Abstract
RecA protein catalyzes DNA strand exchange, a basic step of homologous recombination. Upon binding to single-stranded DNA (ssDNA), RecA protein forms a helical nucleoprotein filament. Normally, this nucleoprotein filament binds double-stranded DNA (dsDNA) and promotes exchange of base pairs between this dsDNA and the homologous ssDNA that is contained within this filament. Here, we demonstrate that this bound dsDNA can be activated by interaction with a heterologous RecA nucleoprotein filament for a novel type of strand exchange with homologous ssDNA that is external to, and, therefore, not within, the filament. We refer to this novel DNA strand exchange as being in trans. Thus, the RecA nucleoprotein filament is a protein scaffold that activates dsDNA for strand exchange with ssDNA either within the filament or external to it. This new property demonstrates that the RecA nucleoprotein filament makes dsDNA receptive for DNA strand exchange, and it defines an early step of the homology recognition mechanism.
Keywords: Genetic recombination, homology recognition, protein–DNA interactions
Recombination between homologous DNA is catalyzed by the ubiquitous family of RecA-like proteins (Ogawa et al. 1993; Kowalczykowski and Eggleston 1994; Eggleston and West 1997; Bianco et al. 1998). The members of this family are conserved among Bacteria, Eucarya and Archaea (Aboussekhra et al. 1992; Bishop et al. 1992; Shinohara et al. 1992; Sung 1994; Seitz et al. 1998). In vitro, RecA protein promotes DNA strand exchange, a central step of homologous recombination (Shibata et al. 1979; Cox and Lehman 1981). DNA strand exchange is a multi-step process that initiates with the binding of RecA protein to single-stranded DNA (ssDNA), resulting in formation of a helical nucleoprotein filament (Stasiak et al. 1984). The RecA nucleoprotein filament can interact with double-stranded DNA (dsDNA; Tsang et al. 1985) to initiate a search for homologous dsDNA; upon finding DNA sequence homology, an exchange of complementary strands between ssDNA and dsDNA occurs within the environment of this elaborate protein–DNA structure. Because the exchange of base pairs occurs within this intricate complex of protein and homologous DNA molecules, simple analysis is precluded.
Of all the steps in DNA strand exchange, the most mysterious is the manner by which DNA sequence homology is recognized. How the RecA nucleoprotein filament makes the normally stable dsDNA receptive for DNA strand invasion remains a key question. As first postulated by Howard-Flanders and co-workers, the binding of nonhomologous dsDNA by the RecA nucleoprotein filament is a necessary prerequisite for homology recognition (Howard-Flanders et al. 1984). To permit a rapid search for homology, this binding is, by necessity, weak (Tsang et al. 1985; Conley and West 1990; Mazin and Kowalczykowski 1996). Nevertheless, the transient unwinding of heterologous dsDNA can be detected upon interaction with the filament (Cunningham et al. 1979; Rould et al. 1992). It was hypothesized that, as a result of such unwinding, dsDNA becomes stretched and partially unstacked. However, the role of this unwinding in the homology search remains undefined.
To learn more about the nature of the dsDNA that participates in this homologous pairing step, we asked whether dsDNA, which is bound to a heterologous nucleoprotein filament, is capable of pairing with free homologous ssDNA that is outside of the filament. The results demonstrate that the heterologous RecA nucleoprotein filament activates dsDNA, rendering it receptive for interaction with ssDNA, either within or external to the filament. We designated this novel type of DNA strand exchange with ssDNA exogenous to the filament as being in trans, in contrast to the conventional DNA strand exchange that occurs within the filament and is here called DNA strand exchange in cis.
Results
The RecA nucleoprotein filament promotes two distinct DNA strand exchange reactions: in cis and in trans
Conventional DNA strand exchange occurs between ssDNA that is contained within the presynaptic filament and dsDNA that interacts with this filament; we refer to this conventional reaction as DNA strand exchange in cis. But by using dsDNA that is not homologous to the ssDNA within the presynaptic filament, exogenous naked ssDNA could be used to probe the receptiveness of the dsDNA, bound to this heterologous filament, to DNA strand exchange in trans, as we refer to it. If dsDNA is activated for DNA strand exchange by the heterologous RecA nucleoprotein filament, then DNA strand exchange with free DNA may occur in trans (Fig. 1). To avoid the complications introduced by branch migration, short oligonucleotide substrates were used (Table 1).
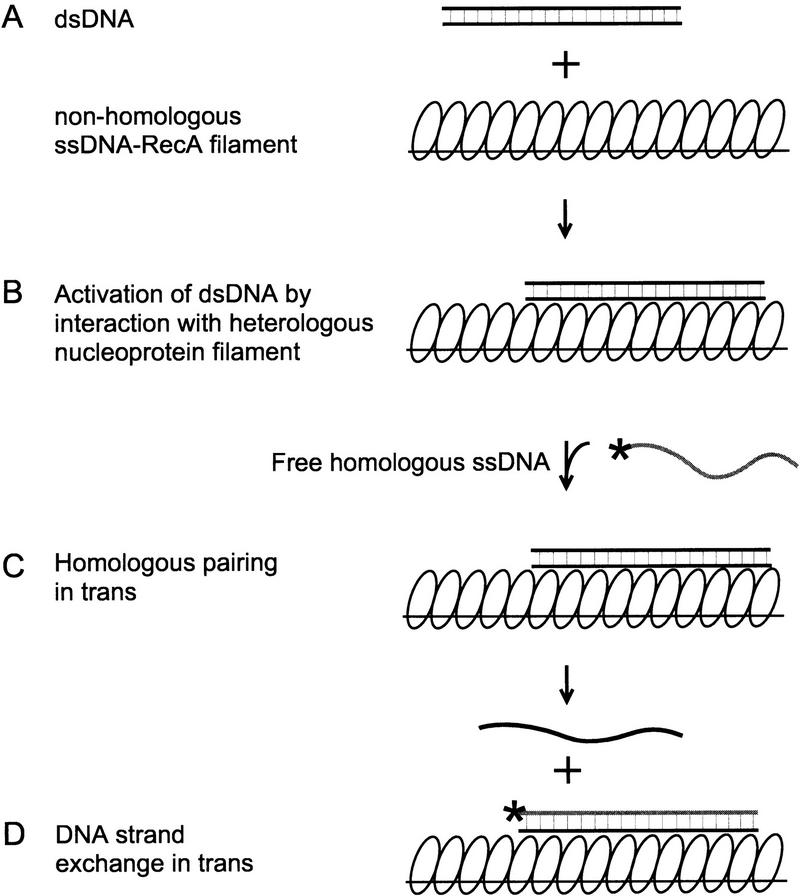
Figure 1.
DNA strand exchange in trans. A RecA nucleoprotein filament is assembled on ssDNA in the presence of ATPγS (A). This filament is introduced to heterologous dsDNA to form a nonhomologously bound complex (B). To probe the receptiveness of the bound dsDNA, exogenous 32P-labeled ssDNA that is complementary to this dsDNA is added to the complex (C). DNA strand exchange in trans between homologous 32P-labeled ssDNA and the unlabeled dsDNA results in 32P-labeled dsDNA and unlabeled ssDNA products.
Table 1.
