Summary
The transcription factor GATA3 plays an essential role during T cell development and T helper 2 (Th2) cell differentiation. To understand GATA3-mediated gene regulation, we identified genome-wide GATA3 binding sites in ten well-defined developmental and effector T lymphocyte lineages. In the thymus, GATA3 directly regulated many critical factors, including Th-POK, Notch1 and T cell receptor subunits. In the periphery, GATA3 induced a large number of Th2 cell-specific as well as Th2 cell non-specific genes, including several transcription factors. Our data also indicate that GATA3 regulates both active and repressive histone modifications of many target genes at their regulatory elements near GATA3 binding sites. Overall, although GATA3 binding exhibited both shared and cell-specific patterns among various T cell lineages, many genes were either positively or negatively regulated by GATA3 in a cell type-specific manner, suggesting that GATA3-mediated gene regulation depends strongly on co-factors existing in different T cells.
INTRODUCTION
GATA3, a zinc-finger transcription factor, plays a critical role in both early and late T cell differentiation (Collins et al., 2009; Hattori et al., 1996; Ho et al., 2009; Zhang et al., 1997; Zheng and Flavell, 1997; Zhu et al., 2010). Conditional deletion of Gata3 in CD4+ T cells abolishes T helper type 2 (Th2) cell differentiation (Pai et al., 2004; Zhu et al., 2004), while forced expression of GATA3 results in Th2 cell differentiation even in the absence of IL-4-STAT6 signaling (Lee et al., 2000; Ouyang et al., 2000; Zhang et al., 1997). Beyond its role in Th2 cells, GATA3 is also critical during multiple stages of T cell development (Hattori et al., 1996; Pai et al., 2004; Ting et al., 1996). Although GATA3 is not required for hematopoietic stem cell (HSC) development or maintenance, it is required for the development of the most primitive T lineage progenitors (Hosoya et al., 2009). Deletion of Gata3 in the CD4-CD8 double-positive (DP) stage of T cell differentiation disrupts the development of the CD4 single positive (SP) lineage (Pai et al., 2003; Zhu et al., 2004). GATA3 acts as a CD4 lineage-determining factor in part by up-regulating expression of the transcription factor Th-POK; however, a Th-POK transgene failed to rescue the CD4+ T cell development defect of Gata3-deficient thymocytes, suggesting that GATA3 regulates other important factors during CD4+ T cell development (Wang et al., 2008).
Despite the critical function of GATA3 at different cell developmental stages, a limited number of its target genes in Th2 and DP cells have been identified (Ansel et al., 2006; Kishikawa et al., 2001; Lavenu-Bombled et al., 2002; Lee et al., 2001; Wang et al., 2008; Zhu et al., 2010). A study using ChIP-chip, reported that GATA3 binds to hundreds of genes in both Th1 and Th2 cells and may function to oppose T-bet activity during differentiation of these T helper cells (Jenner et al., 2009). However, genome-wide analyses of GATA3 binding and GATA3-mediated gene regulation in Th2 cells have not been reported. Furthermore, the transcriptional programs regulated by GATA3 across most of the T cell lineages are largely unknown. For example, GATA3 is known to regulate the expression of Th-POK, which is required for CD4+ lineage commitment. However, other GATA3-regulated genes that are also important for CD4+ cell differentiation have not been identified. In addition to CD4 lineage decision and Th2 cell differentiation, it remains unclear whether GATA3 functions in other Th cells and in CD8+ T cells, where GATA3 is expressed in low amounts.
To investigate the function of GATA3 in various Th cells and during thymic development, we have identified GATA3 binding sites genome-wide, using ChIP-Seq, in naïve CD4+, Th1, Th2, Th17, iTreg, nTreg, NKT, CD8+ cells as well as CD4-CD8 double positive (DP) and CD3-negative CD4-CD8 double-negative (DN) thymocytes. In addition, we investigated changes in gene expression that resulted from deletion of Gata3 in CD3loCD4+CD8int-hiCD69+ (CD3loDP), CD3hiCD4+CD8int-hiCD69+ (CD3hiDP) thymocytes that undergo positive selection and in various Th cells.
Our data indicate that, although there are many GATA3 binding sites shared among different T cells, GATA3 regulates strikingly different transcriptional programs depending on cellular context. GATA3 directly regulates many critical factors, including Th-POK, Notch1 and T cell receptor subunits in the thymus; it directly or indirectly induces a large number of Th2 cell-specific as well as Th2 cell non-specific genes including several transcription factors that may be involved in the GATA3 regulatory network during Th cell differentiation. Finally, we found that GATA3 regulates both active and repressive histone modifications, H3K4me1,2 and H3K27me3, respectively, at the enhancer regions of many of its target genes, including Il4 and Ifng in Th2 cells.
RESULTS
Genome-wide mapping of GATA3 binding sites in various T cell types
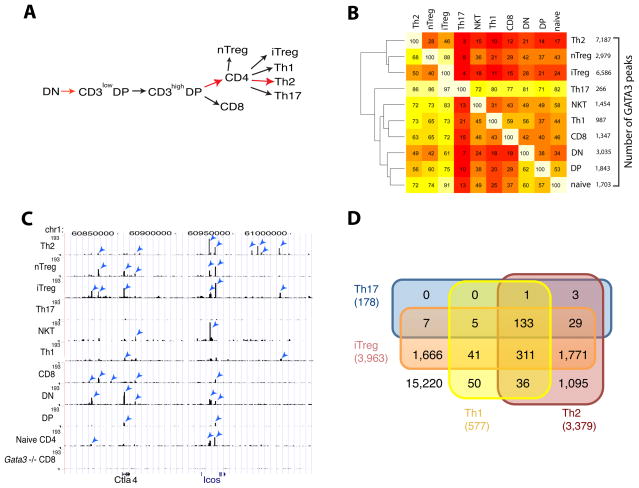
T cells develop in the following order in the thymus: DN, CD3loCD4+CD8int-hi (CD3lowDP), CD3hiCD4+CD8int-hi (CD3hiDP) and single positive (CD4+ or CD8+) cells. Naïve CD4+ T cells are induced to differentiate to Th1, Th2, Th17 and iTreg cells in vitro depending on the cytokine milieu using previously established conditions (Zhu et al., 2009) (Figure 1A and Figure S1). The differentiation steps from DN to DP, from DP to CD4+ T cells and from naïve CD4+ T cells to Th2 cells are known to require GATA3 (highlighted by the red arrows in Figure 1A).
Figure 1. Genome-wide GATA3 binding patterns in various T cell lineages.
A. Cartoon showing various development and differentiation stages of T cells analyzed in this study. GATA3-mediated steps are highlighted in red.
