Abstract
The highly conserved spliceosomal protein Prp8 is known to cross-link the critical sequences at both the 5′ (GU) and 3′ (YAG) ends of the intron. We have identified prp8 mutants with the remarkable property of suppressing exon ligation defects due to mutations in position 2 of the 5′ GU, and all positions of the 3′ YAG. The prp8 mutants also suppress mutations in position A51 of the critical ACAGAG motif in U6 snRNA, which has been observed previously to cross-link position 2 of the 5′ GU. Other mutations in the 5′ splice site, branchpoint, and neighboring residues of the U6 ACAGAG motif are not suppressed. Notably, the suppressed residues are specifically conserved from yeast to man, and from U2- to U12-dependent spliceosomes. We propose that Prp8 participates in a previously unrecognized tertiary interaction between U6 snRNA and both the 5′ and 3′ ends of the intron. This model suggests a mechanism for positioning the 3′ splice site for catalysis, and assigns a fundamental role for Prp8 in pre-mRNA splicing.
Keywords: Pre-mRNA splicing, Prp8, U6 snRNA, yeast
The removal of introns from eukaryotic genes to generate mRNA is catalyzed by the spliceosome, a complex RNA–protein machine composed of small nuclear RNAs (snRNAs), and at least 60 proteins. The pre-mRNA splicing reaction consists of two sequential transesterification steps: The first step results in cleavage of the 5′ splice site (5′SS) and formation of a branched (lariat) intermediate; the second step results in ligation of the 5′ and 3′ exons. The existence of self-splicing group-II introns, which utilize a similar two-step transesterification mechanism, has led to the hypothesis that pre-mRNA splicing is catalyzed by RNA (Sharp 1985; Cech 1986). Indeed, mutational and cross-linking analyses have revealed a network of RNA–RNA interactions, which have been proposed to form the catalytic core of the spliceosome (Newman 1994; Nilsen 1994).
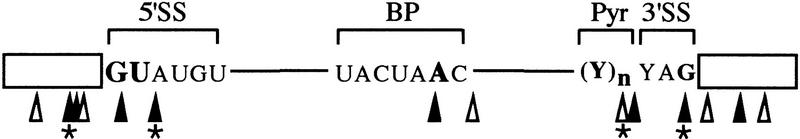
Whether any of the spliceosomal proteins make direct structural or chemical contributions to the active site remains unknown. Numerous cross-linking studies have placed one spliceosomal protein, Prp8, at or near the catalytic core. As summarized in Figure 1, Prp8 has been observed to cross-link to RNA residues within each critical sequence component of the intron: The consensus sequences that define the 5′SS, the branch-point (BP), and the 3′ splice site (3′SS) (MacMillan et al. 1994; Teigelkamp et al. 1995; Umen and Guthrie 1995a; Chiara et al. 1996, 1997; Reyes et al. 1996). Prp8 also forms extensive cross-links to residues in both the 5′ and 3′ exons (Wyatt et al. 1992; Teigelkamp et al. 1995; Chiara et al. 1996) and to U5 snRNA (Dix et al. 1998), which interacts with the 5′ and 3′ exons (Newman and Norman 1992; Sontheimer and Steitz 1993). Prp8 is distinguished by its unusually high evolutionary conservation, that is, 62% sequence identity from yeast to man over the entirety of its ∼2500 residues (Hodges et al. 1995; Lucke et al. 1997; Luo et al. 1999). Prp8 is thus hypothesized to play a fundamental role at the active site. Unfortunately, the sequence of this large protein provides no clues to its domain organization or its potential biochemical activities.
Figure 1.
Prp8 forms extensive cross-links to RNA residues in the intron consensus sequences, and in the 5′ and 3′ exons. Intron consensus sequences that define the 5′SS, BP, and 3′SS are indicated. For most yeast introns, a stretch of Pyr, functions with the 3′SS YAG in definition of the 3′SS. Triangles denote the location of Prp8 cross-links observed in either mammalian (▴), or yeast (▵) systems. For all studies except those with an asterisk (*), the location of the cross-linked site is determined through the use of a site-specific photoreactive substitution or by RNase fingerprinting. (*) Approximated sites of cross-linking within an RNase T1 fragment.
We have taken a genetic approach, in the budding yeast Saccharomyces cerevisiae, to identify functional correlates to Prp8’s cross-linking interactions with RNA. This approach exploits the ACT1–CUP1 splicing reporter system, through which the splicing of mutant introns can be measured in vivo by growth on copper (Lesser and Guthrie 1993a). This system has been used previously to provide functional support for Prp8’s observed interactions with the 3′SS (Umen and Guthrie 1996). Through screens of randomly mutagenized prp8 alleles, mutants were identified (prp8-121 through prp8-125) that suppress the splicing defect of ACT1–CUP1 reporters containing mutations in the 3′SS YAG consensus sequence. This finding suggested a role for Prp8 in recognition of this sequence during the exon ligation step. A separate class of prp8 mutants suggested a role for Prp8 in recognition of the pyrimidine (Pyr) tract that precedes most 3′SS. It was known from in vivo studies of a construct containing two 3′SS in competition that the 3′SS following a Pyr tract is preferentially used (Patterson and Guthrie 1991); the prp8-101 through prp8-107 alleles were isolated in a screen for loss of this preference. Interestingly, prp8 mutations that give rise to the two classes of phenotypes, loss of Pyr tract preference, and 3′SS YAG suppression, map to distinct regions of the protein and are functionally nonoverlapping. This suggests that different domains of PRP8 can be mutated to uncover separable functions.
Our current study was motivated by the identification of a very strong UV cross-link between the human Prp8 homolog, p220, and the invariant GU dinucleotide at the 5′ end of the intron (5′SS GU) (Reyes et al. 1996). Our goals were twofold. First, we sought functional verification of a role for Prp8 in binding the 5′SS GU, by looking for suppression of mutations at the 5′SS. Second, we wanted to know whether the protein location of such 5′SS suppressors would implicate a new functional PRP8 domain.
We have identified alleles of prp8 that suppress the splicing of mutations in position 2 of the 5′SS GU. Suppression arises from viable mutations in at least four regions of the protein, including the two regions implicated previously in 3′SS YAG and Pyr phenotypes. Surprisingly, these alleles concomitantly suppress mutations in the 3′SS YAG. Moreover, we found that all of the originally identified 3′SS YAG suppressor alleles also suppress 5′SS position 2 mutations. This newly defined prp8 phenotype of 5′SS and 3′SS suppression does not extend to other mutations in the 5′SS or in the branchpoint consensus sequences. However, mutations in A51 of the critical U6 ACAGAG motif, which cross-links to 5′SS position 2 (Sontheimer and Steitz 1993; Kim and Abelson 1996), are also suppressed. All of the mutations suppressed confer a strong defect to exon ligation, which is partially rescued by the prp8 alleles. We suggest that suppression of the distinct subset of RNA mutations could occur through a loosening of an active site structure comprised of a previously unrecognized tertiary interaction. The proposed interaction, whose components are conserved from yeast to man, and from U2- to U12-dependent spliceosomes, could serve to position the 3′SS for catalysis. Our findings are consistent with the view that the spliceosomal catalytic core is fundamentally comprised of RNA and that the highly conserved protein, Prp8, binds a critical RNA structure at the spliceosomal active site.