Oligodeoxyribonucleotides used in this study
| Number (#)
|
Length (nucleotides)
|
Sequence
|
Complementary to (#)
|
||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 63 | A | C | A | G | C | A | C | C | A | G | A | T | T | C | A | G | C | A | A | T | T | A | A | G | C | T | C | T | A | A | G | C | 2 | |
| C | A | T | C | C | G | C | A | A | A | A | A | T | G | A | C | C | T | C | T | T | A | T | C | A | A | A | A | G | G | A | |||||
| 2 | 63 | T | C | C | T | T | T | T | G | A | T | A | A | G | A | G | G | T | C | A | T | T | T | T | T | G | C | G | G | A | T | G | G | 1 | |
| C | T | T | A | G | A | G | C | T | T | A | A | T | T | G | C | T | G | A | A | T | C | T | G | G | T | G | C | T | G | T | |||||
| 5 | 32 | C | C | A | T | C | C | G | C | A | A | A | A | A | T | G | A | C | C | T | C | T | T | A | T | C | A | A | A | A | G | G | A | 6 | |
| 6 | 32 | T | C | C | T | T | T | T | G | A | T | A | A | G | A | G | G | T | C | A | T | T | T | T | T | G | C | G | G | A | T | G | G | 5 | |
| 9 | 61 | T | A | T | C | T | A | G | A | C | C | A | C | C | A | G | C | C | A | G | C | G | C | G | T | G | T | C | C | A | C | C | — | ||
| A | G | C | T | C | A | G | C | A | T | C | G | A | C | C | A | C | C | A | G | C | T | C | G | A | G | T | G | C | A | ||||||
| 19 | 32 | A | A | A | T | A | A | G | A | A | A | A | A | A | T | G | A | A | A | T | A | T | T | A | T | A | A | A | A | A | G | G | A | 20 | |
| 20 | 32 | T | C | C | T | T | T | T | T | A | T | A | A | T | A | T | T | T | C | A | T | T | T | T | T | T | C | T | T | A | T | T | T | 19 | |
| 21 | 32 | C | C | T | T | C | C | G | C | T | T | T | T | T | T | G | T | C | C | T | C | T | T | T | T | C | T | T | T | T | G | G | T | — | |
| 39 | 32 | G | C | G | T | G | T | G | T | G | G | T | G | G | T | G | T | G | C | G | C | G | T | G | T | G | T | G | G | T | G | G | T | 40 | |
| 40 | 32 | A | C | C | A | C | C | A | C | A | C | A | C | G | C | G | C | A | C | A | C | C | A | C | C | A | C | A | C | A | C | G | C | 39 | |
| 43 | 33 | A | T | T | C | A | C | A | A | A | C | G | A | A | T | G | G | A | T | C | C | T | C | A | T | T | A | A | A | G | C | C | A | G | 72 |
| 71 | 94 | C | T | T | T | A | G | C | T | G | C | A | T | A | T | T | T | A | C | A | A | C | A | T | G | T | T | G | A | C | C | T | A | C | — |
| A | G | C | A | C | C | A | G | A | T | T | C | A | G | C | A | A | T | T | A | A | G | C | T | C | T | A | A | G | C | C | A | T | |||
| C | C | G | C | A | A | A | A | A | T | G | A | C | C | T | C | T | T | A | T | C | A | A | A | A | G | G | A | ||||||||
| 72 | 33 | C | T | G | G | C | T | T | T | A | A | T | G | A | G | G | A | T | C | C | A | T | T | C | G | T | T | T | G | T | G | A | A | T | 43 |
| 90 | 90 | C | G | G | G | T | G | T | C | G | G | G | G | C | T | G | G | C | T | T | A | A | C | T | A | T | G | C | G | G | C | A | T | C | 91 |
| A | G | A | G | C | A | G | A | T | T | G | T | A | C | T | G | A | G | A | G | T | G | C | A | C | C | A | T | A | T | G | C | G | |||
| G | T | G | T | G | A | A | A | T | A | C | C | G | C | A | C | A | G | A | T | G | C | G | T | ||||||||||||
| 91 | 90 | A | C | G | C | A | T | C | T | G | T | G | C | G | G | T | A | T | T | T | C | A | C | A | C | C | G | C | A | T | A | T | G | G | 90 |
| T | G | C | A | C | T | C | T | C | A | G | T | A | C | A | A | T | C | T | G | C | T | C | T | G | A | T | G | C | C | G | C | A | |||
| T | A | G | T | T | A | A | G | C | C | A | G | C | C | C | C | G | A | C | A | C | C | C | G | ||||||||||||
| htr 1 | 32 | G | T | G | A | G | T | C | G | A | C | A | A | G | C | C | T | G | A | C | T | C | A | A | C | A | T | T | A | T | C | C | T | — | |
A RecA nucleoprotein filament was assembled on ssDNA (a 33-mer oligonucleotide, #43) with a molar ratio of three nucleotides of DNA per RecA protein monomer. To prevent dissociation of the initial RecA nucleoprotein filament, a virtually nonhydrolyzable analog of ATP, ATPγS, was used in its assembly (Menetski and Kowalczykowski 1985). Finally, to ensure that no unbound RecA protein remained, nonhomologous ssDNA (htr#1) was added to sequester any free RecA protein. Heterologous dsDNA (#5 and #6) was then allowed to interact with this nucleoprotein filament. The ability of this dsDNA, which is bound to the heterologous nucleoprotein complex, to engage in DNA strand exchange with free ssDNA was then assayed by addition of exogenous 32P-end-labeled ssDNA (#5) that was homologous to the dsDNA (complementary to #6).
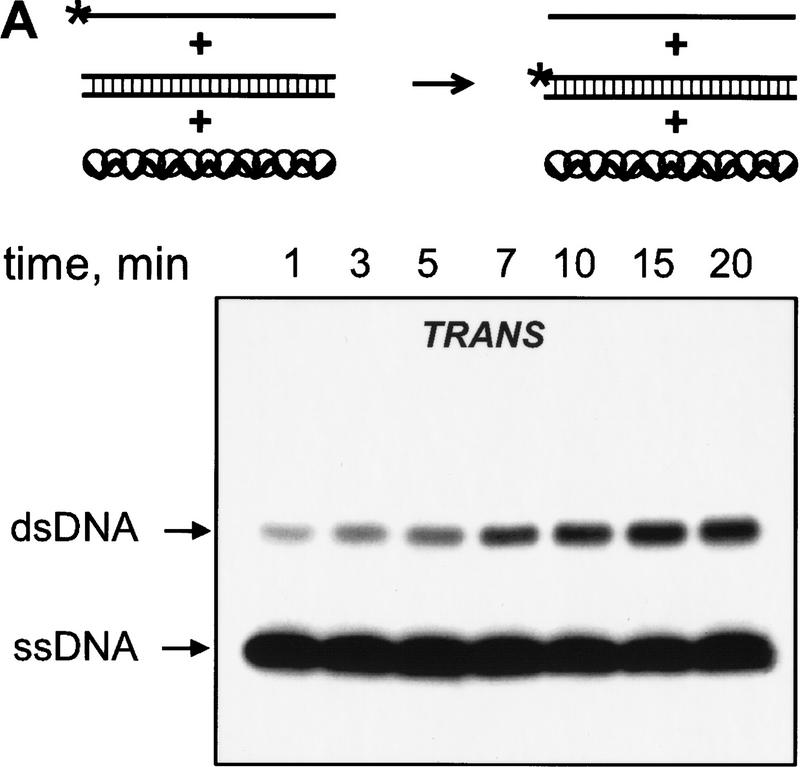
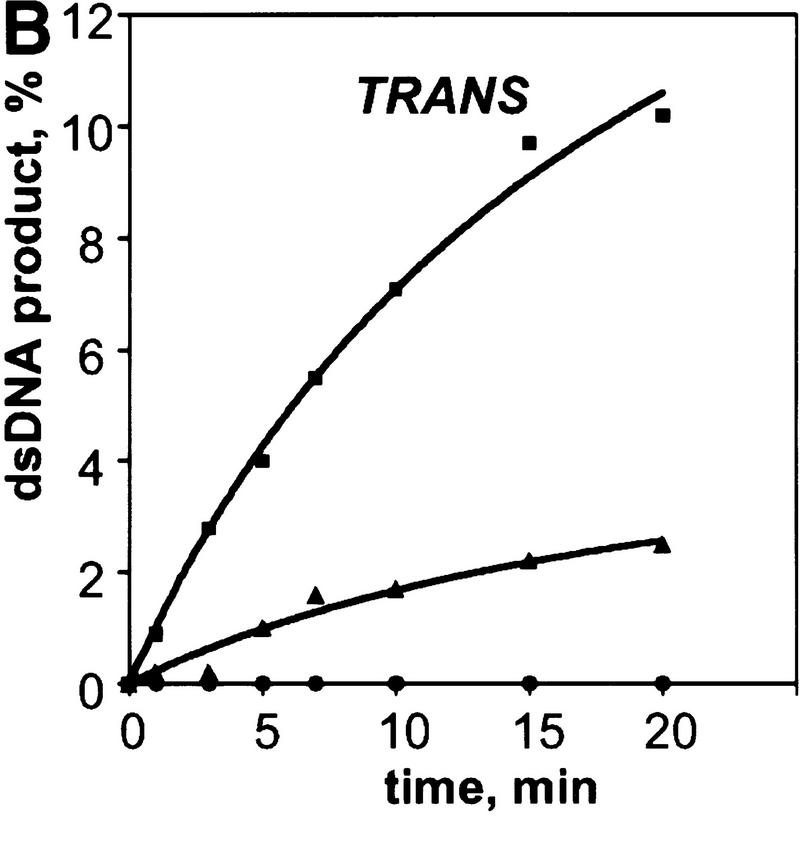
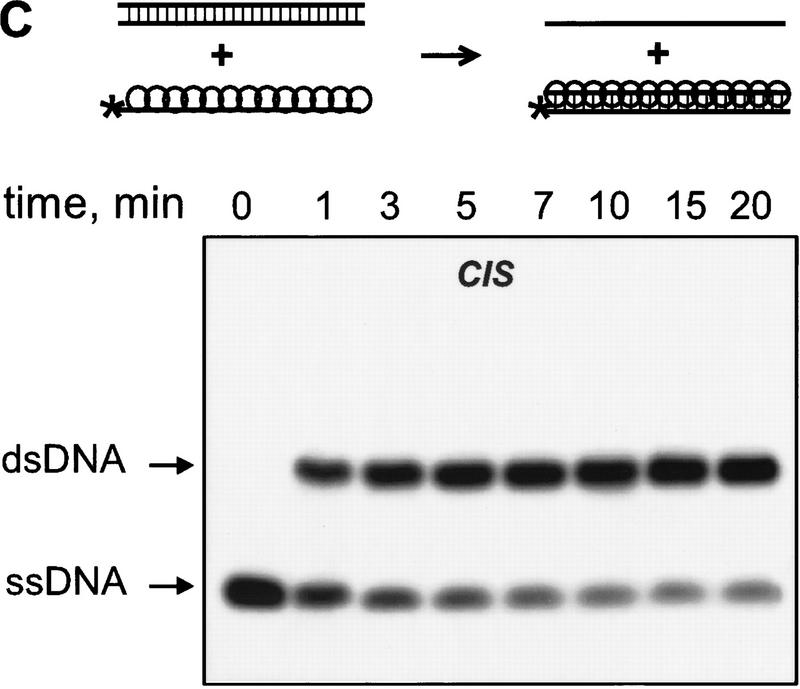
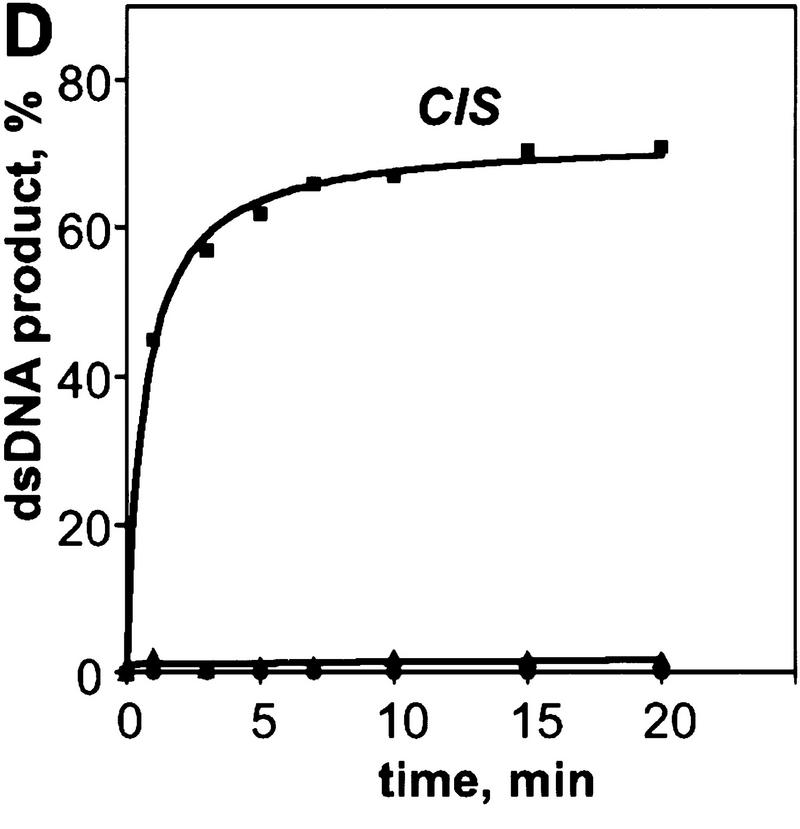
We observed the RecA protein-dependent appearance of the 32P-labeled duplex DNA that resulted from exchange of strands between the dsDNA and the exogenous ssDNA (Fig. 2A,B). In the absence of RecA protein, significantly less exchange occurs, indicating that spontaneous thermal exchange accounts for only a small fraction of the observed DNA strand exchange (Fig. 2B). The reaction is also homology-dependent (Fig. 2B); however, the kinetics of exchange are distinctly slower than for DNA strand exchange occurring in cis between a homologous RecA nucleoprotein filament and dsDNA under identical conditions (Fig. 2C,D).
