of stress-responsive gene expression in A. thaliana.
Results and Discussion
Multiple pCREs Implicated in Regulating Abiotic and Biotic Stress-Responsive Transcription Belong to Two Motif Superfamilies.
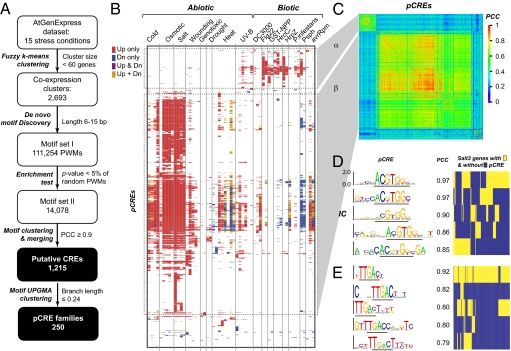
The assumption that genes with similar expression patterns are likely coregulated and have the same CREs has been applied to identify plant CREs (25, 26). Therefore, we set out to identify pCREs involved in regulating stress-responsive transcription on the basis of coexpression patterns of A. thaliana genes under 16 abiotic/biotic stress conditions (SI Methods and Table S1). Each A. thaliana gene was categorized as up-regulated, down-regulated, or not changed under each condition, and a motif discovery pipeline was used to identify 1,215 pCREs from putative promoter regions (Fig. 1A, SI Methods, and Dataset S1). The sites where these pCREs were mapped are preferentially found in the promoters of stress-responsive genes (up- and/or down-regulated) compared with promoters of genes without significant changes under stress (Fig. 1B and Dataset S2 A and B). Among these pCREs, 346 are highly similar [Pearson's correlation coefficient (PCC) ≥ 0.9] (SI Methods) to 52 known CREs (Dataset S2C). To obtain a lower-bound estimate of how many distinct CREs may be represented by these 1,215 motifs, we collapsed the pCREs into 250 motif families (Dataset S2D), using a stringent PCC threshold that errs on the side of collapsing truly distinct motifs into one family. For example, 10% of the mouse high-mobility group family and 48% of budding yeast transcription factor binding sites were collapsed erroneously (SI Methods and Fig. S1). Among the 250 motif families, 225 (containing 517 pCREs) do not have pCREs that are highly similar (PCC ≥ 0.9) to known plant CREs. Thus, we have identified hundreds of motifs that are previously unknown components of plant stress cis-regulation.
Fig. 1.
Putative CRE (pCRE) identification pipeline and pCREs relevant to differential gene expression under various stress conditions. (A) pCRE identification pipeline. P value, Fisher's exact test; PCC, Pearson's correlation coefficient; PWM, position weight matrix. (B) pCREs significantly enriched in the promoters of differentially regulated genes for each condition and treatment duration (latter not labeled). The pCREs were ordered according to results of complete linkage clustering of the enrichment patterns across conditions. Up only, pCREs enriched among up-regulated genes; down (Dn) only, pCREs enriched in down-regulated genes; Up & Dn, pCRE enriched in both; Up + Dn, pCRE enrichment only when up- and down-regulated genes are jointly considered. Dotted rectangles: two major pCRE clusters α and β. (C) PCC matrix indicating the degrees of similarity between pCRE PWMs. The ordering of pCREs is the same as in B. The gray areas indicate the correspondence of the α- and β-clusters. (D) Sequence logos of example pCREs and their similarities (PCC) to ABRE (Left and Middle). IC: information content. (Right) Presence of example pCREs in the promoters of salt-3h up-regulated genes. Each column represents one gene and whether its promoter contains the pCRE in question (yellow) or not (blue). Only genes containing one or more example pCREs are shown. (E) Sequence logos and similarities of example pCREs to W-box (Left and Middle). (Right) Presence of example pCRE sites in the promoters of flagellin-1h up-regulated genes.