Results
Screen for prp8 alleles that affect fidelity for the 5′SS GU
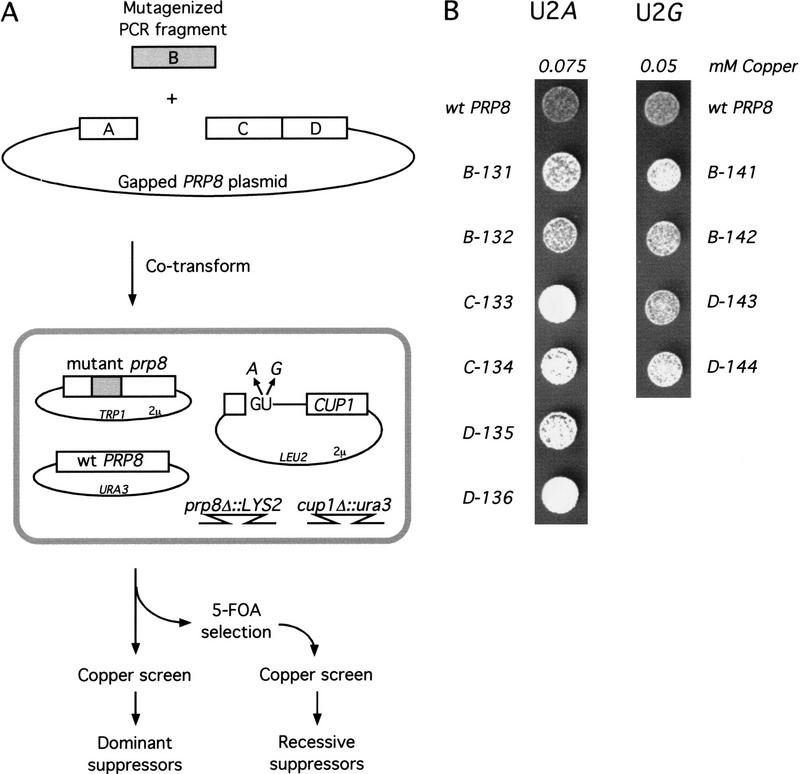
To determine whether Prp8 functions at the 5′SS GU, we screened for prp8 alleles that relax or change the requirement for these residues in splicing. Using ACT1–CUP1 reporters (Lesser and Guthrie 1993a) that contained mutations in 5′SS position 2, we mutagenized PRP8 and screened for alleles that confer growth on higher concentrations of copper, reflecting an increased efficiency of splicing. PRP8 mutagenesis was conducted with Mn2+-enhanced PCR (Leung et al. 1989), and a gap repair strategy was used to introduce plasmid-borne mutagenized prp8 into strains, as depicted in Figure 2A (Muhlrad et al. 1992). To reduce the frequency of null mutations, and to facilitate the mapping of mutations, we separately mutagenized each of four equal fragments (A–D) of the PRP8 coding region (Umen and Guthrie 1996). For each fragment, ∼4000 mutant transformants were screened. The strain used was deleted for the chromosomal PRP8 and wild-type PRP8 was supplied on a counterselectable URA3-marked plasmid. After loss of this plasmid by growth on 5-FOA, ∼60% of the prp8 mutant transformants were unable to support viability. We screened all transformants before 5-FOA passage for dominant suppression and the surviving 40% after 5-FOA passage for recessive suppression. The screens yielded prp8 suppressors of two 5′SS position 2 mutations, U2A and U2G, from the B, C, and D mutagenized fragments. These new prp8 alleles confer a modest but reproducible increase in copper resistance: a two- to fourfold increase for U2A and barely a twofold increase for U2G (Fig. 2B). All of the prp8 alleles identified exhibit dominant suppression and are haploviable.
Figure 2.
Identification of prp8 alleles that suppress mutations in position 2 of the 5′SS. (A) Strategy for isolation of new prp8 alleles. The coding region of PRP8 was divided into four equal fragments (A,B,C,D). Each fragment was amplified by mutagenic PCR and cotransformed with an appropriately gapped plasmid (as shown for B), to generate mutant alleles of prp8 by in vivo gap repair. The resulting strain, which is deleted for its chromosomal copy of PRP8 and CUP1, is diagrammed. Mutant prp8 transformants were screened for the ability to confer increased copper growth on ACT1–CUP1 reporters containing U2A or U2G mutations in the 5′SS, both before and after loss of wild-type PRP8 by 5-FOA selection. (B) Suppression of the copper growth phenotype of mutations in 5′SS position 2 by the newly identified prp8 alleles. Each column shows growth on the concentration of copper, 0.075 mm and 0.05 mm, that is limiting for 5′SS U2A and U2G reporters, respectively. Equal numbers of log phase cells from each strain were spotted onto the same copper-containing plate.
Mapping the GU suppressor alleles
Whereas Prp8’s domain structure is not known, the distinct location of mutations that confer the previously characterized Pyr and 3′SS YAG phenotypes suggested the existence of separable functional domains. Thus, we were curious whether 5′SS position 2 suppression arises from mutations in similar regions of Prp8, or whether another distinct functional domain could be defined. Of particular interest was whether any of these suppressor mutations localize to the site of 5′SS GU cross-linking on Prp8 (Reyes et al. 1999).
The 5′SS position 2 suppressors lie in four regions of Prp8, indicated in solid bars in Figure 3. One cluster of suppressor mutations, identified from screens with the D mutagenized fragment, lie 100 residues from the site of 5′SS GU cross-linking. The clustering of mutations is unlikely to be due to bias in the mutagenesis procedure, because many alleles contained additional mutations outside these regions that did not contribute to the suppression phenotype (data not shown). Interestingly, the cluster of 5′SS position 2 suppressor alleles from the D-mutagenized fragment overlaps in location with the 3′SS Pyr alleles, and another cluster from the C-mutagenized fragment overlaps in location with the 3′SS YAG suppressor alleles.
Figure 3.
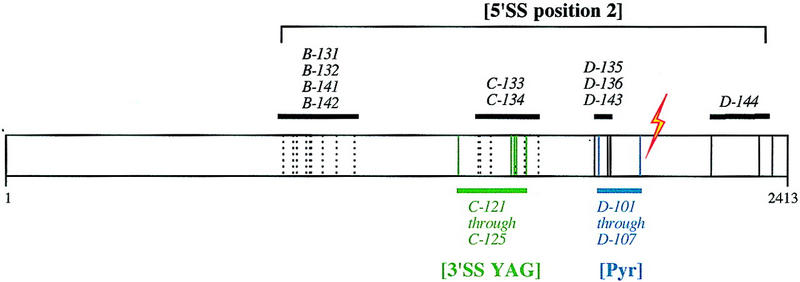
5′SS position 2 suppressor mutations cluster in four regions of PRP8. The location of mutations that confer 5′SS position 2 suppression are indicated in black; green and blue indicate the location of previously characterized 3′SS YAG suppressors and Pyr alleles, respectively (Umen and Guthrie 1996). The corresponding location of the 5′SS GU cross-link in mammalian Prp8 is indicated by the lightning bolt. Solid lines indicate the location of single mutations that confer suppression. Broken lines indicate mutations that confer suppression together but that have not been assayed on their own.