Figure 2.
The RecA nucleoprotein filament catalyzes DNA strand exchange both in cis and in trans. For the reactions in trans, presynaptic filaments were assembled on ssDNA (#43); these filaments were mixed with dsDNA (#5 and #6) and 32P-labeled ssDNA (#5) that were reciprocally homologous, but heterologous to the ssDNA within the filament. For the reactions in cis, the same presynaptic filaments, but containing 32Plabeled ssDNA (#43), were mixed with homologous dsDNA (#43 and #72). Both reactions were carried out at 23°C. Gels showing DNA strand exchange in trans and in cis are presented in A and C, respectively. The analysis of these reactions (█), as well as of control reactions containing either no RecA protein (▴) or heterologous 32P-labeled ssDNA (#19) in place of the homologous ssDNA (#5 for the in trans and #43 for the in cis reaction) (●), are shown in B and D, respectively.
Potentially, a contaminating exonuclease could resect one of the dsDNA ends and facilitate annealing of the partially degraded dsDNA with the complementary ssDNA; subsequent spontaneous branch migration would complete DNA strand exchange. To exclude such an artifact, the integrity of the (identical) DNA strand displaced by DNA strand exchange in trans was analyzed by gel electrophoresis. We found that the displaced DNA strand retained its original length; no traces of exonuclease activity were detected (Fig. 3).
Figure 3.

The strand displaced during the DNA strand exchange in trans is intact. The 32P-labeled DNA strand (#1) that was displaced from dsDNA (#1 and #2) during DNA strand exchange in trans was isolated (see Materials and Methods) and analyzed by electrophoresis on a 10% polyacrylamide gel containing 50% urea (displaced ssDNA), along with the original 32P-labeled oligonucleotide (#1; substrate ssDNA). 32P-labeled DNA migration markers (marker) were obtained by cleavage of the oligonucleotide #2 at adenine and guanine residues according to Maxam and Gilbert (1980).
Previously, we determined that in the presence of ATPγS, as was used here, ssDNA bound to the primary site of RecA protein remains stably associated and that this bound RecA protein does not transfer to ssDNA added subsequently, even in the presence of a high excess of free ssDNA (Menetski and Kowalczykowski 1987; Mazin and Kowalczykowski 1998). As a further precaution and as noted above, all reactions included an excess of nonspecific DNA to titrate any free RecA protein. Thus, the observed DNA strand exchange in trans does not occur as a result of transfer of RecA protein to the added ssDNA; rather, it involves free ssDNA as one of the participants.
DNA strand exchange in trans occurs with different DNA substrates
To ensure that the observed DNA strand exchange in trans is not a peculiar characteristic of the DNA substrates employed, several other substrate pairs were examined. Table 2 shows that DNA strand exchange in trans is not restricted to certain DNA sequences, as it occurs with different DNA substrates. The rates of these reactions vary considerably for these different DNA substrates but, significantly, they do not correlate with the melting temperatures of the dsDNA, confirming that simple denaturation is not responsible for the reaction in trans. Moreover, the rates are notably different when the two different complementary ssDNA strands react with the same dsDNA (Table 2, see rows #40 and #39), demonstrating that the nature of the ssDNA is also a determinant of this pairing reaction. Additionally, increasing the length of the homologous DNA substrates from 32 (#5) to 63-mers (#1) does not significantly affect the rate of the protein-promoted reaction (Table 2), although the rate of spontaneous, thermal DNA strand exchange is decreased. Finally, DNA strand exchange in trans occurs with DNA substrates that are 90 nucleotides in length; the reaction rate was somewhat lower than for 63-mer #1, but it was not lower than for two other 32-mer substrates.
Table 2.
DNA strand exchange in trans is independent of dsDNA melting temperature
| ssDNA substrate (#)
|
Length (nucleotides)
|
dsDNA substrate (#)
|
ssDNA within filament (#)
|
Initial rate (%/min)a
|
Tm (°C); in 1 m NaCl
|
|---|---|---|---|---|---|
| 40 | 32 | 39, 40 | 43 | 2.5 ± 0.3 | 89 |
| 43 | 33 | 43, 72 | 5 | 1.9 ± 0.2 | 78.7 |
| 1 | 63 | 1, 2 | 9 | 1.1 ± 0.1 | 86.7 |
| 5 | 32 | 5, 6 | 43 | 1.0 ± 0.2 | 78.7 |
| 91 | 90 | 90, 91 | 71 | 0.2 ± 0.1 | 94.4 |
| 20 | 32 | 19, 20 | 43 | 0.1 ± 0.5 | 67.2 |
| 39 | 32 | 39, 40 | 43 | 0.1 ± 0.04 | 89 |
The initial rate during the first 1–2 min (10–15 min for ssDNA #20, #39, and #91) of DNA strand exchange in trans was obtained by subtracting the rate of spontaneous thermal DNA strand exchange for each pair of DNA substrates, from the observed rate of RecA protein-promoted reaction. The spontaneous reaction rates (%/min) were 0.06, 0.12, 0.05, 0.1, 0.02, 0.13, and 0.03 for oligonucleotides #40, #43, #1, #5, #91, #20, and #39 respectively.
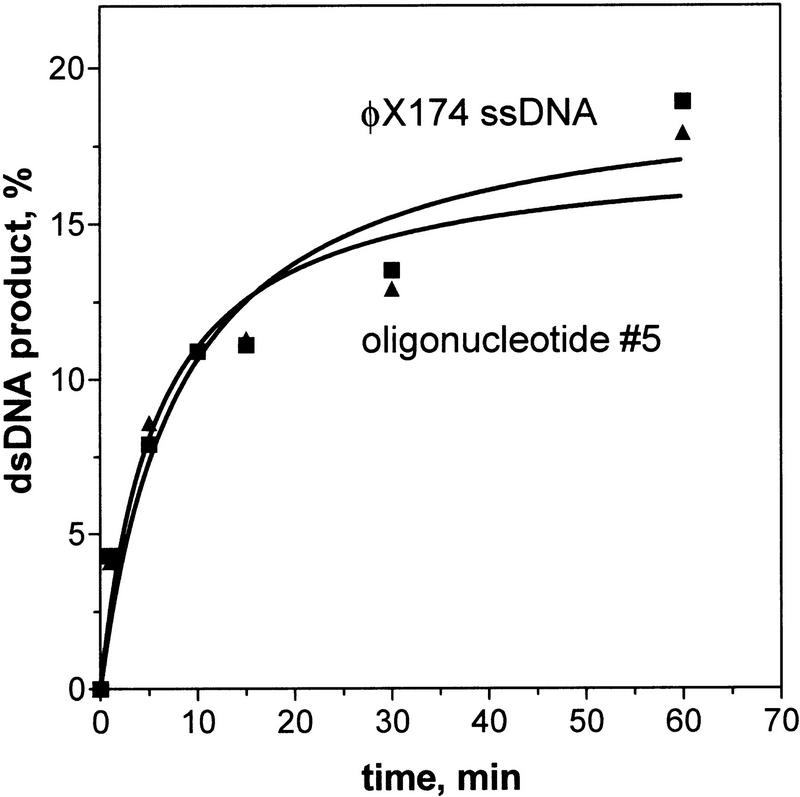
Similarly, the rate of DNA strand exchange in trans is independent of the type and sequence of ssDNA that is within the RecA nucleoprotein filament. Thus, the RecA nucleoprotein filament assembled on φX174 circular ssDNA can promote DNA strand exchange in trans at the same rate as the filament assembled on a 32-mer oligonucleotide (Fig. 4). Furthermore, the large size of the RecA nucleoprotein filaments assembled on φX174 ssDNA enabled the use of gel-filtration chromatography to remove any potentially unbound RecA protein. We found that the DNA strand exchange in trans was unaltered after this purification step (data not shown), again demonstrating the free RecA protein is not responsible for this reaction.