The pCREs enriched among abiotic and biotic responsive genes form two major clusters, α and β (Fig. 1B, dotted rectangles), suggesting that many of the pCREs we have identified are specifically involved in regulating either abiotic or biotic stress responses. In addition, pCREs within each cluster are more similar to each other than to pCREs in different clusters (Fig. 1C). Thus, abiotic and biotic responses, at least among the conditions tested, are mostly mediated by distinct α and β “superfamilies” of CREs and, potentially, distinct families of transcriptional regulators and/or regulatory mechanisms. Consistent with this notion, in the abiotic stress pCRE superfamily, 19.6% of motifs contain a core sequence (ACGT) found in the ABRE (3, 4). Likewise, among pCREs relevant to biotic stress response, 12% have a core sequence (TTGAC) that is identical to the W-box consensus (27) bound by WRKY transcription factors. Nonetheless, these related pCREs have additional conserved signatures flanking the core sequences and are mapped to overlapping but distinct sets of stress-responsive genes (Fig. 1 D and E). Thus, pCREs within the α or the β superfamily are likely involved in regulating distinct but overlapping sets of target genes and may be bound preferentially by distinct transcription regulators. This result is consistent with a study of five WRKY transcription factors that demonstrated that the positions flanking the W-box core sequence are important for binding specificity (28).
pCREs Have Properties of Authentic CREs from Plants and Other Model Systems.
Several lines of evidence indicate that the pCREs identified in this study are authentic. All pCREs were found on the basis of significant enrichment in the promoters of stress-responsive genes and many are similar to known CREs. In addition, may known CREs regulating stress responses are recovered. There is also a significant positional bias of pCREs (Fig. 2) that is similar to experimentally established motifs in yeast (29), humans (30), and plants (31, 32). We found that pCREs are enriched throughout putative promoter regions of stress-responsive genes, particularly within ∼300 bp upstream of the transcriptional start site (TSS) (Fig. 2A). In addition, such pCRE positional bias is, in large part, observed only for genes responsive to conditions under which the pCREs were originally identified (Fig. 2B). This location bias also holds for pCRE families (Fig. S2). Another characteristic that supports pCRE authenticity is the significantly higher degree of evolutionary conservation of sites where pCREs were mapped compared with sites mapped by randomized pCREs (Fig. 2 C and D). When the degree of conservation was assessed through a comparison of putative orthologous regions between A. thaliana, Arabidopsis lyrata, and poplar genomes (SI Methods and Dataset S2E), 80% of pCREs were found to have significantly different conservation score distributions from randomized pCREs (Mann–Whitney test, P value <1e-5). Our findings are consistent with the fact that known CREs tend to evolve at a slower rate compared with functionally neutral sites (33, 34).
Fig. 2.
Positional bias and conservation of pCREs. (A) Log ratio (base 2, y axis) between the number of times that a pCRE is present in promoters of genes responsive to the condition in question in a 100-bp bin (ObsR, observed responsive) and the number of occurrences of X in random sequences generated on the basis of the nucleotide composition in the same bin (ExpB, expected in a bin). The log-ratio value was generated for each pCRE enriched among genes responsive to a particular condition at a particular time. The x axis indicates regions up to 1 kb upstream and 500 bp downstream of the TSS. Black line: the median log ratios for all pCREs. Gray area: the first and third quartiles of the log-ratio values. (B) Log ratios between the observed occurrence of pCREs in genes not responsive to any condition (ObsNR, observed nonresponsive) and the expected number of occurrences of pCREs in random sequences in each location bin. (C) Conservation score distributions of the sites of an example pCRE (sequence logo shown in Inset; IC, information content) and its randomized counterpart (rpCRE). (D) Distribution of log ratios between the median conservation scores (MCS) of pCRE sites and sites where their randomized counterparts (rpCREs) are located.