Allele specificity of 5′SS and 3′SS suppression
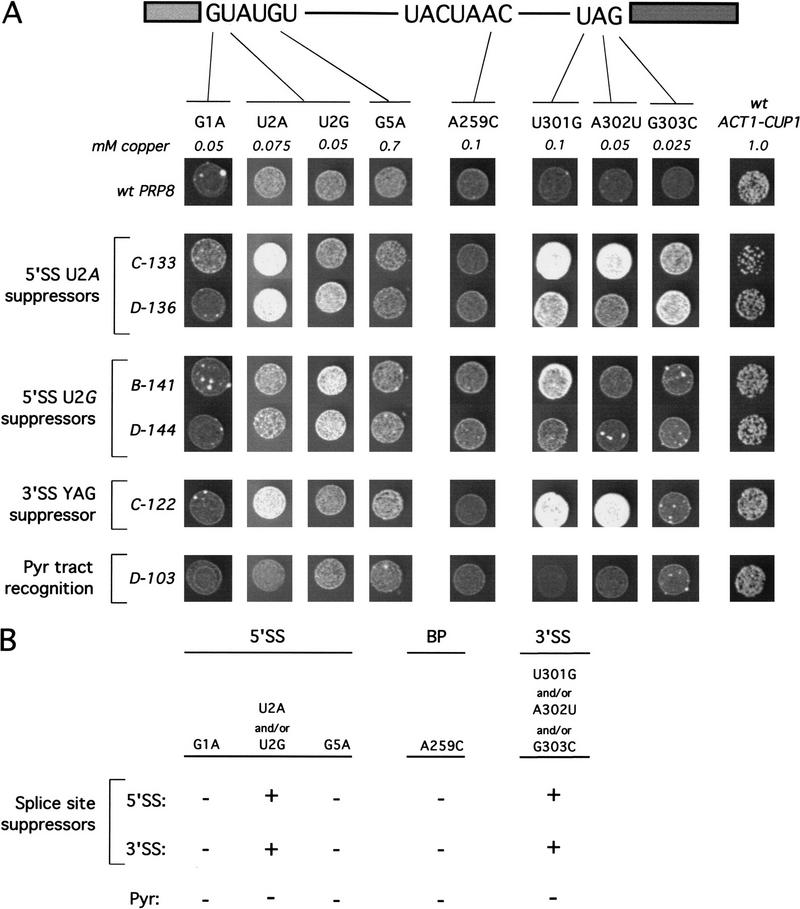
To address whether suppression of mutations in the 5′SS consensus sequence and suppression of mutations in the 3′SS consensus sequence are separable or overlapping phenotypes, we analyzed the allele specificity of suppression. Each prp8 allele was tested for effects on a number of different mutations in the 5′SS and 3′SS sequences. A mutation in the intron residue that becomes the nucleophile for 5′SS cleavage, within the so-called BP consensus sequence, was also tested for suppression by prp8 alleles. Copper growth conferred on strains bearing a series of mutant ACT1–CUP1 reporters are shown for representative prp8 alleles in Figure 4A. The phenotypes for all alleles are summarized in Figure 4B.
Figure 4.
Allele specificity analysis reveals overlapping phenotypes at 5′SS and 3′SS. (A) prp8 alleles identified in different screens were tested for effects on reporters containing various mutations at the 5′SS, BP, or 3′SS. Each column shows growth on the concentration of copper that is limiting for each reporter in a wild-type PRP8 strain. Equal numbers of log phase cells for each mutant strain were spotted onto the same copper containing plate. The prp8 alleles do not confer strong effects on growth when the ACT1–CUP1 reporter is wild type, (C-133, the exception, is temperature sensitive). Not visible, some but not all of the alleles from each of the screens were observed to confer a mild enhancement of copper resistance to the wild-type ACT1–CUP1 reporter. Because some Pyr alleles display this puzzling effect, it does not appear to correlate with the 5′SS and 3′SS suppression phenotype. (B) Phenotypes for prp8 alleles identified in three independent screens are summarized. (+) Intron mutations that are suppressed; (−) intron mutations that are not suppressed. Alleles identified through independent screens for 5′SS position 2 and 3′SS YAG suppression display the same splice site suppression phenotype.
Interestingly, we found that all of the prp8 alleles that suppress 5′SS position 2 also suppress at least one 3′SS YAG mutation. Conversely, the previously identified 3′SS YAG suppressor alleles all suppress the 5′SS U2A mutation. Importantly, suppression by these prp8 alleles does not extend to all mutations in the intron, because the 5′SS mutations G1A and G5A, as well as the BP mutation A259C, are not suppressed. Thus, the effect of the prp8 alleles on splicing is specific to a discrete, although unanticipated, subset of mutations.
Whereas some of the 5′SS position 2 suppressor alleles contain multiple mutations, the phenotypes at the two sites do not arise from separate mutations. For some of the 5′SS suppressors (Table 1) and all of the 3′SS suppressors (Umen and Guthrie 1996), a single mutation that confers suppression has been identified. We conclude that 5′SS position 2 and 3′SS YAG suppression are actually the same phenotype, reflecting a single function for Prp8 at both sites. This phenotype, which we now simply call splice site suppression, is distinct from the previously characterized Pyr phenotype, because prp8 alleles that are defective for Pyr tract recognition do not suppress any intron mutation (Fig. 4; data not shown). Likewise, none of the splice-site suppressor alleles affect recognition of the Pyr tract (Umen and Guthrie 1996; data not shown).
Table 1.
Identity of 5′SS position 2 suppressor mutations in prp8
|
PRP8 allele
|
Mutation(s)
|
|---|---|
| B-131 | Y923C, E942G |
| B-132 | I857T, E888G, Y923C, E935G, E942G, Q976R, D1075G |
| C-133 | I1444V, T1565A, V1621A |
| C134 | Q1455P, D1485G, H1592R |
| D-135 | E1817G |
| D-136 | N1869D |
| B-141 | I857T, Y923C, E942G |
| B-142 | I857T, S894G Y923C, E942G, S1018P |
| D-143 | K1864E |
| D-144a | F2176S, Q2313R, T2364A |
Most of the suppressors contained additional mutations, not listed, which did not affect the suppression phenotype (see Materials and Methods). Single mutations that confer the suppression phenotype are indicated in boldface type.