Figure 4.
RecA nucleoprotein filaments assembled on either ssDNA oligonucleotides or φX174 ssDNA promote DNA strand exchange in trans with the same efficiency. Presynaptic filaments were assembled on oligonucleotide #5 (12 μm, ▴) or φX174 ssDNA (12 μm, █). These filaments were used to mediate DNA strand exchange in trans between the mutually homologous DNA substrates: dsDNA (#43 and #72) and 32P-labeled ssDNA (#43).
Therefore, we conclude that the DNA strand exchange in trans is a general characteristic of the RecA nucleoprotein filament and that it is independent of DNA sequence and type.
DNA strand exchange in trans requires a functional RecA nucleoprotein filament
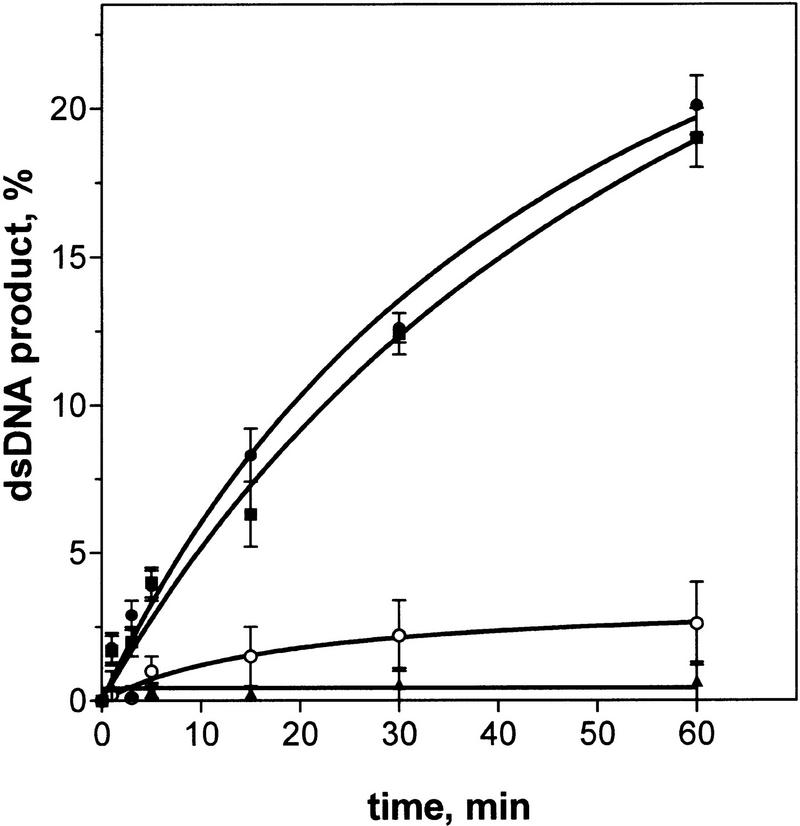
Although RecA protein can bind ssDNA in the absence of ATP to form an inactive filament, formation of the active RecA nucleoprotein filament that is proficient in DNA strand exchange in cis and in LexA repressor cleavage requires ATP or one of its analogs (Kowalczykowski and Eggleston 1994). Consequently, we wished to determine whether the nucleotide cofactor requirements of DNA strand exchange in trans parallel those of the in cis reaction. Because ATP hydrolysis by RecA protein would promote dissociation of RecA protein from the original filament and its transfer to the ssDNA added in trans, DNA strand exchange in trans could not be examined unambiguously in the presence of ATP. Instead, however, we used ATPγS, an ATP analog that is hydrolyzed very slowly, and ADP · AlF4−, a nonhydrolyzable ground state ATP analog. Both of these analogs support active filament formation, DNA strand exchange in cis, and LexA repressor cleavage (Moreau and Carlier 1989; Yu and Egelman 1990; Kowalczykowski and Krupp 1995). In the presence of ATPγS or ADP · AlF4−, DNA strand exchange in trans was equally robust (Fig. 5). In contrast, in the presence of ADP or in the absence of any cofactor, the reaction was quite poor, showing that only the active RecA nucleoprotein filaments can efficiently promote DNA strand exchange in trans.
Figure 5.
The DNA strand exchange in trans requires an active RecA nucleoprotein filament. Presynaptic filaments were assembled by incubation of RecA protein (1.3 μm) with ssDNA (#43.4 μm) in the presence of 1 mm ATPγS (█), 5 mm ADP/10 mm NaF/0.4 mm Al(NO3)3 (●), 5 mm ADP (○), or in the absence of cofactor (▴) under standard conditions. For each filament, the DNA strand exchange in trans was initiated by addition of a pair of mutually homologous DNA substrates: dsDNA (#5 and #6; 8 μm) and 32P-labeled ssDNA (#5; 4 μm).
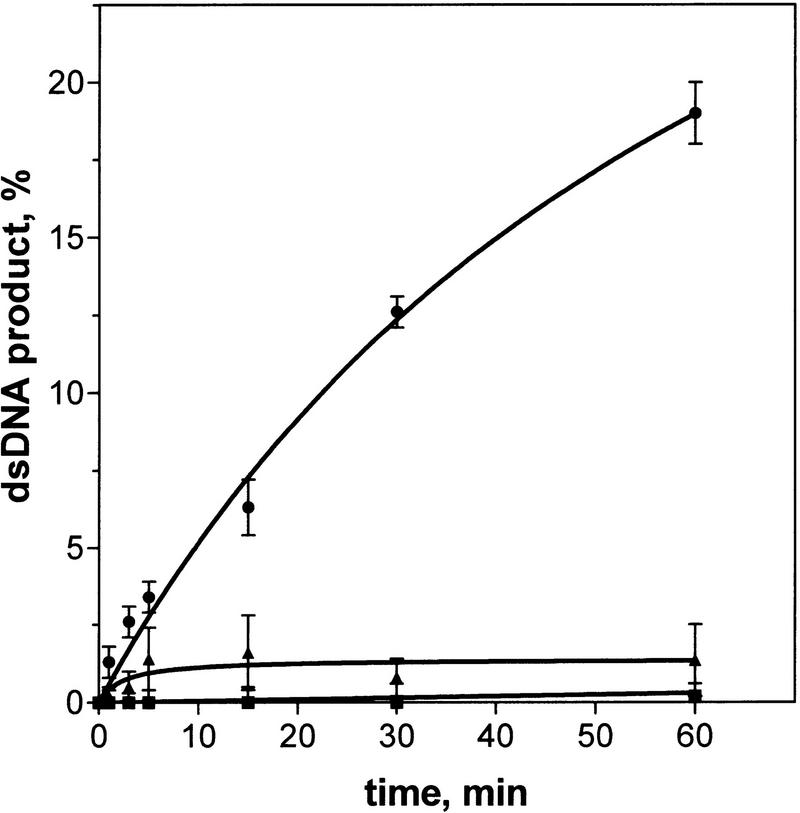
As a further demonstration that DNA strand exchange in trans required the same functional form of the RecA nucleoprotein that is required for the in cis reaction, mutant RecA proteins were tested (Fig. 6). Some defective RecA proteins, such as RecA56 and RecA1, can form filaments with ssDNA, however, these filaments cannot promote DNA strand exchange in cis (Lauder and Kowalczykowski 1993). As shown in Figure 6, these proteins also fail to promote DNA strand exchange in trans.
Figure 6.
Defective RecA56 and RecA1 proteins do not promote DNA strand exchange in trans. Presynaptic filaments were assembled by incubation of RecA wild type (●), RecA56 (█), or RecA1 protein (▴; 1.3 μm, for each protein) with ssDNA (#43; 4 μm) under standard conditions. For each filament, DNA strand exchange in trans was initiated by addition of a pair of mutually homologous DNA substrates: dsDNA (#5 and #6; 8 μm) and 32P-labeled ssDNA (#5; 4 μm).
Therefore, there is an absolute correspondence between the ability of a RecA nucleoprotein filament to promote DNA strand exchange in trans and in cis, and this behavior also correlates with the biological activity of RecA protein.
DNA strand exchange in trans requires binding of DNA to the secondary DNA-binding site of RecA protein
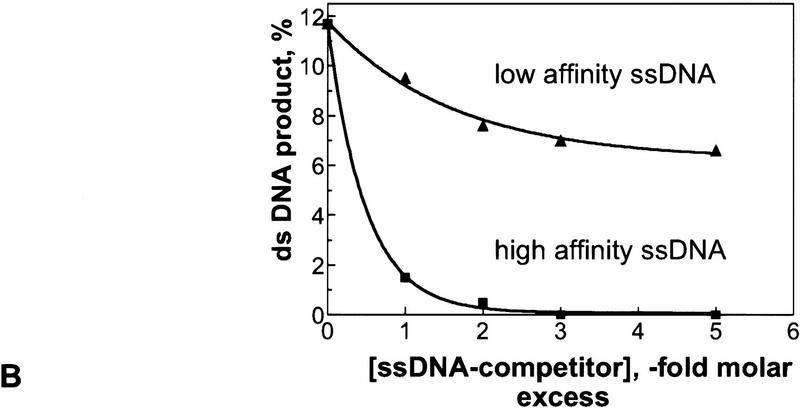
Conventional DNA strand exchange in cis requires binding of the dsDNA to the secondary DNA-binding site of the RecA nucleoprotein filament; therefore, we asked whether DNA strand exchange in trans had the same requirement. Previously, we characterized and defined the functions of the secondary DNA-binding site within the RecA nucleoprotein filament (Mazin and Kowalczykowski 1996, 1998). This site has a higher affinity for ssDNA over dsDNA and has a marked base compositional preference in ssDNA binding. Reactions in trans were performed in the presence of competitor ssDNA that has either a high affinity (#21) or a low affinity (htr#1) for the secondary site (Mazin and Kowalczykowski 1996). The results demonstrate that ssDNA with high, but not low, affinity for the secondary site strongly inhibits the reaction (Fig. 7). Moreover, one molar equivalent of high-affinity ssDNA, the amount required to saturate the RecA protein secondary site (Mazin and Kowalczykowski 1996, 1998), was sufficient to inhibit the reaction in trans by 90% (Fig. 7B). These results argue that DNA strand exchange in trans requires binding of at least one of the two DNA substrates to the secondary site of the RecA nucleoprotein filament.