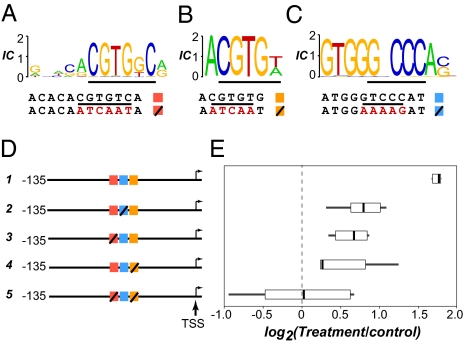
To verify pCRE authenticity experimentally, we carried out promoter deletion studies to determine whether three pCREs overrepresented among salt-responsive genes were necessary for salt-induced expression of ANAC019 (At1g52890), an NAC transcription factor that is significantly up-regulated under high salinity. Among the pCREs targeted, two are similar to ABRE (Fig. 3 A and B), and the third is not similar to any known plant CRE (Fig. 3C). Mutations in any of the three sites resulted in significant reduction in salt-induced expression (constructs 1–4; Fig. 3 D and E), indicating that all three pCREs are necessary for full salt-induced expression. Taken together, the significant enrichment of pCREs in stress-responsive gene promoters, their position bias and conservation, and the demonstration that three pCREs are functional are consistent with our interpretation that many of these pCREs are likely authentic motifs involved in controlling stress-responsive transcription.
Fig. 3.
Experimental verification of the contribution of two pCREs similar to ABRE and one previously unknown pCRE to ANAC019 salt-responsive transcription. (A) The sequence logo is shown for SamNSmyACGTGkCr, a pCRE similar to ABRE (Dataset S2). Alignments below the logo indicate the original and modified (in red) sequences in truncated promoter–β-glucuronidase fusion constructs. Construct 1: original genomic sequences −135 bp to the TSS. Constructs 2–5: construct 1 with modified pCRE sites. (B) The sequence logo and original/modified sequences of ACGTGw, another pCRE similar to ABRE (Dataset S2). (C) The sequence logo and original/modified sequences of a previously unknown pCRE, GTGGGNCCCAS. (D) Schematic representations of five truncated promoter–reporter fusions (not drawn to scale). The colored and checked boxes indicate the original and modified pCRE sites, respectively, following the color keys next to the alignments in A, B, and C. (E) Log-ratio (base 2) boxplots of β-glucuronidase activities between salt-treated and control samples.
Presence or Absence of pCREs Can Be Used to Predict Stress-Responsive Expression.
Our current understanding is that the expression pattern of a gene is influenced not only by the presence of individual CREs but also by combinatorial controls (13). Computational approaches jointly considering CRE properties and patterns of gene expression allow the identification of a cis-regulatory code that stipulates how CREs control gene expression. To uncover a global plant stress cis-regulatory code, we asked how well the presence and absence of pCREs explains up-regulation of genes under a particular stress condition. Initially we focused on salt stress treatment for 3 h (Salt3). The presence of a CRE will be a perfect predictor for Salt3-responsive transcription if the response is controlled by only one CRE and there is no other level of regulation. Contrary to this naive scheme, among Salt3-responsive genes, only 13.9% contain a pCRE (GTCGGTs, reverse complement: wACCGAC) highly similar to the DRE (ACCGAC) in their promoters. Thus, the recall, i.e., the proportion of truly responsive genes that were correctly predicted, is 0.139 (Fig. 4A). On the other hand, among genes containing DREs in their promoters, 16.5% are Salt3 responsive. Therefore, the proportion of responsive genes that were correctly predicted (precision) is 0.165 (Fig. 4A). Similarly, the precision and recall for an ABRE-like pCRE (rmSACGTGkmt) for Salt3 response are also low (Fig. 4A), but still higher than expected by chance; random guesses of Salt3 response would have a precision of 0.067 because 6.7% of A. thaliana genes are Salt3 responsive (Fig. 4A, dotted line). Although taking into account the presence of either ABRE or DRE significantly improves the precision compared with that of random guesses, other CREs and/or other regulatory mechanisms are apparently necessary to fully explain Salt3 response.
Fig. 4.
Performance of stress-response predictive models. (A) The precision-recall curves of Salt3 response predictive models based on known CRE presence/absence (known P/A), pCRE presence/absence (pCRE P/A), and pCRE combinatorial rules (pCRE CR). Arrows: precision and recall based on presence or absence of DRE-like or ABRE-like elements. (B) Precision-recall curves of predictive models for UV1. (C) Precision-recall curves of predictive models for Flg1. (D) The SVM weight of each location bin considering all pCREs jointly in predicting Salt3-responsive gene expression. The higher the weight is, the more important a location bin is in predicting high-salinity–induced expression. (E) Distribution of SVM weights generated from models considering only presence or absence (black bar) and copy number (white bars). (F) Precision-recall curves of predictive models for Salt3-responsive genes up-regulated by two- to three- (blue), three- to six- (orange), and more than sixfold (green). In all plots except E, the thick lines represent the mean and the whiskers represent ±SD in 10 SVM runs.