D-144 contains three mutations, each of which confer a weak suppression phenotype on their own but confer stronger suppression when combined. For other alleles, the mutations listed have not been separated from one another to identify those responsible for suppression.
That the same mutations on prp8 give rise to phenotypes at both the 5′SS and 3′SS suggests that the same part(s) of the protein functions at both sites. Whether the location of the suppressor mutations reflect the location of protein–RNA contacts is not known. Two observations, in addition to the striking specificity, suggest that suppression is not simply due to indirect effects on splice-site residues. One observation is that the degree of suppression for different 3′SS YAG mutations is distinct for each prp8 allele (Fig. 4A; Umen and Guthrie 1996). Another observation in the companion study is that prp8 mutations in residues very close to the site of 5′SS GU cross-linking give rise to the same splice site suppression phenotype (Siatecka et al. 1999).
The dual suppression by the same prp8 mutations also suggests that position 2 of the 5′SS interacts with the 3′SS YAG. Consistent with this notion, strong evidence for an interaction between 5′SS position 1 and the last residue of the 3′SS has been reported (Parker and Siliciano 1993; Chanfreau et al. 1994; Deirdre et al. 1995). However, there are no previous data implicating 5′SS position 2 in an interaction with any residues in the 3′SS YAG. In fact, evidence for an interaction with the 3′SS penultimate residue has been sought, but not found (Ruis et al. 1994), conceivably due to additional constraints imposed by Prp8.
prp8 alleles suppress the exon ligation defect of both 5′SS position 2 and 3′SS YAG mutations
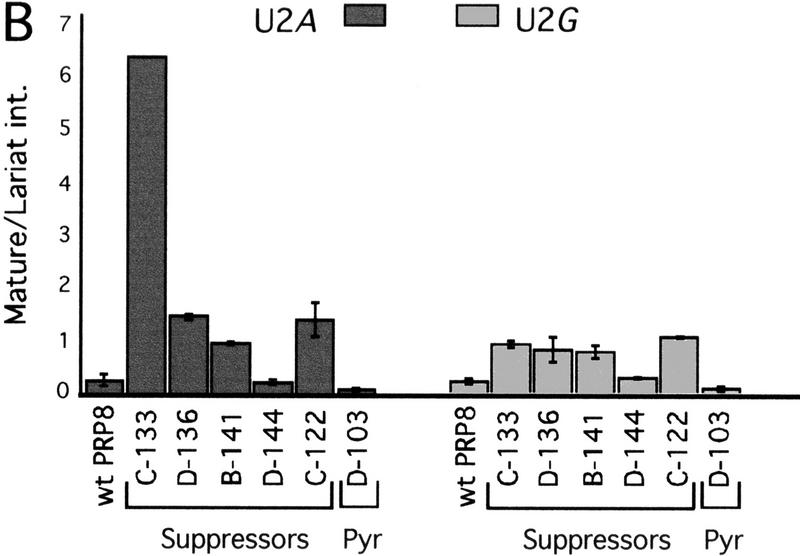
The suggested interaction might explain the longstanding observation that mutations in 5′SS position 2 confer a strong defect to exon ligation, the result of the second catalytic step of splicing, (Aebi et al. 1986, 1987; Fouser and Friesen 1986), as do mutations in the 3′SS YAG (Reed and Maniatis 1985; Ruskin and Green 1985; Vijayraghavan et al. 1986; Fouser and Friesen 1987; Parker and Siliciano 1993; Chanfreau et al. 1994). Because the prp8 splice-site suppressor alleles rescue the exon ligation defect of 3′SS YAG mutations (Umen and Guthrie 1996), they might concomitantly rescue the exon ligation defect of 5′SS position 2 mutations. To test this hypothesis, we assayed the in vivo efficiency of the exon ligation step for the U2A and U2G mutant reporters by primer extension analysis (Fig. 5). The prp8 splice-site suppressor alleles allowed an increase in the steady-state levels of mRNA. From PhosphorImager analysis, the ratio of spliced (mature) RNA to lariat-intermediate RNA was calculated to estimate the efficiency of the exon ligation step (Pikielny and Rosbash 1985; Fouser and Friesen 1986). For all alleles except D-144 (see below), this efficiency was increased for both U2A and U2G splicing from 2- to 20-fold (Fig. 5B; data not shown). The increase in the efficiency of exon ligation could reflect a direct effect on the second catalytic step of splicing, or on any substep in the spliceosomal rearrangements that must occur between the first and second catalytic steps.
Figure 5.

The exon ligation defect of 5′SS U2A and U2G splicing is suppressed. (A) Primer extension analysis of ACT1–CUP1 RNA splicing. Products generated from precursor, mature, lariat-intermediate, and U14 control species are denoted at left of the gel. Other bands are prominent primer extension stops derived from the longer precursor species. (Right) Splicing of a wild-type ACT1–CUP1 reporter is shown, to highlight the splicing defect conferred by the U2A mutation and the modest extent of suppression by prp8 alleles. Note that this experiment used less total RNA, as indicated by the reduced U14 levels. (B) The efficiency of the second step is estimated by calculating the ratio of levels of mature spliced ACT1–CUP1 RNA to the lariat intermediate species (Pikielny and Rosbash 1985; Fouser and Friesen 1986). The values were determined through PhosphorImager analysis of triplicate samples, with the exception of C-133 (for U2A). The near background levels of lariat intermediate in this strain resulted in large deviations for the (Mat/Lar int.) measurement. Shown is the lowest estimate for this ratio. (C) The total splicing efficiency is estimated by the ratio of levels of mature spliced ACT1–CUP1 RNA to unspliced precursor ACT1–CUP1 RNA.
The 5′SS position 2 mutations also confer a modest defect to steps preceding and possibly including 5′SS cleavage (Fouser and Friesen 1986; Aebi et al. 1987; Konforti and Konarska 1994; Siatecka et al. 1999). Notably, the estimated overall efficiency of U2A and U2G splicing by the prp8 alleles (Fig. 5C) correlates, as expected, with the degree of copper resistance observed for these reporters (Fig. 4A), but does not exactly correlate with the estimated efficiency of exon ligation (Fig. 5B). The differences between the estimates in Figures 5C and B could be due to additional effects on other steps of splicing, such as those preceding or including 5′SS cleavage.