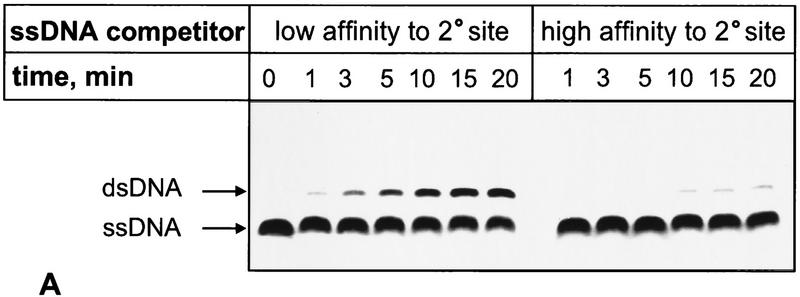
Figure 7.
ssDNA with a high-affinity for the secondary site inhibits DNA strand exchange in trans. (A) Time course of the DNA strand exchange in trans carried out in the presence of a low- or high-affinity ssDNA inhibitor. A standard reaction mixture was divided in two. Each was supplemented with competing ssDNA (120 μm) that had either high (#21) or low (htr#1) affinity for the secondary site of RecA protein. DNA strand exchange in trans was initiated by the addition of mutually homologous dsDNA (#5 and #6) and 32P-labeled ssDNA (#5) to both mixtures. (B) Concentration dependence of the inhibition of the reaction in trans by the same competitors. The indicated amounts of ssDNA were added to the reaction mixtures prior to addition of homologous ssDNA and dsDNA substrates. The reactions were carried out for 15 min.
The secondary DNA-binding site of RecA protein activates dsDNA for DNA strand exchange
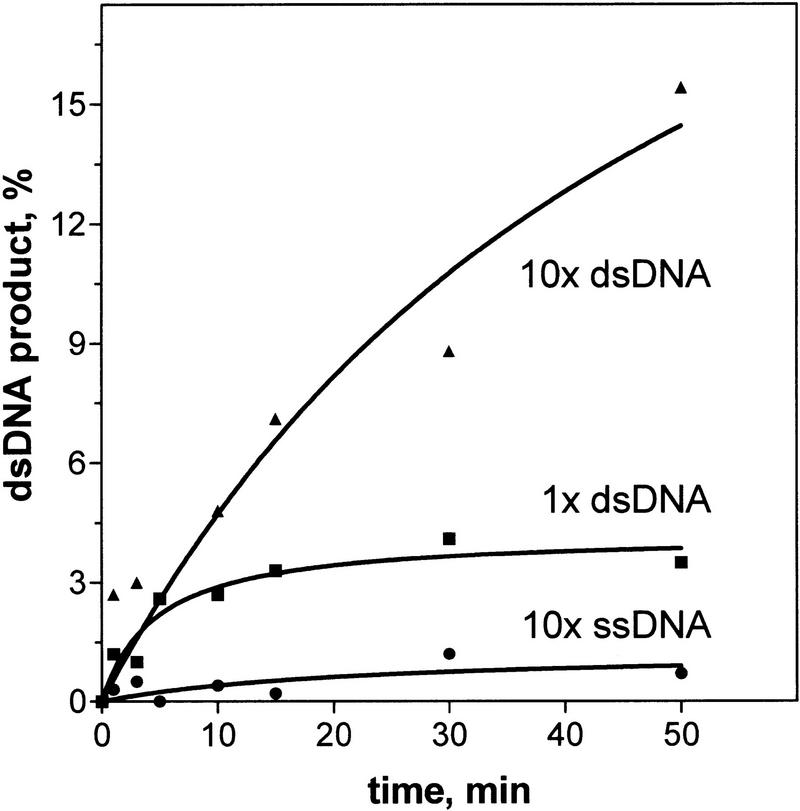
Because the secondary site can bind either ssDNA or dsDNA, the high-affinity ssDNA inhibitor that was used in Figure 7 could block DNA strand exchange in trans by competing with both of these substrates. To ascertain whether binding of dsDNA or ssDNA was essential for DNA strand exchange, the concentration of each was varied separately. When we tested whether this inhibition was overcome by increasing the concentration of dsDNA, we found that an increase in dsDNA concentration did restore DNA strand exchange (Fig. 8). In contrast, an increase in the homologous ssDNA concentration exacerbated the inhibition (Fig. 8). These results demonstrate that the binding of dsDNA to the secondary site is an essential step of DNA strand exchange in trans as it is for strand exchange in cis. Binding of dsDNA that is heterologous with respect to the ssDNA within the RecA nucleoprotein filament, nevertheless, permits homologous interaction with protein-free complementary ssDNA in solution and, simultaneously, activates the dsDNA for DNA strand exchange in cis. Thus, the reaction is occurring as diagrammed in Figure 1.
Figure 8.
An excess of dsDNA alleviates the inhibition of DNA strand exchange in trans caused by ssDNA. RecA nucleoprotein filaments were incubated with a competitor ssDNA (#21; 12 μm) that had a high affinity for the secondary DNA-binding site. DNA strand exchange in trans was initiated by addition of mutually homologous dsDNA (#5 and #6) and 32P-labeled ssDNA (#5). Three different concentrations of these substrates were used: 12 μm of ssDNA and 24 μm dsDNA (1× dsDNA); 12 μm of ssDNA and 240 μm dsDNA (10× dsDNA); or 120 μm ssDNA and 24 μm of dsDNA (10× ssDNA). The product yield was normalized relative to the DNA substrate present at the lowest concentration in the reaction.
It has been shown that RecA protein can provoke some limited strand separation of very short duplexes, shorter than 30 bp, in the presence of ATP, but not ATPγS (Bianchi et al. 1985; Villani et al. 1993). Accordingly, we did not observe DNA strand separation (i.e., the detection of ssDNA by gel electrophoresis) by the RecA nucleoprotein filament under conditions of our DNA strand exchange reactions in trans, in the presence of ATPγS (data not shown); furthermore, ssDNA was not detected even when single strand binding (SSB) protein was added to trap any that might transiently form (data not shown). Likewise, incubation with the nucleoprotein filament did not render the heterologous dsDNA sensitive to reaction with KMnO4, an ssDNA-specific reagent (data not shown).
Thus, DNA strand exchange in trans likely occurs through collision between an activated dsDNA that is bound to the secondary site of RecA nucleoprotein filament and ssDNA that is free in solution. Consistent with this inference, DNA strand exchange in trans is dependent on dsDNA concentration, as shown by the reaction half-time measurements in Table 3.
Table 3.
The rate of DNA strand exchange in trans varies with the dsDNA concentration
DNA strand exchange in trans reactions were conducted using standard conditions at the indicated dsDNA concentrations.
The reaction half-time was determined by fitting the reaction time course to a single-exponential function.
The binding of DNA in the secondary site of the RecA nucleoprotein filament is very dynamic. As we reported previously, equilibration among different DNA species that can bind to the secondary site of the RecA nucleoprotein filament is very rapid, occurring in <1 min (Mazin and Kowalczykowski 1998). Consistent with this observation, the observed rates of DNA strand exchange in trans did not depend on the order with which the homologous ssDNA and dsDNA substrates were bound to the RecA nucleoprotein filament (data not shown).
The heteroduplex dsDNA product of the DNA strand exchange in trans is weakly associated with the RecA nucleoprotein filament
Conventional DNA strand exchange in cis occurs between ssDNA bound to the primary site of the RecA filament and homologous dsDNA bound to the secondary. After exchange of DNA strands, the heteroduplex dsDNA product of this reaction remains associated with the RecA nucleoprotein filament, presumably, occupying the tight-binding primary site (Ullsperger and Cox 1995). In contrast, our results indicate that DNA strand exchange in trans occurs between dsDNA bound to the secondary DNA-binding site and ssDNA that is free in a solution. Because the primary site is occupied by nonhomologous, nonexchangable ssDNA, the product of DNA strand exchange in trans could remain loosely associated only with the weaker secondary DNA-binding site of the RecA nucleoprotein filament. Indeed, it is known that nonhomologous dsDNA binds only weakly to the secondary site of the filament, producing complexes that hardly survive gel electrophoresis (Muller et al. 1990) and that can be detected only by sensitive, though indirect, assays (Chow and Radding 1985; Tsang et al. 1985; Conley and West 1990; Rould et al. 1992).
Exploiting the known differences in DNA binding affinities displayed by the two DNA-binding sites of RecA protein, we wanted to ascertain more directly whether the dsDNA products of the in cis and in trans reactions are associated with different RecA protein binding sites, that is, with the primary and the secondary sites, respectively. Therefore, the nondeproteinized products of the DNA strand exchange in cis and in trans were analyzed by polyacrylamide gel electrophoresis.