To test whether the inclusion of additional CREs would improve Salt3 response prediction, we first constructed a predictive model for Salt3 responses on the basis of known plant CREs with Support Vector Machine (Methods). As shown in Fig. 4A, considering more known CREs led to marked improvement in precision and recall. In addition, we observed moderate improvement in Salt3 response prediction when pCREs were used (Fig. 4A). This improvement is expected because pCREs include previously unknown motifs and are in the form of PWMs that provide binding site degeneracy information. When we expanded our analysis to two other conditions, 1-h treatments of UV-B (UV1) and flagellin (Flg1), we found that known CRE-based models allow only marginally better predictions than random CREs (Fig. 4 B and C). However, building UV1 and Flg1 predictive models on the basis of newly identified pCREs led to significant improvements compared with known motif-based models (Fig. 4 B and C), lending support to the hypothesis that pCREs are authentic components of plant cis-regulation. We also built pCRE family-based models, but they do not perform as well as models based on individual pCREs (Fig. S2D). Therefore, individual pCREs likely include more specificity information and may better resemble authentic cis-elements than pCRE families that are likely more general descriptions of binding sites.
Our findings indicate that presence and absence of motifs are important predictors of stress-responsive transcription, and models based on pCREs in general lead to moderate (Salt3) or significant (UV1, Flg1) improvement over those built with known CREs. The rather modest improvement in Salt3 response prediction with pCRE-based models is likely due to more extensive knowledge of cis-regulatory mechanisms for salt and related cold, drought, and osmotic stress conditions (35–37). Regardless, the models based on motif presence and absence are clearly insufficient because responses of many genes were not correctly predicted. In addition, despite the fact that we found 851 pCREs enriched in the promoters of Salt3 up-regulated genes, only the top 100 ranked pCREs were needed to build predictive models with similar performances to models built with all pCREs (Fig. S3). These findings raise the question whether the lower-ranked pCREs are involved in Salt3 response, if they play a minor role, potentially as low-affinity sites, or if they are important only in combinations.
Considering Combinatorial Controls Leads to Further Improvement in Stress Cis-Regulatory Models.
Because condition-specific expression is likely controlled by one or more transcription factors (13), we next tested the hypothesis that considering binary pCRE combinations would further improve the performance of expression prediction models. Using a classification algorithm that integrates association rule mining (Methods), 274, 357, and 271 pCRE combinatorial rules with above threshold precision and recall were identified for Salt3, UV1, and Flg1, respectively (Dataset S3A). Similar to earlier studies of yeast combinatorial control (13), some pCREs appear to be hub-like, working in combination with multiple distinct CREs (Fig. S4). In addition, predictive models built from these combinatorial rules led to substantial improvement in stress-response predictions for all three conditions (Fig. 4 A–C, red line; and Fig. S5A). For example, when recall is 20%, the combinatorial rule-based models led to 25–31% better precision than models based on pCRE presence or absence.
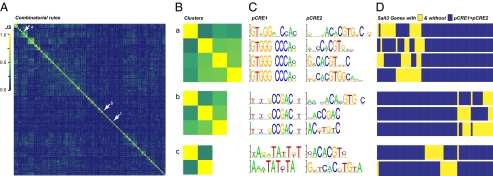
We emphasize that no single combinatorial rule has >5% recall (Dataset S3), indicating that instead of one or a few master regulatory rules that control the majority of responsive genes, multiple regulatory rules exist, each controlling a small number of genes under a stress condition. Consistent with this, distinct combinations relevant to a stress condition tend to be found in overlapping but mostly distinct sets of genes (Fig. 5A). A minority of combinatorial rules is found in similar sets of genes (Fig. 5 A and B, clusters a–c). Within these small clusters, the member pCREs tend to be similar to each other (Fig. 5 B and C). Nonetheless, there is little overlap in putative regulatory targets between clusters (Fig. 5D). In addition, although combinations with high similarity tend to contain similar pairs of pCREs (e.g., in cluster a, the second, third, and fourth combinations, Fig. 5D), there are substantial differences in the gene sets to which these pCREs are mapped.