B. Overlaps of GATA3 binding sites in various T cell types. The number of binding sites identified in each cell type is indicated on the right of the heatmap. Number within each cell (i, j) in the heatmap and the corresponding intensity indicate the percentage of sites in cell type i (row) overlapping those in cell type j (column). The cluster pattern shown on the left was generated based on the overall binding similarity.
C. GATA3 binding pattern at the genomic region containing Ctla4 and Icos genes. Blue arrowheads indicate the peaks identified by MACS with a P value of ≤10−13.
D. Overlap of GATA3-bound genes in Th1, Th2, Th17 and iTreg cells. GATA3 was considered associated with a gene if a MACS peak was located anywhere from 10 kb upstream of transcription start site to 3 kb downstream of transcription end site.
GATA3 expression can be detected at different amounts in various T cell populations (Figures S1B–E). To elucidate the transcriptional programs regulated by these different amounts of GATA3 during T cell development from DN to the various differentiated Th cells, we mapped GATA3 binding sites genome-wide using ChIP-Seq in each of these developmental stages and differentiated lineages (Table S1). The CD3loCD4+CD8int-hi and CD3hiCD4+CD8int-hi were used as a single population (CD4+CD8int-hi, DP) for the ChIP-Seq analysis. To expand the spectrum of cell types studied, we also analyzed GATA3 binding in a closely related T lineage, NKT cells.
To confirm that the antibody used is specific for the GATA3 protein and set criteria for specific peak identification from the ChIP-Seq data, we performed ChIP-Seq using the same antibody on CD8+ T cells in which the Gata3 gene was absent as a result of Cre-mediated deletion of loxP-flanked Gata3 alleles. As expected, many ChIP-Seq peaks were found in wild type CD8+ T cells but not in the Gata3-deleted cells (Figure S1F. To define a peak as significant, we applied a p-value of 10−13 for peak identification using MACS (Zhang et al., 2008). We chose this as the most stringent parameter that allowed us to still identify the known GATA3 target genes, Il4, Il5 and Il13, as GATA3-bound genes in the ChIP-Seq data from Th2 cells. Using this criterion, we identified 266 and 7,187 GATA3 sites in Th17 and Th2 cells, respectively, while only 82 sites were identified in the Gata3-deleted CD8+ cells. Other cells contained intermediate numbers of GATA3 binding sites (Figure 1B, listed on the right of the panel). GATA3 binding was highly enriched in the region 10kb upstream of TSS and 5′ UTR in all the cell types studied (Figure S1G). Substantial overlaps of the GATA3 peaks were observed between biological replicate experiments (93% and 71% in DP and Th2 cells, respectively) (Figures S1H and S1I). Most of the apparent non-overlapping peaks in these replicate samples showed enrichment in GATA3 binding in both replicates by visual inspection using the UCSC Genome Browser, suggesting that they may be true GATA3 peaks although they did not pass our stringent statistic criteria to be identified as significant peaks in one of the two experiments. Indeed, more than 90% of the peaks identified in one Th2 cell library using a cutoff P value of 10−13 overlapped with the peaks identified in the other Th2 cell library with a cutoff P value of 10−6, the latter cutoff still only detected <300 peaks in GATA3-deficient cells. However, we kept this stringent criterion (P < 10−13) in peak identification in order to provide a set of highly confident GATA3 binding sites.
Examination of the ChIP-Seq data revealed both conserved and cell-specific binding of GATA3 among various T lineages, as exemplified by the genomic region containing the Ctla4 and Icos genes, which showed the most GATA3 peaks in Th2 cells and the fewest in Th17 and Th1 cells (Figure 1C). As mentioned above, some peaks identified by visual inspection did not pass our statistical criterion for peak identification (arrows in the figure indicate peaks identified by MACS).
Pair-wise comparisons of GATA3 binding sites between any two of these cell types revealed overlaps of 3%–97% as indicated by the heatmap (Figure 1B), which is asymmetric because of the directional nature of the binary overlap, i.e. X in Y vs. Y in X. We can draw two conclusions from this hierarchical clustering analysis: (1) the number of binding sites appears to be related to the amounts of GATA3 expression; (2) GATA3 binding exhibits marked cell specificity, although there are substantial overlaps of binding sites among different cells. Even cells with similar number of binding sites, for example, nTreg and DN cells, exhibited very different binding patterns reflected by their distant placement in the hierarchical clustering pattern (Figure 1B), suggesting that distinct co-factors in these cells that play critical roles in the differential GATA3 binding. Interestingly, even within the same gene cluster, GATA3 binding patterns were different in different cell types. For example, GATA3 was bound to many more sites at the Icos locus in Th2 cells than in nTreg or iTreg cells, whereas several sites to which GATA3 bound at the Ctla4 locus in nTreg and iTreg cells were not bound by GATA3 in Th2 cells (Figure 1C).
We illustrated the shared and lineage-specific GATA3-bound genes among the Th cells with a Venn diagram (Figure 1D). Our analysis indicated that only 133 genes are bound by GATA3 in all helper T cells (Table S2). While Th1, Th2 and iTreg cells each expressed a distinct set of GATA3-bound genes, a large fraction (75%) of the GATA3-bound genes in Th17 cells belong to the group shared among all the helper T cells, suggesting that GATA3 binding in Th17 cells is mainly limited to the group of core target genes, presumably containing high affinity binding sites in an open chromatin environment.
Deletion of GATA3 has distinct outcomes in different lineages
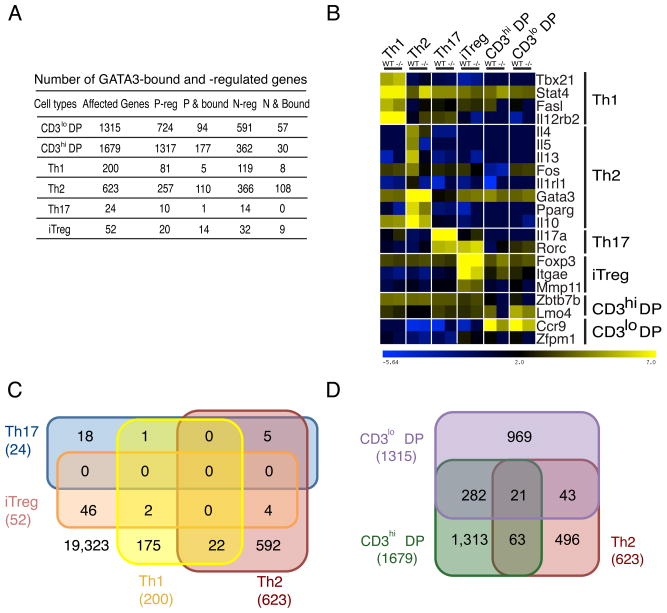
GATA3 has critical functions in DP and Th2 cells. The thousands of binding sites we identified in other cell types suggest that it could have potentially important functions in those cells as well. To directly test this, we deleted Gata3 in Th1, Th2, Th17 and iTreg cells that had been generated in vitro from naïve CD4+ T cells obtained from Gata3fl/fl mice by infecting the differentiating cells with a retrovirus expressing GFP-Cre. The mRNA profiles in both wild type (WT, GFP-negative) and Gata3-deleted (−/−, GFP-positive) cells were determined by RNA-Seq. To understand the transcriptional programs regulated by GATA3 during CD4+ T cell development in the thymus, we also analyzed the mRNA profiles in WT and Gata3-deleted CD69+CD3loDP and CD69+CD3hiDP thymocytes, which were directly purified from WT and Gata3 conditional deficient (−/−) mice (Gata3fl/fl-CD4Cre). Analysis of the biological replicate RNA-Seq data from these cells revealed that deletion of Gata3 resulted in a change in expression of different genes, and of different numbers of genes in these cells, ranging from 24 genes in Th17 cells to 1,679 genes in CD3hiDP cells (Figure 2A, Table S3).