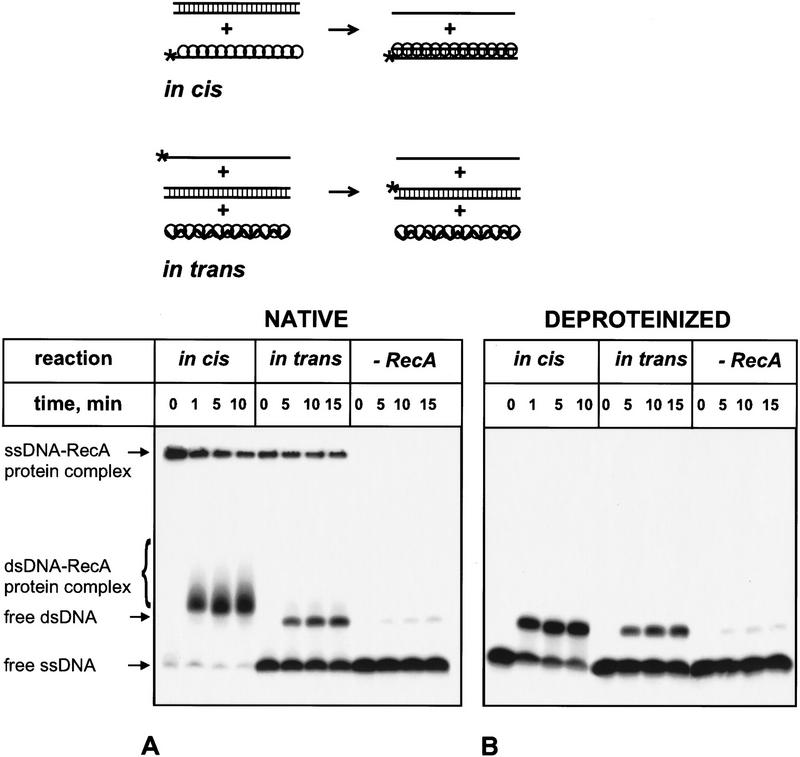
Binding of ssDNA by free RecA protein results in complexes that migrate slowly in the polyacrylamide gel (Fig. 9A; reaction in cis, time 0). Previously, we showed that ssDNA is accommodated in the primary site of these RecA protein–ssDNA complexes (Mazin and Kowalczykowski 1996, 1998). As expected, the dsDNA heteroduplex product of DNA strand exchange in cis was retarded in the gel, indicating its tight association with the primary site of RecA protein (Fig. 9A; reaction in cis). In contrast, binding of ssDNA by the RecA nucleoprotein filament produces complexes in which ssDNA is bound to the secondary site of RecA protein (Mazin and Kowalczykowski 1996, 1998). These complexes, shown in Figure 9A (reaction in trans, time 0), differ from the complexes containing ssDNA in the primary site by virtue of their high sensitivity to competing ssDNA (Mazin and Kowalczykowski 1996, 1998). Importantly, the dsDNA heteroduplex product of the DNA strand exchange in trans migrated almost unretarded in the gel, showing lack of an association with the RecA nucleoprotein filament (Fig. 9A; reaction in trans). The amount of dsDNA heteroduplex that is found as free, unassociated dsDNA in Figure 9A is 14.8% 19.4%, and 23% for the 5, 10, and 15 min of the reaction, respectively. In comparison, deproteinized samples from the same reaction in trans yielded 17.4%, 22.8%, and 26.7% of dsDNA heteroduplex product for the same times, respectively (Fig. 9B), showing that only about 15% in trans dsDNA product was associated with the RecA nucleoprotein filament. A similar comparison of the deproteinized and nondeproteinized samples for the DNA strand exchange in cis shows that 95%–100% of dsDNA product was associated with the filament.
Figure 9.
The heteroduplex dsDNA product of DNA strand exchange in trans is associated weakly with the RecA nucleoprotein filament. For the reactions in trans, presynaptic filaments were assembled on ssDNA (#43); these filaments were mixed with dsDNA (#5 and #6) and 32P-labeled ssDNA (#5). For the reactions in cis, the presynaptic filaments were formed with 32P-labeled ssDNA (#5) and mixed with homologous dsDNA (#5 and #6) in the presence of heterologous ssDNA (#43). Both reactions were carried out at 37°C. The products of these two reactions were resolved on a polyacrylamide gel, both as native complexes (A) and after deproteinization (B).
In agreement with our expectations, these results demonstrate an important distinction between DNA strand exchange in cis and in trans: The dsDNA products of these reactions have notably different degrees of association with the RecA nucleoprotein filament, suggesting that they are produced in, and are associated with, the two different DNA-binding sites.
Discussion
The results presented in this paper show that the RecA nucleoprotein filament can promote two types of DNA strand exchange reactions, which we call in cis and in trans. Both our present and previous results (Mazin and Kowalczykowski 1996, 1998) demonstrate that each of these reactions requires binding of dsDNA to the secondary site of the RecA protein–ssDNA filament. For the in cis reaction, the homology search is a transient process that is coupled to, or immediately followed by, DNA strand exchange. However, the use of heterologous dsDNA disrupts this coupling and reveals an activated form of dsDNA that exists at the time of homologous recognition.
Our results indicate that the binding of dsDNA to the secondary DNA-binding site of RecA protein plays an essential role in DNA strand exchange in trans. The ssDNA competitor with high, but not low, affinity for the secondary site completely inhibits the reaction in trans. Furthermore, only one molar equivalent of the high affinity ssDNA competitor is sufficient both to saturate the secondary site (Mazin and Kowalczykowski 1996, 1998), and to fully inhibit DNA strand exchange in trans. However, an excess of the dsDNA substrate could restore the reaction, demonstrating the reversible nature of this inhibition. These findings are consistent with our previous demonstration of a competition between ssDNA and dsDNA for binding to the secondary site (Mazin and Kowalczykowski 1996).
Previously, it was shown that the RecA nucleoprotein filament topologically unwinds dsDNA upon its binding (Cunningham et al. 1979; Rould et al. 1992). Our results argue that this unwinding renders dsDNA accessible and highly receptive for the interaction with complementary ssDNA, even when this ssDNA is not within the nucleoprotein filament (Fig. 1). Thus, binding to the secondary site of the RecA nucleoprotein filament activates dsDNA for both homologous recognition and DNA strand exchange by locally disrupting DNA structure. These structural changes may involve the local melting of dsDNA that is short lived and, therefore, undetected by conventional methods. Alternatively, the dsDNA that is complexed to the RecA nucleoprotein filament may adopt a novel and unique structure with an enhanced ability to interact with ssDNA. In either case, our results demonstrate that this activated dsDNA, which is bound to the secondary site, collides with homologous ssDNA, which is free in solution. This interpretation is substantiated by our finding that DNA strand exchange in trans is apparently second order with respect to dsDNA, occurring as depicted in Figure 1.
Although the dsDNA is structurally activated for DNA strand exchange, macroscopic strand separation is not occurring. Though the binding of short (<30 bp) dsDNA to the primary site of RecA protein in the presence of ATP, but not ATPγS, results in a slight formation of ssDNA (Bianchi et al. 1985; Villani et al. 1993), DNA strand exchange in trans occurs (1) in the presence of ATPγS; (2) without detectable strand separation; and (3) is not limited to such short duplex DNA. DNA strand separation by the RecA nucleoprotein filament did not occur under conditions of the DNA strand exchange in trans, even when measured in the presence of SSB protein, nor did incubation with the nucleoprotein filament render the heterologous dsDNA sensitive to reaction with an ssDNA specific reagent, KMnO4. Finally, the possibility that DNA strand exchange in trans could occur via RecA protein-dependent denaturation of dsDNA followed by its spontaneous reannealing with complementary ssDNA is not substantiated by our experimental observations. This possible explanation implies that the rates of DNA strand exchange in trans would be similar for the reactions of dsDNA with each of its two individual complementary ssDNA strands. However, this expectation is not supported by our results, which show a significant difference in the reaction rate for each complementary ssDNA strand with the same dsDNA homolog. Thus, we conclude that DNA strand exchange in trans occurs in the absence of any detectable ssDNA as an intermediate.
Our data show that the nature of the ssDNA is an important determinant of the in trans reaction. We noticed that ssDNA with high affinity for the secondary site (e.g., #39 or #20), which consequently competed effectively with its dsDNA homolog for binding to the secondary site, was a poor substrate for DNA strand exchange in trans. This result underscores further the importance of dsDNA binding to the secondary site as a prerequisite for the in trans reaction. However, the inverse correlation between affinity of ssDNA for the secondary site and its participation for the in trans reaction was not absolute. For instance, the affinity of ssDNA #40, a substrate that showed the highest rate of exchange in trans, was higher than that of ssDNA #43, the second most effective substrate (data not shown). Although the ssDNA substrates were selected for this study to be free from conventional secondary structure, this imperfect correlation indicates that some unidentified properties of the DNA affect rates of DNA strand exchange in trans. Alternatively, the two strands of the dsDNA may interact asymmetrically with the secondary DNA binding site and thus display different reactivities with the ssDNA. The effects of DNA base composition on DNA strand exchange in trans are complex and required further exploration.
Several observations argue that DNA strand exchange in trans is a novel, specific process rather than a nonspecific aggregation-mediated reaction. First, only fully functional RecA nucleoprotein filaments promote DNA strand exchange in trans. Neither the condensed, inactive filaments of wild-type RecA protein formed in the absence of an ATP analog, nor the inactive filaments of the RecA56 and RecA1 mutant proteins supported this reaction. This result points to the common limitation of each of these inactive filaments, which is their inability to induce an extended or unwound form of DNA (Lauder and Kowalczykowski 1993). Second, the highly specific inhibition by ssDNA with high affinity for the secondary site rules out the possibility that DNA strand exchange in trans is caused by a nonspecific aggregation of DNA. Finally, the finding that nondeproteinized dsDNA product of the in trans reaction migrates in gel electrophoresis as free dsDNA further corroborates this notion.