Fig. 5.
Similarities between Salt3 pCRE combinatorial rules. (A) Pairwise similarity between combinatorial rules. For each rule specifying a pair of pCREs, a vector was generated consisting of presence (1) and absence (0) of sites of both pCREs in the promoters of Salt3-responsive genes. Using these presence/absence vectors, pairwise Jaccard similarities (JS) between combinatorial rules were calculated (Methods) and used for hierarchical clustering. The x and y axes contain pCRE combinatorial rules in the same order. Yellow, a complete overlap between genes containing distinct binary combinations; deep blue, no overlap. Arrows: example clusters: a, previously unknown element + ABRE-like; b, DRE-like + ABRE-like; c, EVENING element-like + ABRE-like. (B) Jaccard similarities between combinatorial rules in the example clusters (a, b, and c as indicated in A). The heat maps represent magnified views as in A. (C) The sequence logos of pCREs found in the example binary combinations. Each row represents one unique pCRE combination in the same order as in B. (D) Genes with (yellow) and without (blue) a particular binary combination (in the same order as in B). Each column represents the same gene. Only genes with one or more combinations in cluster a, b, or c are shown.
Supporting the validity of the computational predictions, the combinatorial rules include the experimentally established ABRE-DRE (18) and ABRE-EVENING (19) CRE combinations (Fig. 5C). Furthermore, our experiments confirmed the computationally predicted combinatorial rule between two ABRE-like pCREs and a previously unknown pCRE (Fig. 3); mutations in any of these three pCREs led to significantly lower levels of salt-induced expression compared with constructs with intact sites (two-sample t tests, all with P values <0.001). Thus, the results demonstrate the necessity of the ABRE-like and the unique pCREs in combination for proper salt-induced expression. Considering the performance of the combinatorial rule-based models, the identification of known combinations, and our proof-of-concept validation experiments, it is likely many of the combinatorial rules identified in this study are relevant to the control of stress-responsive transcription.
pCRE Copy Number and Location Are Important but Their Incorporation Does Not Significantly Improve Model Accuracy.
In addition to presence or absence of and combinatorial relationships between CREs, motif location (e.g., refs. 13 and 21) as well as copy number (e.g., ref. 20) is important for cis-regulatory control. To assess the importance of motif location, we asked first how well pCRE-mapped sites in each location bin from −1 kb to the TSS predict Salt3 response. Note that here we are interested in finding out how important each location bin is if we considered all pCRE sites collectively. We found that pCRE sites located from −200 bp to the TSS have significantly better power to predict Salt3 response (Fig. 4D), consistent with pCRE location bias (Fig. 2 A and B). We next asked which location bins are more important for predicting Salt3 response for each pCRE. Similar to considering pCREs in a bin jointly, pCREs located in regions proximal to the TSS tend to have higher weights (Fig. S5B). However, despite the importance of motif location, the model incorporating pCRE location performs similarly to the simpler presence/absence model [area under receiver–operating characteristic curves (AUC-ROCs) 0.781 and 0.789, respectively].
We next considered the importance of the number of pCRE sites in predicting Salt3 response (Fig. 4E). Similar to pCRE location, we found that although the number of pCRE sites is important, the model based on pCRE copy number does not outperform the model based solely on pCRE presence or absence (AUC-ROCs are 0.783 and 0.789, respectively). In addition to pCRE location and copy number, we also explored a more complicated model for predicting levels of up-regulation instead of predicting just up-regulation. We found that more highly differentially expressed genes are better predicted (Fig. 4F); however, this model does not perform as well as the model that simply classifies stress responses into up-regulation and no significant change (Fig. 4A).