Figure 2. GATA3 regulates distinct transcription programs in different cells.
A. Number of genes that undergo change in expression and are bound by GATA3. “Affected genes” indicates all genes that show changed expression in a specific T cell stage-lineage by deleting the Gata3 gene. “P-reg” indicates the genes that are positively regulated by GATA3. “P & bound” indicates the genes that are positively regulated and bound by GATA3. “N-reg” indicates the genes that are negatively regulated by GATA3. “N & bound” indicates the genes that are negatively regulated and bound by GATA3.
B. Deletion of Gata3 results in changes in expression of cell-specific genes. The expression pattern of a set of genes characteristic of each cell type (indicated on the right) is shown. Heatmap intensity denotes the normalized expression (log2 RPKM values).
C. Overlap of GATA3-regulated genes among all Th cells.
D. Overlap of GATA3-regulated genes among both DP subsets and Th2 cells. Genes regulated by GATA3 in each cell type were determined by the overlaps of two independent biological replicates.
Deletion of Gata3 led to changed expression of 623 genes in Th2 cells, including the previously reported Th2 cell-specific genes Il4, Il5, Il10, Il13, and Il1rl1, as shown for selected lineage specific genes in Figure 2B and in a complete list in Tables S3, consistent with our previous report (Zhu et al, 2004). In contrast, Th1cell-specific genes including Tbx21, Fasl and Il12rb2 were slightly up-regulated in Th2 cells after Gata3 deletion (Figure 2B; Tables S3). We also examined the effects of Gata3 deletion in Th2 cells on Th17 cell-specific genes and found that Rorc was modestly up-regulated as the result of Gata3 deletion; a number of iTreg-specific genes including Cd83 and Tnfsf11 were up-regulated in these Gata3-deleted Th2 cells (Figure 2B; Tables S3). These data suggest that GATA3 is involved in repression of many non-Th2-specific genes in Th2 cells.
There are far fewer genes affected by deletion of Gata3 in other Th cells (Figure 2A and Table S3). Deletion of Gata3 in Th1 or Th17 cells did not significantly affect expression of Th1- or Th17 cell-specific genes (Figure 2B and Table S3). Gata3 deletion in iTreg cells affected expression of 52 genes including the iTreg-specific genes Itgae and Mmp11; Foxp3 expression was modestly affected (Figures 2A and 2B; Table S3). Interestingly, Ccr8 expression was reduced whereas Batf3 was up-regulated after Gata3 deletion in both Th2 and iTreg cells (Table S3).
Function of GATA3 is dependent on cellular and lineage context
Among the thousands of GATA3 binding sites we identified, many are shared among different lineages. As already noted, 133 genes are bound by GATA3 in all four Th cell subtypes (Table S2). However, examination of the RNA-Seq expression data revealed that none of the genes affected by deletion of Gata3 is shared among all Th cells (Figure 2C). Although, Th1 and Th17 cells shared 138 genes bound by GATA3, only one differentially expressed gene (Ubtd1) caused by Gata3 deletion was shared between these two lineages and GATA3 did not bind to this gene. Similarly, only a minor fraction of the genes affected by deletion of Gata3 were shared between DP and Th2 cells (Figure 2D). These data suggest that GATA3-mediated gene regulation in different cell types requires cell type-specific co-factors.
GATA3 binding is associated with primary and secondary motifs
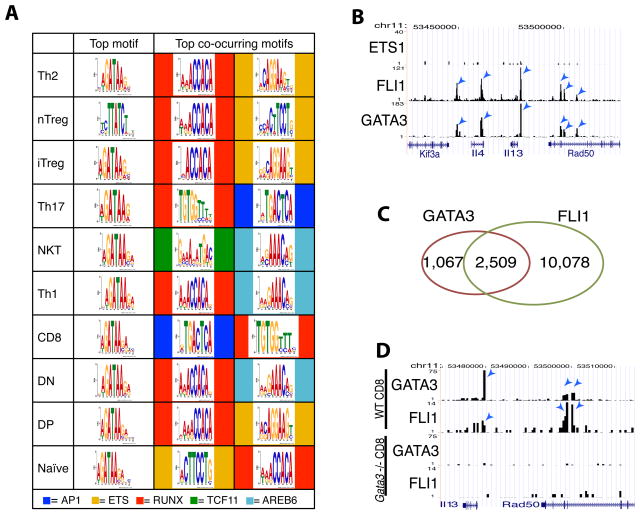
To understand the lineage specificity of GATA3 binding and function, we performed a motif enrichment analysis of the GATA3 binding sites identified in various cell types. It has been shown previously that the zinc finger domain of the GATA family proteins recognizes the 5′-(A/T)GATA(A/G)-3′ motif in vitro (Evans et al., 1988). Using the in vivo binding sites identified in this study, we found WGATAA to be the top motif in all GATA3-bound sites in each cell type (Figure 3A). Interestingly, WGATAA is much more prevalent than WGATAG in our study suggesting the conserved recognition site by GATA3 in vivo is somewhat different from that in vitro. In addition to WGATAA, a number of other enriched motifs were identified in the GATA3-bound regions, both in those that contain a WGATAA motif and those that do not. These included motifs of several transcription factors known to be involved in T cell differentiation and function, including the Ets (AGGAAG), Runx (ACCACA), and AP1 (TGACTCA) families of transcription factors. The frequency with which these associated secondary motifs appeared varied with cellular context. For example, while the Runx motif was found as the top secondary motifs in all T helper cells, the AP1 and TCF11 motifs were the top secondary motifs in CD8+ and NKT cells, respectively. The presence of the Runx motif in GATA3-bound sites in Th1 and Th2 cells is consistent with our recent report that GATA3 protein and Runx3 protein can form a complex to regulate Th1 and Th2 cell differentiation (Yagi et al., 2010).