In our in vivo analysis, one splice site suppressor allele, prp8-144, does not appear to suppress the exon ligation step. Consistent with only a weak effect on exon ligation, this allele exhibits barely detectable suppression of 3′SS YAG mutations (Fig. 4A). Because this prp8 allele does increase the overall efficiency of 5′SS U2G splicing (Fig. 5C), we suspect that the distinct location of prp8-144’s mutations allows for suppression at steps prior to but not including exon ligation.
prp8 alleles suppress a mutation in U6 snRNA
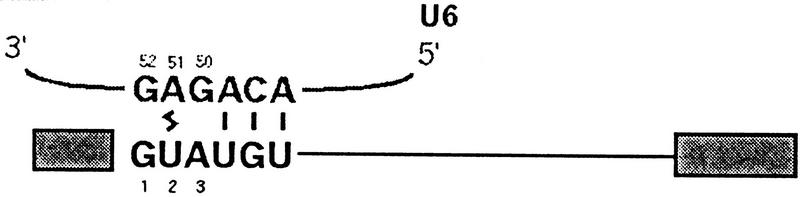
The proposed interaction between 5′SS position 2 and the 3′SS YAG could include other spliceosomal components in addition to Prp8. Excellent candidates for such an interaction are residues in the invariant ACAGAG motif of U6 snRNA. Base-pairing interactions between U6 (positions 47–49) and the 5′SS consensus sequence (positions 4–6), (Kandels and Séraphin 1993; Lesser and Guthrie 1993b), is thought to bring U6 G50, U6 A51, and U6 G52 close to 5′SS positions 1–3 (Table 2). Consistent with this notion, 5′SS position 2 is known to cross-link to U6 A51 (Sontheimer and Steitz 1993; Kim and Abelson 1996). Notably, mutations in the G50, A51, and G52 of the ACAGAG motif result in a severe block to exon ligation (Fabrizio and Abelson 1990; Madhani et al. 1990), reminiscent of the requirements for the 5′SS GU and 3′SS YAG residues at this step.
Table 2.
Genetic interactions between a prp8 splice site suppressor allele and U6 snRNA
 | ||||
|---|---|---|---|---|
|
|
+ Wild-type U6
|
− Wild-type U6
|
||
| wild-type PRP8 | mutant prp8 | wild-type PRP8 | mutant prp8 | |
| Wild-type U6 | +++ | +++ | +++ | +++ |
| G50A | +++ | +++ | — | — |
| C | +++ | +++ | + | + |
| U | +++ | +++ | + | + |
| A51C | + | ++ | — | — |
| G | +++ | +++a | — | — |
| U | — | + | — | — |
| G52A | +++ | +++ | — | — |
| C | +++ | +++ | — | — |
| U | +++ | +++ | + | + |
Mutations in U6 snRNA residues of the ACAGAG motif were tested for genetic interactions with the prp8 suppressor allele C-122. Growth (at 30°C) was assayed in either in the presence or absence of an extra copy of wild-type U6. The effects conferred by C-122 upon mutant U6 are shaded. (—) No growth; (+) or (++) intermediate growth; (+++) wild-type growth rate.
Although effects on growth were not detected, a 30% increase in splicing efficiency was measured for a wild-type ACT1–CUP1 reporter by primer extension assays.
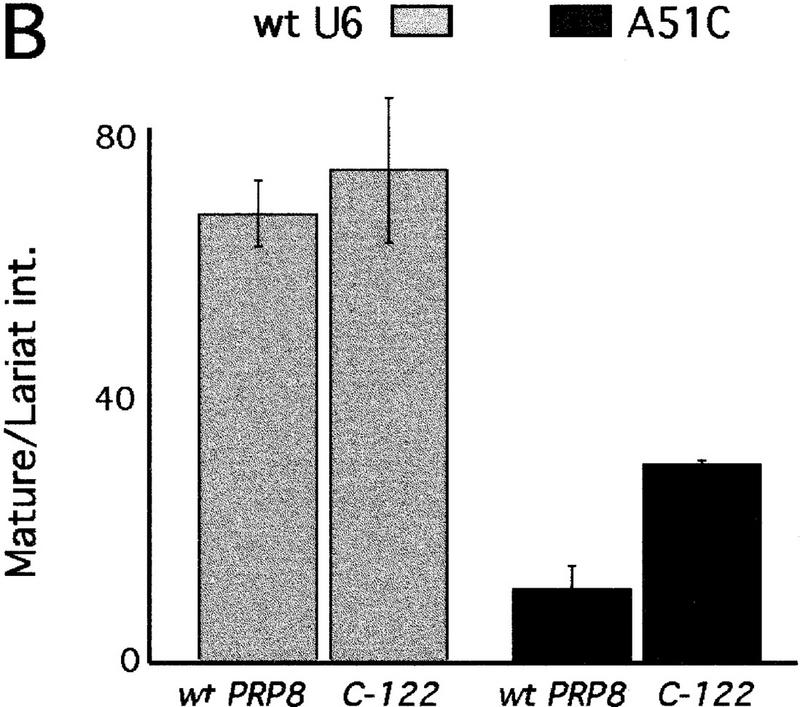
We thus investigated whether the effect of prp8 suppressor alleles extended to the candidate 5′SS-interacting residues in U6. We assayed the growth phenotypes of mutations in U6 G50, A51, and G52 in the presence of wild-type PRP8 or C-122, a representative splice site suppressor allele of prp8 (Table 2). Recessive U6 mutant phenotypes are very severe [inviable or very sick (Madhani et al. 1990)]; mutations in U6 A51, but not adjacent residues, are dominant-negative (Luukkonen and Séraphin 1998a). The presence of the prp8 suppressor allele (C-122), has no effect on the recessive growth phenotypes. Notably, however, the dominant-negative effect of mutations in A51 is suppressed (Table 2; Fig. 6).
Figure 6.

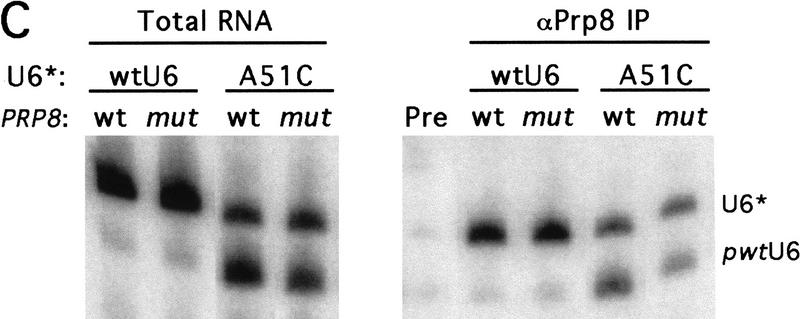
Splice site suppressor alleles of prp8 suppress the dominant-negative phenotype of mutations in U6 A51. (A) Growth (at 30°C) conferred by each prp8 allele in strains containing wild-type or A51C mutant U6 on a plasmid, in addition to a wild-type chromosomal copy of U6 snRNA. Equal numbers of log phase cells were spotted onto medium lacking histidine to require retention of the HIS3-marked plasmid containing dominant-negative U6. (B) The exon ligation defect conferred by dominant-negative mutations in U6 A51 is partially suppressed by prp8 alleles. Primer extension analysis of wild-type ACT1–CUP1 reporter RNA was conducted in triplicate for wild-type PRP8 and C-122 strains. The efficiency of the exon ligation step of splicing was estimated as described in Fig. 5. (C) prp8 suppressor allele C-122 allows for expression and association of U6 A51C. Strains containing mutant or wild-type prp8, and a shorter, pseudo-wild-type version of U6 snRNA (pwtU6), were transformed with an additional plasmid containing full-length U6*: either wild-type or A51C. Total RNA and RNAs that coimmunoprecipitation with polyclonal α-Prp8 were resolved on a denaturing gel and subjected to Northern analysis with an oligo probe that hybidizes to both pwtU6 and U6*(Madhani et al. 1990). The polyclonal antibodies immunoprecipitate Prp8 with a very low efficiency, (data not shown), but the critical comparison is the ratio of mutant U6*/pwtU6. This ratio is not significantly different between C-122 (mut) and wild-type (wt) PRP8. (Lane Pre) Background levels of RNA precipitated by preimmune sera.