In this study, we evaluated model performance by cross-validation, dividing our data into training and validation sets. We found that models considering pCRE location, copy number, and level of differential expression were likely overfitted because these models do not lead to further improvement in precision and recall. These more complicated models may explain the training data very well but not the validation data. Thus, there are likely limitations to how much information one can extract from the expression dataset in building cis-regulatory models. Nonetheless, our findings highlight the relative importance of combinatorial regulation compared with other cis-regulatory features; despite the very large parameter space (large number of possible pCRE combinations), it still outperforms the model considering motif numbers, location, or level of expression.
Conclusion
Our studies led to the discovery of 1,215 pCREs with multiple properties that resemble experimentally identified cis-elements. In addition, we provided a comprehensive first look at plant stress cis-regulatory codes on the basis of presence or absence of pCREs, their combinatorial relationships, locations in putative promoters, and copy number. Our ability to use pCREs to make reasonable expression predictions provides additional support for the notion that the pCREs identified computationally are likely authentic cis-elements. Furthermore, prediction accuracies of regulatory models based on binary relationships are much higher compared with those of presence/absence-based models. There are very few documented binary interactions between plant CREs. Thus, the combinatorial rules we have uncovered provide prioritized targets for experimentation.
Despite the importance of motif location and copy number in transcriptional regulation, considering these cis-regulatory features does not lead to better performing models. Thus, there is clearly room for improving the stress cis-regulatory model. Aside from optimizing the parameters of the analysis steps, performance would be further improved if additional information is incorporated, including transcription factor–CRE relations, transcription factor level, site affinity, or epigenetic states under stress conditions. For example, it would likely be very informative to consider information about how the expression patterns of transcription factors are affected by stress (38) and which transcription factors bind to pCREs on the basis of genome-wide one-hybrid studies (39). Further iterations of model building considering the above information in conjunction with experimental verification will be necessary for a more detailed global stress cis-regulatory code.
Methods
Putative CRE Identification, Assessing Location Bias, and Conservation.
The overall procedure for identifying pCREs among stress-responsive genes is shown in Fig. 1A. See SI Methods, section 1 for details on pCRE identification, pCRE mapping, random sequence generation, and statistical analysis of positional bias. Dataset S1 contains information on the pCREs identified. Information on pCREs identified in this study is also available in The Arabidopsis Information Resource (TAIR). To evaluate the conservation of pCREs, we first identified constrained sites in aligned genomic sequences among 1-to-1-to-1 syntenic orthologs in A. thaliana, A. lyrata, and Populus trichocarpa. A pCRE was defined as conserved between species if it mapped to constrained sites significantly more frequently than its randomly shuffled counterpart. The constrained sites were identified through comparisons of observed substitution patterns in each aligned putative promoter site against a neutral evolution model derived from substitution rates of fourfold degenerate sites in orthologous coding sequences (SI Methods, section 1.4).
Building pCRE-Based Models for Predicting Stress-Responsive Transcription.
To determine how well the presence or absence of pCREs explains stress-responsive transcription, we used the Support Vector Machine (SVM) (40) algorithm to generate classifiers for predicting expression with twofold cross-validation. SVM was used to generate expression prediction models on the basis of (i) only motif (known or pCRE) presence or absence, (ii) pCRE location, (iii) pCRE copy number, and (iv) levels of differential expression. To assess the importance of pCRE location, a “weight” that reflects the importance of pCRE sites in a given location bin was calculated (SI Methods, sections 2.1 and 2.2). The importance of copy number was examined by comparing the predication model considering only presence or absence of pCREs against a model based on the copy number of each pCRE (SI Methods, section 2.3). In prediction models considering levels of differential expression, high-salinity up-regulated genes were classified into three classes (two- to three-, three- to six-, and more than sixfold; SI Methods, section 2.4). Combinations of pCREs that regulate stress response were identified using an association rule mining method, Classification Based on Associations (41) (SI Methods, section 2.5). Combinatorial rule information is also available from TAIR.
Experimental Validation of pCRE and Combinatorial Rules.