Figure 3. GATA3 binding is associated with primary and various secondary motifs.
A. Motif enrichment analysis of GATA3 binding sites in various T cells. The co-occurring motifs associated with the primary WGATAA motif in the GATA3 binding sites were also analyzed and the top two secondary motifs (indicated at the bottom) from each cell were displayed.
B. Genome Browser image showing that Fli1 but not Ets1 co-localized with GATA3 at the Th2 cytokine locus in Th2 cells.
C. Genome-wide co-localization of GATA3 and Fli1 binding sites in Th2 cells.
D. Genome Browser image showing that Gata3 deletion abrogated Fli1 binding to the Th2 cytokine locus in CD8+ T cells.
GATA3 co-localizes with Fli1
The association of the WGATAA motif with the Ets motif in the GATA3-bound regions suggests that GATA3 may be co-localized with one or more Ets family members in the genome. Indeed, our ChIP-Seq analyses indicate that Fli1 but not Ets1 co-localized with GATA3 as exemplified in Figure 3B; Fli1 bound to 70% of all GATA3-bound sites in Th2 cells (Figure 3C). It has been suggested previously that another member of the GATA family of transcription factors, GATA1, physically interacts with Fli1 and that this interaction facilitates their mutual binding to their target sites (Eisbacher et al., 2003). Thus, GATA3 binding may be recruited or stabilized by Ets family proteins and vice versa. While we have not been able to demonstrate the recruitment of GATA3 by Fli1 because of difficulties in knocking down Fli1 efficiently, we found that Fli1 binding to the Il13 and Rad50 loci was abrogated in Gata3-deleted T cells (Figure 3D). Globally, 75% of the shared GATA3 and Fli1 sites lost Fli1 binding upon deletion of Gata3.
Taken together, these data suggest that transcription factors exhibit different target specificity depending on the existence of co-occurring motifs and/or the presence of interacting factors in different cellular contexts. The regulatory complexity and potential exhibited by a fixed set of transcription factors in any genome can be dramatically expanded by such a mechanism.
GATA3 regulates critical factors of cell signaling and cell fate determination in the thymus
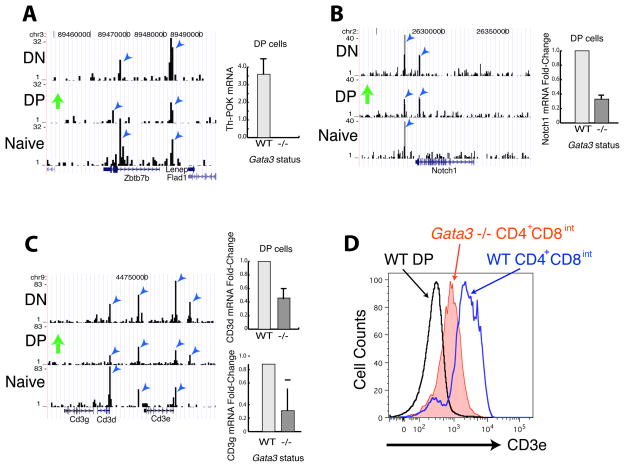
GATA3 has been suggested to regulate the expression of Th-POK (Zbtb7b), a transcription factor that is essential for the development of CD4+ T cells (He et al., 2005; Sun et al., 2005; Wang et al., 2008). Even though Th-POK is not expressed at the DN stage (He et al., 2005; Sun et al., 2005), the Zbtb7b gene was already bound by GATA3 at this stage (Figure 4A, left panel), suggesting that GATA3 binding may prepare the gene to be expressed later. Indeed, deletion of Gata3 abrogated Th-POK expression in post-selection CD69+DP cells (Figure 4A, right panel), suggesting an essential function of GATA3 in Th-POK expression. However, ectopic expression of Th-POK fails to rescue CD4+ T cell differentiation defects caused by GATA3 deficiency (Wang et al., 2008), suggesting other GATA3 targets are also involved in CD4+ T cell development.
Figure 4. GATA3 regulates critical components of signaling and transcription during T cell development in the thymus.
A–C. Genome Browser image showing that the Thpok gene (zbtb7b) (A), the Notch1 gene (B) and Cd3d and Cd3e (C) were bound by GATA3 in DN, DP and naïve CD4+ T cells (left panel). Gata3 deletion abolished their expression in CD69+CD3hiCD4+CD8int-hi DP cells (right panel).
D. Gata3 deletion resulted in a decrease of surface T cell receptor complex in CD4+CD8int cells by FACS analysis.
Runx proteins function to mediate silencing of CD4 expression at different stages of T cell differentiation and facilitate CD8+ T cell differentiation (Collins et al., 2009). Our data indicate that GATA3 binding exists around all three Runx family members (Runx1, Runx2 and Runx3) in DN, DP and naïve CD4+ cells (data not shown). Deletion of Gata3 decreased Runx1 expression in CD69+DP cells (data not shown). Our ChIP-Seq and RNA-Seq data also indicated that GATA3 binds to and regulates two dozens of other transcription factors, including Irf8, Fli1, Rorc, Smad7, Tcf3, Tcf7 and Zfpm1, whose functions during T cell development remain to be tested (Table 3).
In addition to Th-POK and many other transcription factors, signaling molecules that are involved in T cell development are also regulated by GATA3. Notch signaling plays an essential role in T cell fate specification and may also be involved in the CD4-CD8 lineage decision (Laky and Fowkes, 2008). We found that GATA3 bound to the Notch1 locus in DN, DP and naïve cells (Figure 4B, left panel) and deletion of Gata3 diminished its expression in CD69+DP cells (Figure 4B, right panel). GATA3 may also regulate T cell differentiation by directly controlling the expression of TCR components. Our ChIP-Seq analysis revealed strong binding of GATA3 to the Tcra, Tcrb, Cd3d and Cd3e loci (Figure 4C, left panel and data not shown). The mRNA expression of Cd3d and Cd3e decreased about 50% in Gata3-deleted CD69+DP cells (Figure 4C, right panels). Cell surface expression of the T cell receptor complex indicated by CD3ε staining is reduced on CD4+CD8int thymocytes (Figure 4D) upon Gata3 deletion. Since signal strength plays such a central role in thymic selection (Hogquist, 2001) and Th cell differentiation (Tao et al., 1997), GATA3 regulation of TCR expression could directly or indirectly affect T cell development and differentiation. Although deletion of Gata3 did not completely abolish TCR expression in these thymocytes, we cannot rule out the possibility that GATA3 plays an essential role in TCR expression during the early phase of T cell commitment.