We were fully aware that an excess of RecA protein or its transfer from the original filament to the in trans ssDNA could cause DNA strand exchange in cis that might be mistakenly interpreted as the in trans reaction. Therefore, extra precautions were taken to exclude these possibilities. First, all reactions were performed in the presence of nonspecific ssDNA. Second, purification of the RecA nucleoprotein filament by gel chromatography to remove unbound RecA protein did not affect DNA strand exchange in trans. Third, all reactions were carried out in the presence of ATPγS or ADP·AlF4−. Previously, we showed, that in the presence of ATPγS, RecA protein remains tightly associated with the ssDNA bound to its primary site; the transfer of RecA protein does not occur (Mazin and Kowalczykowski 1996, 1998).
However, most importantly, we found that the reaction in trans displays a qualitative distinction from the DNA strand exchange in cis. The nondeproteinized heteroduplex dsDNA, the product of the in trans reaction, migrates as free dsDNA during polyacrylamide gel electrophoresis. In contrast, the heteroduplex dsDNA product of the in cis reaction, was tightly associated with the RecA nucleoprotein filament. This finding indicates that the dsDNA products of DNA strand exchange in cis and in trans are produced in, and are associated with, the two different RecA protein DNA-binding sites, that is, the primary and the secondary, respectively. This distinction along with other specific characteristics of DNA strand exchange in trans allows us to define this reaction as a novel reaction promoted by the RecA nucleoprotein filament.
DNA strand exchange in trans can be added to the list of RecA protein activities that require ATP binding, but not hydrolysis. These activities include renaturation of ssDNA, DNA pairing, DNA strand exchange in cis, and co-proteolysis of UmuD, LexA repressor, and lambdoid prophage repressors (Kowalczykowski et al. 1994). There are a few instances in which ATPγS cannot substitute for ATP. For example, neither DNA four-strand exchange nor the bypass of DNA heterology occurs in the presence of ATPγS. However, more importantly, there is no activity of RecA protein that occurs in the presence of ATPγS, for which ATP cannot substitute. Thus, as expected, in the case of the in trans reaction, we observed that when the RecA nucleoprotein filament that was assembled on φX174 ssDNA in the presence of ATP (and an ATP-regeneration system) DNA strand exchange between ssDNA and dsDNA substrates also occurred (data not shown). The extent of this reaction was comparable to the in trans reaction occurring in the presence of ATPγS. Moreover, the reaction in the presence of ATP was sensitive to the inhibition by ssDNA with high, but not with low, affinity for the secondary site, indicating that this reaction required involvement of the secondary RecA protein DNA binding site. Although transfer of RecA protein from the original filament onto ssDNA-substrate cannot be ruled out entirely in such experiments, we believe that DNA strand exchange in trans, as with other reactions promoted by RecA protein, can occur in the presence of either ATP or its nonhydrolyzable analogs.
The in trans reaction emphasizes a distinct and essential step of DNA strand exchange: activation of dsDNA for homologous pairing. Though we can speculate about its biological significance, the in vivo role of DNA strand exchange in trans, if any, remains to be fully appreciated. Clearly, DNA strand exchange in trans requires formation of the RecA nucleoprotein filament, and, therefore, is most feasible after extensive DNA damage. Because DNA strand exchange in trans occurs between naked dsDNA- and ssDNA partners, this reaction in vivo would require a situation that generates ssDNA that is free of SSB or RecA proteins. Extensive DNA damage could result in depletion of the unbound pool of SSB protein (Kowalczykowski and Roman 1990). In addition, the degraded ssDNA could be too short to permit the efficient binding of RecA protein. Such limitations clearly exclude DNA strand exchange in trans from being involved in the mainstream mechanisms of repair of DNA double-stranded breaks. Still, DNA strand exchange in trans may have a role in some of the minor pathways of DNA repair. Interestingly, the phenomenon termed indirect stimulation of homologous recombination seems to be the closest that fits the hypothetical requirements for DNA strand exchange in trans in vivo. Golub and Low (1983) found that if a λlacZ phage is coinfected with a second, UV-irradiated phage (phage λ or a λ–φ80 hybrid) which does not carry the lacZ gene, recombination between the unirradiated λlacZ phage and a chromosomal copy of the lacZ gene is enhanced in a SOS-independent manner. Indirect stimulation of recombination required functional helicase II and RecJ exonuclease, whose combined activities could generate ssDNA substrates (Schellhorn and Low 1991). This phenomenon of indirect stimulation of recombination, still has no clear explanation and may potentially involve various underlying mechanisms. DNA strand exchange in trans may contribute to this indirect stimulation of homologous recombination. We imagine that the RecA nucleoprotein filament, which is formed on the damaged λ phage DNA, could serve as a scaffold that stimulates recombination between two mutually homologous DNA partners. The region of shared homology between damaged and recombining phages that is required for indirect stimulation of recombination could help to anchor DNA of the recombining phage on the nucleoprotein filament scaffold via a conventional recombination event which, in turn, would facilitate DNA strand exchange in trans through a pseudo-first order reaction between the lacZ genes.
Both our biochemical interpretations and their possible relationships to recombination in vivo prompt us to speculate on the evolutionary origin of the in trans reaction. Given the remarkable properties of the RecA protein (Kowalczykowski and Eggleston 1994; Eggleston and West 1997; Bianco et al. 1998), it is difficult to imagine the single-step evolution of a protein with the collective capacity to (1) form a helical filament, (2) bind two different DNA molecules, (3) activate the normally stable duplex form of DNA, (4) homologously pair and exchange DNA strands, and then (5) acquire the ability to turnover through an ATPase activity. Instead, our results demonstrate that the RecA filament alone, without ATP hydrolysis, is capable of promoting an effective bimolecular DNA strand exchange reaction. This finding, in turn, suggests that the progenitor of the modern RecA protein was a protein that either self-assembled or assembled in an ssDNA-dependent manner and that this resulting filament possessed the ability to activate dsDNA for the important cellular process of recombination-mediated repair. Subsequently, this early in trans-proficient DNA strand exchange protein acquired the more complex ability to bind two DNA molecules simultaneously that is needed to promote DNA strand exchange in cis, as well as the ability to hydrolyze ATP, an activity that allowed the protein dissociate itself from the heteroduplex product and promote unidirectional branch migration through limited mismatches, all of which are rather sophisticated biochemical processes. The realization that RecA-like proteins are found in all organisms, including the Archaea (Seitz et al. 1998), further argues that this class of proteins is both evolutionarily old and important (Brendel et al. 1997); the finding that an ATP hydrolysis defective variant of Rad51 protein, a eukaryotic RecA-like protein, is active in vivo (Sung and Stratton 1996) also argues that the ATPase activity is dispensable. The discovery of DNA strand exchange in trans offers insight into both the mechanism and the development of the homologous pairing process.
Materials and methods
RecA protein and DNA substrates
RecA wild-type protein was purified from Escherichia coli strain JC12772 by use of a procedure based on precipitation by spermidine acetate (Griffith and Shores 1985; S.C. Kowalczykowski, unpubl.). RecA56 and RecA1 proteins were purified as described previously (Lauder and Kowalczykowski 1993). Virion φX174 ssDNA was purchased from New England Biolabs. Oligodeoxyribonucleotides were purified after their synthesis by electrophoresis in polyacrylamide gels containing 8 m urea. To prepare dsDNA, two complementary oligonucleotides were denatured at 95°C for 3 min, and then annealed in a buffer containing 7 mm Tris-HCl (pH 7.6), 100 mm NaCl, 10 mm Na citrate, 1 mm MgCl2, and 0.5 mm DTT for 1 hr at a temperature optimal for their hybridization. Oligonucleotides were stored at −20°C. DNA concentrations are reported as total nucleotides.
DNA strand exchange in trans and in cis
Unless otherwise indicated, the conditions of DNA strand exchange in trans were the following. Presynaptic filaments were formed by incubation of RecA protein (4 μm) with a ssDNA oligonucleotide (12 μm) (the 33-mer, #43) or with φX174 ssDNA (12 μm) in standard buffer containing 33 mm HEPES (pH 7.0), 1.2 mm magnesium acetate, 2 mm DTT, 100 μg/ml BSA, and 1 mm ATPγS at 37°C for 12 min. Incubation was followed by an increase in the magnesium acetate concentration to 11.2 mm. The unbound RecA protein was sequestered by addition of ssDNA (htr#1; 30 μm), and incubation was continued for 4 min. Alternatively, the unbound RecA protein was removed by gel filtration of the RecA nucleoprotein filaments through Chromaspin-1000 columns (Clontech) equilibrated with standard buffer. In this case, the magnesium acetate concentration was increased to 11.2 mm after the gel filtration. DNA strand exchange in trans was initiated by addition of the 32P-labeled ssDNA (12 μm) and homologous dsDNA (24 μm). To minimize the level of spontaneous DNA strand exchange, the reaction temperature was 23°C. The products of DNA strand exchange were deproteinized with 1% SDS and 50 mm EDTA and resolved by electrophoresis on 10% polyacrylamide gels in TBE buffer [90 mm Tris-borate (pH 8.3), 0.5 mm EDTA]. DNA strand exchange in cis was performed under identical conditions, except the RecA nucleoprotein filament was assembled on 32P-labeled ssDNA. When ADP·AlF4− was used, ATPγS was replaced with 5 mm ADP, 10 mm NaF, and 0.4 mm Al(NO3)3 (Kowalczykowski and Krupp 1995). Gels were quantitated by use of a Storm 840 PhosphorImager (Molecular Dynamics) with ImageQuant software. Unless indicated otherwise, the rate of spontaneous thermal DNA strand exchange was subtracted from the observed rates of RecA protein-promoted reactions.