The ability to suppress the dominant-negative effects of A51 mutants was then tested for all representative prp8 alleles by transformation efficiency (data not shown), growth (Fig. 6A), and copper resistance conferred on a wild-type ACT1–CUP1 (data not shown). The splice site suppressor prp8 alleles suppress the dominant-negative phenotype of A51C by these assays, whereas the Pyr alleles of prp8 do not (Fig. 6A; data not shown). The extent of suppression exhibited by all of the alleles with mutations in the B fragment was not as strong (Fig. 6A; data not shown). Thus, the extent of A51C suppression appears to correlate with the location of the mutation(s) on PRP8.
The prp8 suppressor alleles exerted a weak effect on the transformation efficiency of the severely dominant-negative A51U mutation, but the analysis for A51U was complicated by a high frequency of reversions (data not shown). The weak dominant effects of the A51G mutation were not detectable by growth assays. However, in primer extension assays, the prp8 allele C-122 was observed to allow for a small, ∼30% increase in the splicing efficiency of the wild-type ACT1–CUP1 reporter in the presence of A51G (data not shown).
The exon ligation defect is suppressed
Like the 5′SS position 2 mutations, mutations in U6 A51 confer a strong inhibition to the exon ligation step of splicing (Fabrizio and Abelson 1990). Thus, as for the 5′SS mutations, the exon ligation defect of U6 A51 mutants is predicted to be suppressed by the splice site suppressor prp8 alleles. By primer extension analysis, we see a 2.5-fold increase in the efficiency of exon ligation for wild-type ACT1–CUP1 splicing (Fig. 6B), as well as for the endogenous RP51 transcript (data not shown). Thus, the exon ligation defect of the dominant-negative U6 mutation is partially suppressed by the prp8 mutant.
Splice site suppressor prp8 alleles allow for assembly of A51 mutants into snRNPs
Because the prp8 alleles did not rescue the recessive lethal phenotype of A51 mutations (Table 2), we explored the possibility that the prp8 alleles suppress the dominant-negative phenotype by impairing the ability of mutant U6 snRNA to assemble into snRNPs. This could in turn allow the assembly of more snRNPs containing wild-type U6 snRNA, and thus cause an apparent suppression of the exon ligation defect. Prp8 is a component of the U5 snRNP, which joins the U4/U6 snRNP to form the U4/U5/U6 triple snRNP. It is thus possible that Prp8 forms contacts with or influences U6 in the triple snRNP. We assayed whether the prp8 mutants affected the stability of mutant U6 snRNA, or its ability to coimmunoprecipitate with Prp8. We used strains containing pseudo-wild-type U6 (Madhani et al. 1990), which can be distinguished in size from mutant U6. The pseudo-wild-type U6 is expressed at low levels in the presence of wild-type U6, but is increased when the A51C mutant is the only other copy of U6 in the cell. As shown in Figure 6C, the prp8 mutant C-122 does not affect the ratio of A51C mutant U6 to pseudo-wild-type U6 in either the total pools of snRNAs, or in those immunoprecipitated by α-Prp8 antibodies. These data suggest that the prp8 suppressor allele allows for the assembly of 51C mutant U6 into snRNPs and, probably, into spliceosomes. It is thus likely that suppression of the dominant-negative phenotype of the 51C mutation occurs on the spliceosome, at the exon ligation step. To explain why suppression of the recessive U6 A51C growth phenotype was not observed, we suggest that suppression by these prp8 alleles is simply not strong enough to confer viability to the severe U6 mutant.
Mutations adjacent to U6 A51 are not suppressed
Within U6, suppression by the prp8 alleles appears specific to U6 A51. Combining the prp8 alleles with mutations in U6 nucleotides adjacent to A51 did not result in any growth alterations (Table 2), despite the fact that mutations in G52 and G50, like A51, have been shown to strongly impair exon ligation (Fabrizio and Abelson 1990). For a more quantitative comparison, we conducted the primer extension analysis of ACT1–CUP1 splicing for the viable U6 G52 and G50 mutations. In contrast to what is observed with A51C, neither the efficiency of exon ligation, nor the overall splicing efficiency, was improved (data not shown). Thus, although a number of U6 mutations impede exon ligation, suppression by prp8 appears specific for mutations in A51.
Discussion
A new tertiary interaction between the 5′SS, 3′SS, and U6
Our initial goal was to provide functional correlates for Prp8’s cross-linking interactions to the 5′SS and 3′SS consensus sequences, and to determine whether functions at the two splice sites would map to distinct or to the same functional domains of the protein. We found prp8 alleles that suppress mutations in both the 5′SS GU and in the 3′SS YAG; all prp8 alleles that suppress mutations in the 3′SS YAG also suppress mutations in 5′SS position 2. Furthermore, we found that mutations in A51 of the U6 ACAGAG motif are also suppressed by the prp8 alleles. However, other mutations in intron consensus sequences, and in neighboring residues of U6, are not suppressed. To explain the distinct, but unanticipated pattern of suppression, we suggest that Prp8 influences a specific, previously unrecognized, tertiary interaction between the suppressed residues (Fig. 7A,C).
Figure 7.