The β-glucuronidase (GUS) reporter gene constructs were generated by cloning truncated promoters, both wild-type and mutated variants, into the Gateway cloning vector PMDC163 (42). Mesophyll protoplasts were isolated from fresh A. thaliana rosette leaves, and ∼5 × 104 protoplasts were transformed with the constructs (43). After incubation at room temperature for 13 h under light, the protoplasts were evenly divided into two subsamples and treated for 10 min with water and 250 mM NaCl, respectively. Fluorometric assays of GUS activity were performed using the fluorogenic substrate 4-methylumbelliferyl glucuronide and were normalized with the total protein content (Bio-Rad Laboratories).
Supplementary Material
Acknowledgments
We thank David Arnosti and Melissa D. Lehti-Shiu for for discussion and critical comments on this manuscript, Yang Zhou and Jinfeng Yi for advice on group lasso, and two anonymous reviewers and the editor for very helpful comments and suggestions. This work was partially funded by National Science Foundation Grants MCB-0749634 and DEB-0919452 (to S.-H.S.) and DBI-0701709 (to M.F.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103202108/-/DCSupplemental.
References
- 1.Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw AD. Evolutionary significance of phenotypic plasticity in plants. Adv Genet. 1965;13:115–155. [Google Scholar]
- 3.Marcotte WR, Jr, Russell SH, Quatrano RS. Abscisic acid-responsive sequences from the em gene of wheat. Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mundy J, Yamaguchi-Shinozaki K, Chua NH. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- 7.Rushton PJ, et al. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 8.Davuluri RV, et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003 doi: 10.1186/1471-2105-4-25. 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo AY, et al. PlantTFDB: A comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36(Database issue):D966–D969. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bussemaker HJ, Foat BC, Ward LD. Predictive modeling of genome-wide mRNA expression: From modules to molecules. Annu Rev Biophys Biomol Struct. 2007;36:329–347. doi: 10.1146/annurev.biophys.36.040306.132725. [DOI] [PubMed] [Google Scholar]
- 11.Bonn S, Furlong EE. Cis-regulatory networks during development: A view of Drosophila. Curr Opin Genet Dev. 2008;18:513–520. doi: 10.1016/j.gde.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Segal E, Widom J. From DNA sequence to transcriptional behaviour: A quantitative approach. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 14.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 15.Das D, Nahle Z, Zhang MQ. Adaptively inferring human transcriptional subnetworks. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100067. 2006.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 17.Shen Q, Ho TH. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell. 1995;7:295–307. doi: 10.1105/tpc.7.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narusaka Y, et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 19.Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–339. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- 20.Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell. 2002;14:749–762. doi: 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim CY, et al. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehrotra R, et al. Effect of copy number and spacing of the ACGT and GT cis elements on transient expression of minimal promoter in plants. J Genet. 2005;84:183–187. doi: 10.1007/BF02715844. [DOI] [PubMed] [Google Scholar]
- 23.Palaniswamy SK, et al. AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priest HD, Filichkin SA, Mockler TC. Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol. 2009;12:643–649. doi: 10.1016/j.pbi.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Haberer G, Mayer KF. Discovery of cis-elements between sorghum and rice using co-expression and evolutionary conservation. BMC Genomics. 2009;10:284. doi: 10.1186/1471-2164-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1540. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruyama K, et al. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 32.Tran LS, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moses AM, Chiang DY, Kellis M, Lander ES, Eisen MB. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol Biol. 2003;3:19. doi: 10.1186/1471-2148-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 35.Thomashow MF. PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 37.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brady SM, et al. A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol. 2011;7:459. doi: 10.1038/msb.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joachims T. Making large-scale SVM learning practical. In: Schölkopf B, Burges C, Smola A, editors. Advances in Kernel Methods - Support Vector Learning. Cambridge, MA: MIT Press; 1999. pp. 169–184. [Google Scholar]
- 41.Liu B, Hsu W, Ma Y. Integrating classification and association rule mining. In: Agrawal R, Stolorz O, Piatetsky-Shapiro G, editors. Proceedings of the Fourth International Conference on Knowledge Discovery and Data Mining. New York: AAAI Press; 1998. pp. 80–86. [Google Scholar]
- 42.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu FH, et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009 doi: 10.1186/1746-4811-5-16. 10.1186/1746-4811-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.