Association of the DNA strand exchange products with native RecA nucleoprotein filaments
For the reactions in trans, presynaptic filaments were assembled by binding of RecA protein (8 μm) to ssDNA (#43) (12 μm). The unbound RecA protein was sequestered by addition of ssDNA (htr#1) (60 μm). The filaments were mixed with dsDNA (#5 and #6) (48 μm) and 32P-labeled ssDNA (#5) (12 μm) under standard conditions. For the reactions in cis, the presynaptic filaments were formed by mixing RecA protein (8 μm) with 32P-labeled ssDNA (#5; 12 μm). Heterologous ssDNA (htr#1) (60 μm) was then added to sequester the unbound RecA protein. DNA strand exchange in cis was initiated by addition of homologous dsDNA (#5 and #6; 48 μm). To achieve higher yields, the reactions were carried out at 37°C. After the indicated period of time, aliquots were withdrawn from the reaction mixtures and were either loaded directly on a 10% polyacrylamide gel after mixing with a one-tenth volume of loading buffer (20% Ficoll, 0.1% bromophenol blue) or were deproteinized with 1% SDS and 50 mm EDTA prior to the loading on a gel. The control for the spontaneous thermal DNA strand exchange was performed under conditions identical with those for the DNA strand exchange in trans, except that RecA protein was substituted with RecA protein storage buffer which contained 20 mm Tris-HCl (pH 7.5), 1 mm DTT, 0.1 mm EDTA and 10% glycerol.
Nuclease assays
To control for nuclease activity, the RecA protein preparation was assayed at the standard conditions for DNA strand exchange. A RecA nucleoprotein filament was assembled on ssDNA (#9). Duplex DNA (#1 and #2) and complementary ssDNA oligonucleotide (#1) were used as substrates for DNA strand exchange in trans. The dsDNA was 32P-labeled at the 5′ end of the identical DNA strand (#1) by use of T4 polynucleotide kinase and [γ-32P]ATP. DNA strand exchange was carried out for 1 hr at 23°C. After deproteinization, the products of this reaction were separated by use of a 10% polyacrylamide gel run in TBE. The DNA strand that was displaced from dsDNA was eluted from the gel, and its integrity was confirmed by gel electrophoresis on a 10% polyacrylamide gel containing 50% urea.
Chemical modification of dsDNA with KmnO4
The RecA nucleoprotein filament was formed as for DNA strand exchange in trans, except that the concentration of DTT in the standard buffer was reduced to 0.5 mm. Heterologous dsDNA (#5 and #6) in which either of the strands was 32P-labeled at the 5′ end was added to the filament. The samples were treated further with 2–6 mm KMnO4 for 2–5 min at 23°C. The reactions were quenched with 0.72 m β-mercaptoethanol and 25 mm EDTA. Modified DNA was precipitated by ethanol and cleaved with 10% piperidine for 30 min at 95°C. The products of DNA cleavage were resolved by electrophoresis on a 10% sequencing gel that was run at 1200 V for 2 hr.
Acknowledgments
We thank the following members of the laboratory for both the discussion of our results and the critical reading of the manuscript: Dan Anderson, Piero Bianco, Joel Brockman, Frederick Chedin, Deana Haddox, Frank Harmon, Noriko Kentake, Julie Kleiman, James New, Erica Seitz, Tomohiko Sugiyama, and Eugene Zaitsev. We are also grateful to Andrei Kuzminov and Murty Madiraju for their comments and suggestions. A.M. is thankful to the Institute of Cytology and Genetics, Russian Academy of Science, Novosibirsk, for granting him a leave of absence. This work was supported by National Institutes of Health grant AI-18987.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sckowalczykowski@ucdavis.edu; FAX (530) 752-5939.
References
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M, Riboli B, Magni G. E. coli recA protein possesses a strand separating actvity on short duplex DNAs. EMBO J. 1985;4:3025–3030. doi: 10.1002/j.1460-2075.1985.tb04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Brendel V, Brocchieri L, Sandler SJ, Clark AJ, Karlin S. Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J Mol Evol. 1997;44:528–541. doi: 10.1007/pl00006177. [DOI] [PubMed] [Google Scholar]
- Chow SA, Radding CM. Ionic inhibition of formation of recA nucleoprotein networks blocks homologous pairing. Proc Natl Acad Sci. 1985;82:5646–5650. doi: 10.1073/pnas.82.17.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley EC, West SC. Underwinding of DNA associated with duplex–duplex pairing by RecA protein. J Biol Chem. 1990;265:10156–10163. [PubMed] [Google Scholar]
- Cox MM, Lehman IR. Directionality and polarity in recA protein-promoted branch migration. Proc Natl Acad Sci. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham RP, Shibata T, DasGupta C, Radding CM. Single strands induce recA protein to unwind duplex DNA for homologous pairing. Nature. 1979;281:191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- Eggleston AK, West SC. Recombination initiation: Easy as A, B, C, D... chi? Curr Biol. 1997;7:R745–R749. doi: 10.1016/s0960-9822(06)00394-0. [DOI] [PubMed] [Google Scholar]
- Golub EI, Low KB. Indirect stimulation of genetic recombination. Proc Natl Acad Sci. 1983;80:1401–1405. doi: 10.1073/pnas.80.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J, Shores CG. RecA protein rapidly crystallizes in the presence of spermidine: A valuable step in its purification and physical characterization. Biochemistry. 1985;24:158–162. doi: 10.1021/bi00322a022. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P, West SC, Stasiak A. Role of recA protein spiral filaments in genetic recombinations. Nature. 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Roman LJ. Reconstitution of homologous pairing activity dependent upon the combined activities of purified E. coli RecA, RecBCD, and SSB proteins. In: Richardson CC, Lehman IR, editors. Molecular mechanisms in DNA replication and recombination. New York: Wiley-Liss; 1990. pp. 357–373. [Google Scholar]
- Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Krupp RA. DNA-strand exchange promoted by RecA protein in the absence of ATP: Implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc Natl Acad Sci. 1995;92:3478–3482. doi: 10.1073/pnas.92.8.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder SD, Kowalczykowski SC. Negative co-dominant inhibition of recA protein function. Biochemical properties of the recA1, recA13 and recA56 proteins and the effect of recA56 protein on the activities of the wild-type recA protein function in vitro. J Mol Biol. 1993;234:72–86. doi: 10.1006/jmbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Kowalczykowski SC. The specificity of the secondary DNA binding site of RecA protein defines its role in DNA strand exchange. Proc Natl Acad Sci. 1996;93:10673–10678. doi: 10.1073/pnas.93.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 1998;17:1161–1168. doi: 10.1093/emboj/17.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- ————— Transfer of recA protein from one polynucleotide to another: Kinetic evidence for a ternary intermediate during the transfer reaction. J Biol Chem. 1987;262:2085–2092. [PubMed] [Google Scholar]
- Moreau PL, Carlier MF. RecA protein-promoted cleavage of LexA repressor in the presence of ADP and structural analogues of inorganic phosphate, the fluoride complexes of aluminum and beryllium. J Biol Chem. 1989;264:2302–2306. [PubMed] [Google Scholar]
- Muller B, Koller T, Stasiak A. Characterization of the DNA binding activity of stable RecA-DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J Mol Biol. 1990;212:97–112. doi: 10.1016/0022-2836(90)90307-8. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman EH, Ogawa H. RecA-like recombination proteins in eukaryotes: Function and structures of RAD51 genes. Cold Spring Harbor Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- Rould E, Muniyappa K, Radding CM. Unwinding of heterologous DNA by RecA protein during the search for homologous sequences. J Mol Biol. 1992;226:127–139. doi: 10.1016/0022-2836(92)90129-8. [DOI] [PubMed] [Google Scholar]
- Schellhorn HE, Low KB. Indirect stimulation of recombination in Escherichia coli K-12: Dependence on recJ, uvrA, and uvrD. J Bacteriol. 1991;173:6192–6198. doi: 10.1128/jb.173.19.6192-6198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes & Dev. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, DasGupta C, Cunningham RP, Radding CM. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Stasiak A, Stasiak AZ, Koller T. Visualization of recA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harbor Symp Quant Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Tsang SS, Chow SA, Radding CM. Networks of DNA and recA protein are intermediates in homologous pairing. Biochemistry. 1985;24:3226–3232. doi: 10.1021/bi00334a023. [DOI] [PubMed] [Google Scholar]
- Ullsperger CJ, Cox MM. Quantitative RecA protein binding to the hybrid duplex product of DNA strand exchange. Biochemistry. 1995;34:10859–10866. doi: 10.1021/bi00034a019. [DOI] [PubMed] [Google Scholar]
- Villani G, Cazaux C, Pillaire MJ, Boehmer P. Effects of a single intrastrand d(GpG) platinum adduct on the strand separating activity of the Escherichia coli proteins RecB and RecA. FEBS Lett. 1993;333:89–95. doi: 10.1016/0014-5793(93)80380-d. [DOI] [PubMed] [Google Scholar]
- Yu X, Egelman EH. Image analysis reveals that Escherichia coli RecA protein consists of two domains. Biophys J. 1990;57:555–566. doi: 10.1016/S0006-3495(90)82571-6. [DOI] [PMC free article] [PubMed] [Google Scholar]