Our data indicate that, in addition to regulating Th-POK expression, GATA3 is involved in controlling the expression of several key components of signaling and transcription required for T cell development, therefore explaining the failure of a Th-POK transgene to rescue the defect in CD4+ T cell development in Gata3fl/fl-CD4Cre mice. These data also indicate that GATA3 can bind to target genes at a developmental stage before the genes are expressed (for example, Th-POK gene in DN cells), which may prepare the chromatin for future action; and after the regulation occurred (for example, binding to Cd3d and Thpok in naïve T cells), which may serve as a memory of previous GATA3 function.
GATA3 mediates both activation and repression of transcription
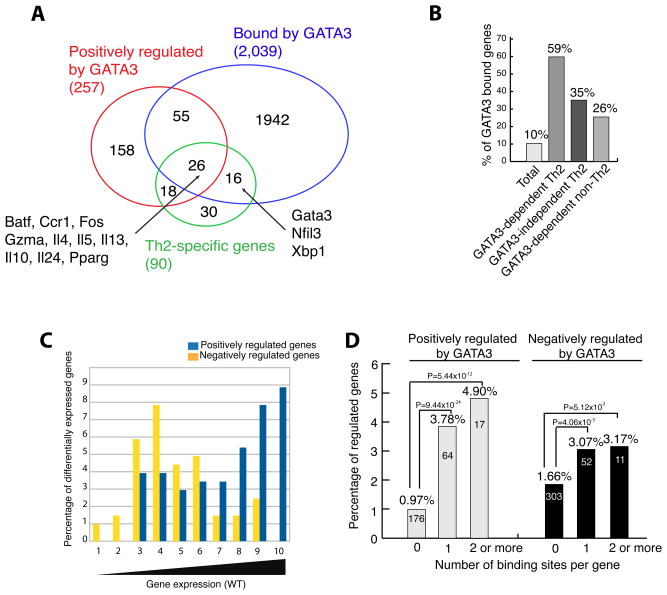
Comparison of expression profiles of the four helper T cell lineages revealed 91, 90, 7 and 43 genes uniquely expressed in Th1, Th2, Th17 and iTreg cells, respectively, using the criteria that a lineage-specific gene should have a RPKM (reads per kilobase of exon model per million mapped reads) ≥ 5 and should be 2-fold higher than in other lineages (Table S4). Since GATA3 is required for Th2 cell differentiation, we examined how GATA3 binding correlated with gene expression in Th2 cells in a greater detail. Among the 90 Th2 cell-specific genes, 44 (49%) were down regulated by deletion of Gata3 (we will hereafter refer to these genes as positively regulated by GATA3), which is much higher than 3% of all genes that were affected in Th2 cells and thus consistent with a key role of GATA3 in regulating Th2 cell differentiation. Most of the Th2 cell specific genes including Batf, Ccr1, Fos, Gzma, Il4, Il5, Il10, Il13, Il24, Pparg, Gata3, Nfil3, and Xbp1 were either bound or positively regulated by GATA3 (Figure 5A). Compared to only 10% of all genes, 59% of GATA3-dependent Th2 cell-specific genes were bound by GATA3 in Th2 cells, while the binding of GATA3 was also detected at 35% of Th2 cell-specific genes that do not require GATA3 for expression in this setting (Figure 5B). Gata3 mRNA (truncated mRNA in the Gata3-deficient cells) did not change after removing GATA3 implying that GATA3 is not required for its own expression in the presence of IL-4-STAT6 signaling. However, GATA3 binds to multiple elements at the Gata3 locus, including a Th2 cell-specific binding to one site ~1 Mb downstream of transcription start site (Data not shown), which suggests that GATA3 could self-regulate when it reaches a threshold level and/or when IL-4-STAT6 signaling ceases or is absent. These data robustly demonstrate a critical role of GATA3 in the Th2 cell differentiation program. Among these 90 Th2 cell-specific genes, there are several GATA3 dependent and independent transcription factors whose functions during Th2 cell differentiation require further investigation (Table S4).
Figure 5. GATA3 binding is correlated with both gene activation and repression in Th2 cells.
A. Venn diagrams showing the overlaps of Th2 cell-specific genes, GATA3-bound genes and the genes positively regulated by GATA3. The locations of some interesting genes are indicated. All the affected and bound genes were determined by the overlaps of two independent biological repeats.
B. The percentages of GATA3-bound genes among various classes of genes in Th2 cells.
C. Differential distribution of genes positively and negatively regulated by GATA3. A total of 2,039 GATA3-bound genes were binned into 10 equal-sized groups according to their expression in wild-type Th2 cells (X axis), and the percentage of differentially expressed genes resulting from the Gata3 deletion was plotted (Y-axis).
D. GATA3 binding is correlated with both activation and repression. Y-axis indicates the fraction of genes that are either positively or negatively regulated by GATA3 in each group of genes separated by the number of GATA3 binding sites in a gene.
To examine how the binding of GATA3 is related to gene expression, we grouped the 2,039 genes bound by GATA3 in Th2 cells into 10 bins according to their level of expression in wild-type cells, and plotted the percentage of differentially expressed genes for each expression bin (Figure 5C). While the genes positively regulated by GATA3 (blue columns) exhibited a bimodal distribution, the genes negatively regulated by GATA3 (orange columns) were highly enriched in lower expression groups.
The above results indicate that GATA3 activated one group and repressed another group of genes. To test whether GATA3 binding is positively correlated with both gene activation and repression, we plotted the number of GATA3 binding sites per gene against the fraction of bound genes that exhibited significant changes when Gata3 was deleted in Th2 cells. Our data indicate that, while 0.97% of genes not associated with GATA3 binding were positively regulated by GATA3, 3.78% of genes with 1 GATA3 binding sites and 4.90% of genes with ≥2 GATA3 binding sites were activated by GATA3 (Figure 5D). Interestingly, it appears that GATA3-mediated repression also correlated with GATA3 binding (Figure 5D). We found that 43% of the genes activated by GATA3 and 30% of the genes repressed by GATA3, respectively, were bound by GATA3 in Th2 cells (Figure 2A). Our data indicate that GATA3 binding may lead to either activation or repression of transcription, which is consistent with the observation that GATA1 and GATA2 also act as both activators and repressors of transcription (Cheng et al., 2009; Fujiwara et al., 2009; Yu et al., 2009). We hypothesize that this equivalence may be due to the upstream role GATA3 plays in the transcription cascade. Gene ontology analysis using KEGG pathways within DAVID tools indicates that both the GATA3-activated and repressed genes are enriched in cytokine-receptor interaction and JAK-STAT signaling pathways (Data not shown).