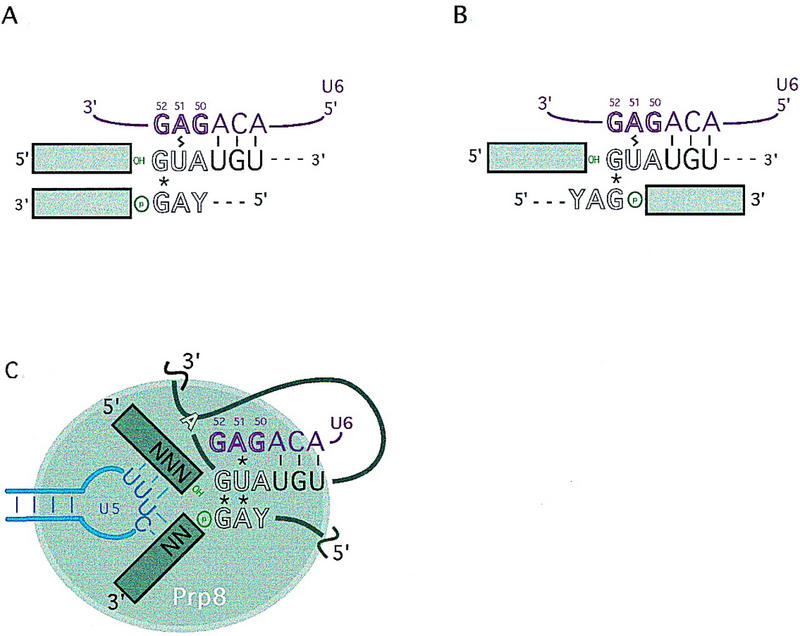
Hypothesized interaction of RNA residues suppressed by prp8 extends the juxtaposition of conserved residues at the 5′ and 3′ ends of the intron. (A,B) The previously implicated non-Watson–Crick base-pairing interaction between the first and last residue of the intron (*), in addition to Watson–Crick (−) and cross-linking (lightning bolt) interactions between the 5′SS and U6 snRNA, equally favor an alignment of the 5′ and 3′ ends of the intron in which additional residues are juxtaposed (A), or an alignment in which they are not (B). Residues that are critical for the second step are denoted in outline. The 3′ hydroxyl of exon 1 and the 3′SS phosphate, reactants for the exon ligation chemical step are colored green. (C) Prp8 could function in conjunction with RNA interactions at the spliceosomal active site for the second step. The known and hypothesized (asterisks) RNA interactions could align the 5′ and 3′ ends of the intron, in addition to the 5′ and 3′ exons, to position the reactants (green) for exon ligation catalysis. Interactions between the invariant loop of U5 snRNA and the 5′ and 3′ exons are shown in blue.
This proposed interaction is consistent with several previous observations. First, mutations in the 5′ and 3′ terminal intron residues can reciprocally suppress each other, with a specificity that suggests that they form a non-Watson–Crick base-pairing interaction (Parker and Siliciano 1993; Chanfreau et al. 1994; Deirdre et al. 1995). Second, 5′SS position 2 cross-links to U6 A51 (Sontheimer and Steitz 1993; Kim and Abelson 1996). In addition to Watson-Crick pairing of U6 with positions 4–6 of the 5′SS (Kandels and Séraphin 1993; Lesser and Guthrie 1993b), genetic interactions between U6 and 5′SS positions 1 and 3 support the hypothesis that the entire 5′SS consensus sequence is juxtaposed to the U6 ACAGAG sequence (Luukkonen and Séraphin 1998b). Third, the U6 G52U mutation suppresses mutations in the terminal intron residue, suggesting that U6 and the 3′SS interact at least indirectly (Lesser and Guthrie 1993b).
Importantly, whereas all of the above previous observations are consistent with an extended interaction between the 5′ and 3′ ends of the intron (Fig. 7A), they have been equally consistent with a configuration (Fig. 7B) in which the sole interaction between the 5′ and 3′ ends of the intron occurs between the first and last nucleotides. Evidence for an interaction between the second and penultimate nucleotides of the intron, predicted by Figure 7A, has not been found by directed mutagenesis (Ruis et al. 1994). The tertiary interaction suggested by our data between 5′SS position 2 and the 3′SS YAG now favors the extended interaction between the 5′ and 3′ ends of the intron, (Fig. 7A,C). An interaction between the second and penultimate residues of the intron is thus likely to occur, and could have eluded previous studies through constraints imposed by additional factors that were not mutagenized in that study, namely U6 and Prp8. Supportive of this idea, an indirect interaction was suggested to occur between 5′SS position 3 and 3′SS position −3, on the basis of the observation that mutations in 5′SS position 3 nonspecifically affect the competition of closely spaced 3′SS YAG sequences (Deirdre et al. 1995). Prp8 and U6 may also affect a tertiary interaction between these positions (see below).
It is striking that participants in the postulated interaction are conserved between the conventional U2-dependent and the divergent U12-dependent spliceosomes. Although introns spliced by the two different pathways vary in whether they contain G-G or A-C in the first and last positions, all contain U in the second and A in the penultimate positions (Dietrich et al. 1997; Sharp and Burge 1997; Shukla and Padgett 1999). Moreover, the residue in U6atac snRNA corresponding to position 51 of yeast U6 is also an A (Tarn and Steitz 1996). Additionally, the three adjacent residues, 5′SS position 3, 3′SS position −3 of the intron, and the position corresponding to U6 G50, are also conserved (Tarn and Steitz 1996; Dietrich et al. 1997; Sharp and Burge 1997). Notably, Prp8 has also been shown to be a component of the two spliceosomes (Luo et al. 1999). This supports the idea that the proposed interaction of the three RNAs and Prp8 is fundamental to the mechanism of pre-mRNA splicing.
Interactions that bridge the two catalytic steps of splicing
The long-standing observation that residues at the 5′ end of the intron are required for the second transesterification step has always seemed paradoxical, as the 5′ end of the intron is no longer a substrate for splicing chemistry after the first step is completed. In the splicing reaction catalyzed by group I introns, the 5′ end of the intron leaves the active site after the first cleavage step, to be replaced by the 3′ end of the intron for the second transesterification step (Cech 1990). In the spliceosome, the 5′ end of the intron could function at the active site for both steps. If the 5′ exon does not stray far from the cleaved 5′ end of the intron, these residues could serve, through the proposed interaction (Fig. 7C), to position the 3′SS for catalysis of exon ligation.
Little is known about how the spliceosome couples 5′SS cleavage to exon ligation. The 5′ exon is thought to be held or carried from the first to the second catalytic step by an invariant loop in U5 snRNA (for review, see Newman 1997). Because Prp8 makes extensive cross-links to the loop, and to the 5′ and 3′ exons, it has been hypothesized that Prp8 acts in conjunction with the U5 loop to position the exons for ligation (Teigelkamp et al. 1995; Dix et al. 1998; O’Keefe and Newman 1998). Alignment of the 5′ and 3′ ends of the intron could help in the alignment of the exons and in the positioning of the 3′SS for catalysis (Fig. 7C). This could be accomplished by the proposed network of RNA interactions and facilitated by Prp8.
The progression of splicing from 5′SS cleavage to exon ligation is known to involve a conformational change in the spliceosome (Schwer and Guthrie 1992; Umen and Guthrie 1995b; Chiara et al. 1996; Chua and Reed 1999) and the construction of a chemically distinct active site (Moore and Sharp 1993; Sontheimer et al. 1997). However, a number of interactions that are critical for exon ligation are established during spliceosome assembly, before 5′SS cleavage. These include the interaction of the 5′SS consensus sequence with the U6 ACAGAG motif (Kim and Abelson 1996), and the 5′ exon with the U5 invariant loop (Newman and Norman 1991, 1992; Wassarman and Steitz 1992; Sontheimer and Steitz 1993; O’Keefe et al. 1996). Prp8 is also observed to cross-link to the 5′ exon (Wyatt et al. 1992; Teigelkamp et al. 1995; Chiara et al. 1996) and to the 5′SS GU before 5′ cleavage (Reyes et al. 1996). In a concurrent study, Siatecka et al. (1999) report evidence that Prp8 functions in recognition of 5′SS position 2 during spliceosome assembly, before 5′SS cleavage, as well as during exon ligation (Siatecka et al. 1999). The maintenance of interactions between the two catalytic steps of splicing suggests ways in which one active site could be altered to undergo both transesterification steps. Through interactions with the 5′SS, U6 and 3′SS, in addition to the 5′ exon, U5, and 3′ exon, Prp8 could perform a critical function in bridging the two steps of splicing.