Distinct chromatin structure at GATA3 sites associated with activated and repressed genes
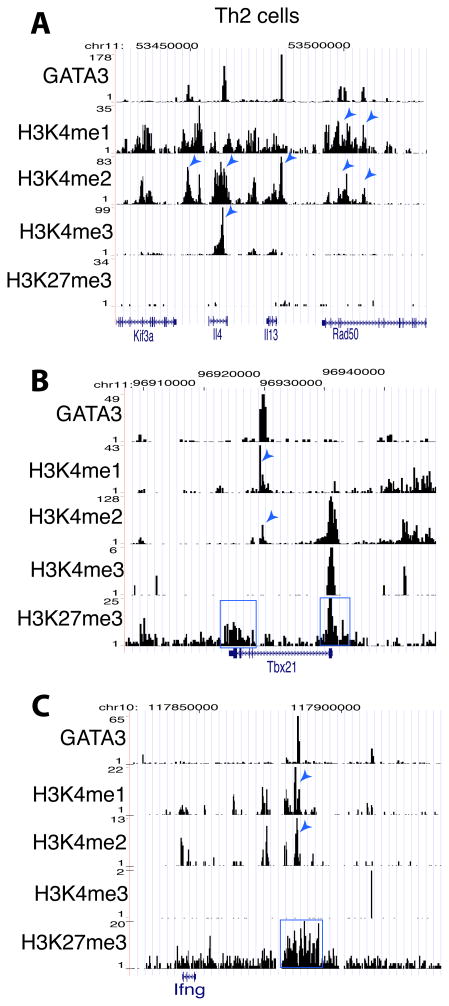
To understand how GATA3 mediates both gene activation and repression, we tested whether GATA3 binding modulates the chromatin structure of its target sites. Our data indicate that the genomic regions surrounding the GATA3 binding sites in the Il4 and Il13 genes, which are activated by GATA3, are associated with either H3K4me1, H3K4me2 and/or H3K4me3 but no notable signals of H3K27me3 in Th2 cells (Figure 6A), while, also in Th2 cells, the GATA3 sites in the Tbx21 and Ifng genes, which are repressed by GATA3, are associated with H3K27me3 modification in addition to H3K4me1 and H3K4me2 modifications (Figures 6B and 6C). These data suggest that GATA3 may influence gene expression by regulating histone methylation at its target sites in Th2 cells.
Figure 6. Active and repressive activities of GATA3 are associated with distinct epigenetic modification patterns.
A–C. Genome Browser image showing the patterns of GATA3 binding and H3K4me1, H3K4me2, H3K4me3 and H3K27me3 modifications at the Il4 and Il13 gene locus (A), at the Tbx21 gene (B) and at the Ifng gene (C) in Th2 cells. The histone H4K4 methylation peaks co-localized with GATA3 peaks are indicated by arrowheads and the regions enriched with H3K27me3 are highlighted by the blue box.
GATA3 regulates H3K4 and H3K27 methylations at its target sites
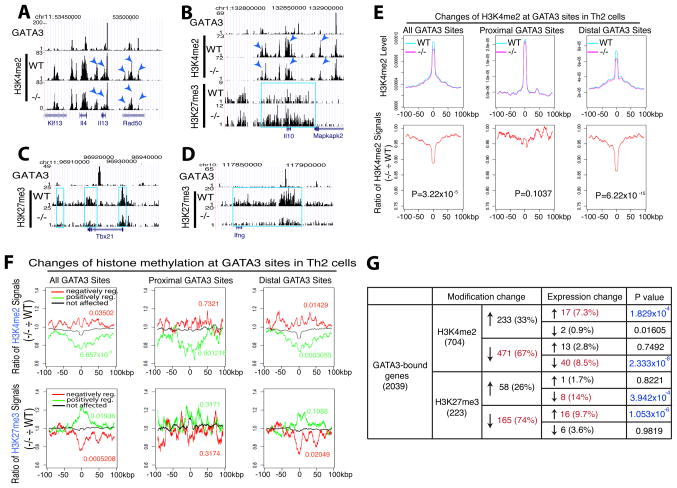
To directly test whether GATA3 regulates histone modification at its target sites, we compared the profiles of H3K4me1, H3K4me2, H3K4me3 and H3K27me3 in wild type and Gata3-deleted Th2 cells. Deletion of Gata3 resulted in substantial decreases in H3K4me2 at a number of specific sites in the Th2 cytokine locus (Figure 7A, compare peaks indicated by arrowheads). On the other hand, while deletion of Gata3 resulted in decreased H3K4me2 (indicated by arrowheads) at GATA3 binding sites and it also increased H3K27me3 (highlighted in blue) across the entire Il10 locus (Figure 7B). Interestingly, Gata3 deletion caused a decrease in H3K27me3 at both the Tbx21 and Ifng loci (highlighted in blue) (Figures 7C and 7D). While it has been previously suggested that GATA3 indirectly suppresses IFNγ production (Yagi et al., 2010), our data here indicate that GATA3 can also directly act on the Ifng gene in Th2 cells to suppress the expression of this cytokine by modulating H3K27me3 marks around GATA3 binding sites. To examine the genome-wide regulation of H3K4me2 at GATA3 binding sites, we plotted the H3K4me2 signals surrounding all GATA3 binding sites in Th2 cells and found that H3K4me2 was highly elevated surrounding the GATA3 sites (Figure 7E, upper left panel). Interestingly, the H3K4me2 signal decreased significantly (P = 3.22×10−15 using the Kolmogorov-Smirnoff test) in GATA3-deleted cells (compare magenta with cyan). It appears that decrease in H3K4me2 mainly occurred at the distal, non-promoter GATA3 sites (compare upper middle and upper right panels in Figure 7E), which is more clearly indicated by normalizing the H3K4me2 signals in Gata3-deficient cells to that in wild type cells (Figure 7E, lower panels). Notably, the H3K4me2 peak surrounding the GATA3 sites spanned a large region of about 15 kb; and interestingly, the H3K4me2 signal across the entire 15 kb region decreased when Gata3 was deleted, suggesting that GATA3 binding was responsible for promoting H3K4me2 methylation. To test whether GATA3 functions similarly in other cells, we have extended this analysis to H3K4me1, H3K4me3 and H3K27me3 in DP cells and found that deletion of Gata3 also decreased H3K4me1 signals, mainly at distal binding sites in DP cells (Figure S2).
Figure 7. GATA3 regulates transcription by modulating H3K4 and H3K27 methylations of enhancers.
A–D. Genome Browser image showing that deletion of Gata3 resulted in changes in H3K4me2 (indicated by arrowheads) and/or H3K27me3 (highlighted in blue box) at or near the GATA3 sites of the Il4/Il13 locus (A), of the Il10 locus (B), of the Tbx21 locus (C) and of the Ifng locus (D) in Th2 cells.
E. Global analysis indicated that deletion of Gata3 resulted in decreases in H3K4me2 signals over the distal GATA3 sites in Th2 cells. Upper panel: the H3K4me2 tag densities (Y-axis) surrounding the total GATA3 sites (left), promoter-proximal (middle) and distal (right) GATA3 sites were plotted for a region of 100kb on each side of the GATA3 binding site. Lower panel: Y-axis shows the ratio of the H3K4me2 signals in Gata3-deficient cells to that in wild type cells surrounding the GATA3 binding sites across the same region as in the upper panel. The P-values for the histone modification differences between the Gata3-deficient and wild type cells are obtained using the Kolmogorov-Smirnoff test and shown in the panels.
F. Gata3 deletion-induced changes in histone methylation correlate with changes in gene expression in Th2 cells. Upper panel: plotted are the ratios of H3K4me2 signals between Gata3-deficient cells and wild type cells surrounding the GATA3 binding sites associated with the genes positively regulated (green), negatively regulated (red) and not regulated (black) by GATA3; lower panel: the same are plotted for H3K27me3. The P-values for the histone modification differences between the regulated genes and non-regulated genes are obtained using the Kolmogorov-Smirnoff test and shown either in red or green.
G. GATA3 regulates both H3K4me2 and H3K27me3 at numerous target genes. The changes in H3K4me2 and H3K27me3 modifications (P<10−5; ratio≥2 or ≤0.5) after Gata3 deletion) and expression level at each individual GATA3-bound gene were summarized. The arrows indicate the direction of changes in histone modification and gene expression after Gata3 deletion. The P values of either increased or reduced gene expression correlated with a change in histone modification are indicated.
To examine whether the changes in H3K4me2 at the GATA3 sites correlate with gene expression, we plotted the ratio of H3K4me2 signals in Gata3-deficient and wild type Th2 cells at the GATA3 sites associated with the genes that are positively or negatively regulated or not affected by GATA3 (Figure 7F, upper panels). This analysis revealed that deletion of Gata3 indeed led to a significant decrease (P=6.857×10−8, Kolmogorov-Smirnoff test) in H3K4me2 surrounding both the proximal (P=0.001216) and distal (P=0.0003055) GATA3 sites in the genes positively regulated by GATA3 (green), although the increase in H3K4me2 in genes negatively regulated by GATA3 was only modest. Interestingly, the region with decreased H3K4me2 methylation extended far beyond the GATA3-binding sites to about 50 kb each side of its binding. On the other hand, decreased and increased H3K27me3 signals were detected in Gata3-deleted cells at the GATA3 sites associated with the genes repressed and activated by GATA3, respectively, mainly surrounding the distal GATA3 sites (Figure 7F, lower panels).
Among the 2,039 GATA3-bound genes, 704 and 233 genes exhibited significant changes (FDR<10−5; ratio≥2 or ≤0.5) in H3K4me2 and H3K27me3 modifications, respectively, after deletion of Gata3 (Figure 7G). Gata3 deletion resulted in more genes with a decrease in H3K4me2 than genes with an increase in H3K4me2 (67% vs. 33%). Interestingly, it also resulted in more genes with a decrease H3K27me3 than genes with an increase in H3K27me3 (74% vs. 26%), suggesting that GATA3 may directly facilitate both H3K4me2 and H3K27me3 modifications at its target genes. In the groups of genes with either increased H3K27me3 or decreased H3K4me2 after Gata3 deletion, there were more genes that displayed reduced expression than enhanced expression in Gata3-deficient “Th2” cells, while in the groups of genes with either increased H3K4me2 or decreased H3K27me3 after Gata3 deletion, many of them represented the genes that are negatively regulated by GATA3 (Figure 7G). These results indicate that GATA3-regulated H3K4me2 and H3K27me3 are highly correlated with activation and repression of its target genes (Figure 7G). Although only a minor fraction of the genes that GATA3 bound to showed a significant change in expression (4.0% and 3.1% are positively and negatively regulated, respectively, by GATA3), Gata3 deletion significantly changed the histone modifications at 46% of its bound genes (11% for H3K27me3 and 35% for H3K4me2), indicating that gene expression change is not required for the observed epigenetic changes caused by GATA3 deletion.
DISCUSSION
Here, we found that there is substantial cell specificity in both GATA3 binding and gene regulation by GATA3 even among the closely related T cell lineages. GATA3 regulates distinct key factors in the thymus and during T helper cell differentiation. Our data indicated that GATA3 facilitates both H3K4me1 and H3K4me2, and H3K27me3 at enhancers to mediate gene activation and repression, respectively.
Among the most striking aspects of our results is the very large number of genes to which GATA3 binds and the fact that it activates some, represses others and appears to have little or no effect on the level of expression of the rest. This suggests that GATA3 acts in a more subtle manner than simply as a “switch” to turn on the Th2 cell phenotype. Instead, it may control the activation of many genes that are critical for Th2 cell function while simultaneously repressing genes that might inhibit Th2 cell function or Th2 cell differentiation.
One challenge is to understand the importance of GATA3 binding to sites at which it does not appear to alter the expression of the bound genes. One possibility is that the local binding of GATA3 may regulate other genes through long-distance chromatin interaction, such as Th2 cytokine locus control region in the Rad50 gene. A second possibility is that these genes are regulated under the various conditions that the Th cells may encounter in vivo but which are not operative in vitro. A third possibility is that GATA3 binding does not directly activate or repress transcription of its target genes; instead, it prepares chromatin so that other factors that collaborate with GATA3 can bind and regulate transcription when they become available. Thus, the effects of deleting Gata3 could not be detected in the absence of these collaborating factors.
Our data indicate that the function of GATA3 exhibits two levels of specificity in different cells: one level at its binding site selection and the second level at its functional regulation of bound genes. At the binding level, GATA3 target genes demonstrate some lineage specificities even among the closely related T helper cell lineages. The lineage specificity of GATA3 binding in different cells cannot be attributed solely to differences in GATA3 expression and number of binding sites among these different cell types.
Since GATA3 is a sequence-specific DNA-binding protein, its cell type-specific binding suggests that GATA3 target recognition is regulated by other mechanisms in addition to the GATA3 sequence motif. Indeed, our motif analysis revealed that in addition to a primary WGATAA motif, GATA3 binding sites contained various secondary motifs including Ets, Runx, AP1, TCF11 and AREB6 motifs or only secondary motifs but lacked the primary WGATAA motif. There are several potential mechanisms that secondary motif-recognizing factors may be used to influence GATA3 function: (1) GATA3 physically interacts with another factor, which stabilizes the binding of both factors or destabilizes the binding of one factor; (2) GATA3 binding facilitates H3K4 methylation and creates a chromatin environment such that another factor can bind; (3) GATA3 binds to a site in open chromatin that is maintained by another factor; (4) GATA3 facilitates H3K27me3 modification at target region to inhibit binding or to suppress the activity of another factor. These non-exclusive ways of action may explain how GATA3 binding can be associated with both gene activation and repression. The functional collaboration between different transcription factors is not unique for GATA3 but has been found with other key regulators, for example, PU.1 in macrophage and EBF1 in B cells (Ghisletti et al., 2010; Treiber et al., 2010).
Lineage-specific cytokine and transcription factor genes are associated with distinct histone modification patterns in different T cell lineages (Ansel et al., 2006; Hatton et al., 2006; Roh et al., 2005; Schoenborn et al., 2007; Wei et al., 2009). Active histone modifications at the Th2 cytokine locus depend on key transcription factors such as STAT6 and GATA3 (Yamashita et al., 2002), suggesting that these factors may regulate transcription by modulating the chromatin structure of their target genes. Our genome-wide analyses indicated that deletion of Gata3 decreased H3K4me1 and H3K4me2 modifications at GATA3 binding sites associated with the genes that were positively regulated by GATA3. By contrast, deletion of Gata3 resulted in a decrease in H3K27me3 at the negatively regulated genes. We found that in Th2 cells, a substantial proportion of GATA3-bound genes had changed their epigenetic modifications but not their gene expression when Gata3 was deleted. These results indicate that the epigenetic changes are directly regulated by GATA3 binding and are not a consequence of transcriptional activation or repression. The modest changes in the histone modifications at GATA3 binding sites upon deletion of Gata3 suggest that other factors co-bound with GATA3 may play redundant roles in recruiting histone methyltransferases.
Histone modification may influence transcription by attracting different transcription co-factors. H3K4 methylation can recruit co-activators, e.g., the ATP-dependent chromatin remodeling complex, NURF (Wysocka et al., 2006) and HATs (Wang et al., 2009), whereas H3K27me3 signals can be recognized by the PRC1 polycomb repressor complex (Fischle et al., 2003), which mediates transcriptional repression in Drosophila and humans. Therefore, our data support the model that GATA3 binding creates a chromatin environment that can make target sites of other transcription factors accessible or can be recognized by co-activators or co-repressors of transcription although it is also possible that in some cases, GATA3 may activate or repress transcription directly.
In summary, we have characterized GATA3 binding in many distinct T lymphocytes and analyzed GATA3-mediated gene regulation at a genome-wide level. Our dataset provides valuable information for further characterization of GATA3 target genes and cis-regulatory elements that GATA3 binds to as well as proteins that collaborate with GATA3 to regulate the differentiation and function of various T cell lineages.
Materials and Methods
Mice
C57BL/6 mice and Foxp3-RFP mice (Line 8374) (Wan et al., 2005) were purchased from the Jackson Laboratory (JAX). Gata3fl/fl and Gata3fl/fl-CD4Cre mice were previously described (Zhu et al., 2004) except that they were further backcrossed to C57BL/6 background for at least 9 generations. All mice were bred and maintained in NIAID specific pathogen free animal facility and the experiments were done when mice were at 8 to 16 weeks of age under protocols approved by the NIAID Animal Care and Use Committee. We used three types of Gata3-deficient cells: DP and CD8+ Gata3-deficient cells were from Gata3fl/fl-Cd4Cre mice; for RNA-Seq analysis of Th1, Th2, Th17 and iTreg cells, Gata3 was deleted by hCre-GFP infection of Gata3fl/fl cells; for histone modification analysis of Th2 cells, we used Gata3fl/fl-dLckCre Th2 cells in which ~70% of cells deleted Gata3 (Yagi et al., 2010). See Supplemental Methods for details about preparation of the cells from wild type and Gata3-deficient mice.
ChIP-Seq and RNA-Seq
ChIP-Seq experiments were performed as described previously (Barski et al., 2007). RNA-Seq experiments were performed using total RNA isolated from both the wild type and Gata3-deficient cells. Binding site identification, motif analysis and differential gene expression analysis were performed as described in Supplemental materials.
Supplementary Material
Acknowledgments
We thank Dr. Warren Leonard for critical reading of the manuscript. We thank Drs. Iouri Chepelev, Gangqing Hu and Dustin Schones for helpful discussions on data analysis. This work was supported by the Division of Intramural Research, National Institute of Environmental Health Sciences (RJ; Project Number Z01ES102625), Division of Intramural Research, National Institute of Allergy and Infectious Diseases, and Division of Intramural Research, National Heart, Lung and Blood Institute, National Institutes of Health, USA. All the ChIP-Seq and RNA-Seq data have been deposited in the NCBI GEO database (GSE20898).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M, Chong BH. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol Cell Biol. 2003;23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Reitman M, Felsenfeld G. An Erythrocyte-Specific DNA-Binding Factor Recognizes a Regulatory Sequence Common to All Chicken Globin Genes. Proc Natl Acad Sci U S A. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering Hematopoietic Mechanisms through Genome-wide Analysis of GATA Factor Chromatin Occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu WH, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA. Signal strength in thymic selection and lineage commitment. Curr Opin Immunol. 2001;13:225–231. doi: 10.1016/s0952-7915(00)00208-9. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U S A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol. 2001;167:4414–4420. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- Laky K, Fowkes BJ. Notch signaling in CD4 and CD8 T cell development. Curr Opin Immunol. 2008;20:197–202. doi: 10.1016/j.coi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenu-Bombled C, Trainor CD, Makeh I, Romeo PH, Max-Audit I. Interleukin-13 gene expression is regulated by GATA-3 in T cells - Role of a critical association of a GATA and two GATG motifs. J Biol Chem. 2002;277:18313–18321. doi: 10.1074/jbc.M110013200. [DOI] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O’Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun GP, Liu XL, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Tao X, Constant S, Jorritsma P, Bottomly K. Strength of TCR signal determines the costimulatory requirements for Th1 and Th2 CD4+ T cell differentiation. J Immunol. 1997;159:5956–5963. [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Zhu JF, Zhang XY, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie HF, Schindler Y, Moran TB, Cheng Y, Yu DN, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-Mediated Gene Activation versus Repression via Genome-wide Chromatin Occupancy Analysis. Molecular Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng WP, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zhu JF, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Guo LY, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Davidson TS, Wei G, Jankovic D, Cui K, Schones DE, Guo L, Zhao K, Shevach EM, Paul WE. Down-regulation of Gfi-1 expression by TGF-beta is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J Exp Med. 2009;206:329–341. doi: 10.1084/jem.20081666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.