In another recent study (Kuhn et al. 1999), a mutation in PRP8, (prp8-201), was found to suppress a U4 mutation, U4–cs1, which compromises the interaction of the U6 ACAGAG motif with the 5′SS consensus sequence. Interestingly, suppression is conferred by a mutation in residue 1861 of Prp8, which is strikingly close to the location of mutations in the splice site suppressor alleles D-135, D-136, and D-143 (Table 1). In fact, prp8-201 suppresses 5′SS U2A and 3′SS YAG mutations (C. Collins and C. Guthrie, unpubl.). However, our splice site suppressor alleles fail to suppress U4–cs1 (A. Kuhn and D. Brow, pers. comm.); thus, the relationship between splice-site suppression and U4–cs1 suppression by prp8 remains to be understood. Notably, U4–cs1 suppression appears to occur prior to 5′ cleavage (Kuhn et al. 1999). The fact that one mutation, in prp8-201, can suppress defects at two different steps suggests, again, that Prp8 forms interactions that are maintained throughout both chemical steps of splicing.
A role for Prp8 at the spliceosomal catalytic core
The model based on our genetic results provides an attractive explanation for the extensive cross-linking interactions that have been observed previously between Prp8 and residues near both the 5′SS and 3′SS. These data suggest that Prp8 actually binds the proposed tertiary RNA interaction. However, it is not known whether the amino acids mutated in the suppressor alleles, which map to four distinct regions of the protein, play a direct role in RNA recognition. Whereas most of these residues are conserved in the known homologs of Prp8, all of the suppressor mutations are viable and confer only a mild, if any, detectable defect to wild-type splicing. The suppressor alleles could serve to relax some of the constraints imposed on the identity of splice-site residues, thereby allowing mutant as well as wild-type sequences to be utilized. In well-studied systems of protein-substrate recognition, mutations that permit a wider range of interactions have been observed to increase the flexibility of the binding interface or pocket (Bone et al. 1989; Morton and Matthews 1995). Such effects are not limited to mutations at the site of binding, and, in fact, may occur at considerable distances (Mace et al. 1995; Gutierrez et al. 1998). It remains formally possible that the suppression observed is mediated by allosteric effects on another protein that binds the 5′ and 3′ ends of the intron. However, despite the numerous proteins that have been found to function during the exon ligation step (Umen and Guthrie 1995b), Prp8 is the only protein that has yet been found to affect the requirement for YAG at the 3′SS.
Whereas spliceosome catalysis is thought to be performed by RNA, many of the same types of observations that suggest a critical role for RNA at the catalytic core have yielded similar results for Prp8, namely, cross-linking to RNA near sites of chemistry, genetic suppression of splice-site mutations, and phylogenetic conservation. What is, then, the specific role of Prp8? Prp8 could conceivably make direct structural or chemical contributions to the spliceosomal active site. Alternatively, Prp8 could serve to add constraints or stability to a structure that is intrinsically comprised of RNA. This latter role for protein in RNP catalysis has been demonstrated for group-I introns (Weeks and Cech 1995a,b, 1996) and Rnase P (Guerrier-Takada et al. 1983). Recent crystal structures of ribosomal L11 protein complexed with a 58-nucleotide domain of 23S rRNA illustrate that a protein component of an RNP can stabilize an unusual RNA fold through direct contact with a critical RNA tertiary interaction (Conn et al. 1999; Wimberly et al. 1999). Ultimately, an ultrastructural analysis of Prp8’s interaction with RNA in the spliceosome is required to understand the contribution of this protein to catalysis.
Materials and methods
Yeast strains and plasmids
Screens and characterization of prp8 alleles were conducted with the strain yJU75 (Umen and Guthrie 1996): MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp8Δ::LYS2 trp1; pJU169 (PRP8 URA3 CEN ARS). ACT1–CUP1 reporters are described in Lesser and Guthrie (1993a) and Umen and Guthrie (1996). For all reporters, the LEU2-marked (pGAC24) version was used. The 5′SS U2A, U2G, and wild-type ACT1–CUP1 reporters are described in Ruis et al. (1994), except that the pGAC14 vector backbone was swapped for pGAC24 (Lesser and Guthrie 1993a), to generate pCC72 (U2A), pCC44 (U2G), and pCC71 (wild type).
For testing genetic interactions between prp8-122 and mutations in U6 snRNA (Fig. 6), a prp8Δ::LYS2 disruption was generated, according to Umen and Guthrie (1996), in a diploid heterozygous for the disruption snR6Δ::LEU2 (Brow and Guthrie 1988). The diploid was then transformed with plasmids (see below) and sporulated. Haploid progeny containing both disruptions, prp8Δ::LYS2 covered by wild-type PRP8 or prp8–C122 on a pRS423 plasmid (2 μ HIS3), and snR6Δ::LEU2 covered by wild-type U6 in pSE360, were identified by nutritional markers, and the presence of both prp8 and snR6 disruptions was confirmed by whole-cell PCR. The resulting U6-shuffle strains were transformed with plasmids containing wild-type (pSX6T) or mutant U6 (Madhani et al. 1990), and assayed for growth before or after counter selection on 5-FOA of the URA3-marked plasmid containing wild-type U6 (Table 2). For the experiments in Figure 6C, a similar strategy was used to generate a double-knockout haploid strain containing plasmid-borne pseudo-wild type U6, and wild-type or mutant prp8 (in pRS423). The resulting strain was then transformed with plasmids (pSE360) containing wild-type or A51C mutant U6.
Screens for suppressors of mutations in the 5′SS GU
PRP8 was PCR mutagenized in four parts, with primers and Mn2+ conditions described in Umen and Guthrie (1996), according to mutagenic PCR conditions described in Leung et al. (1989). prp8 mutants consisting of each of four mutagenized fragments were created by in vivo gap repair (Muhlrad et al. 1992). The four corresponding gapped plasmids were generated from pJU225 (PRP8 TRP1 2μ), as described in Umen and Guthrie (1996).
yJU75 strains containing either 5′SS U2A or U2G mutant ACT1–CUP1 reporters were transformed, and, for each reporter, ∼4000 were replica plated onto medium containing 0.025, 0.05, or 0.1 mm copper sulfate (Lesser and Guthrie 1993a). To look for recessive as well as dominant suppression, transformants that survived passage on 5-FOA were rescreened for increased growth on copper. For each candidate suppressor, the TRP-marked plasmid was recovered, transformed into a fresh yJU75 strain, and retested for the ability to confer increased growth on copper with the 5′SS U2A and U2G ACT1–CUP1 reporters.
Copper growth
Duplicate cultures were grown to log phase in medium lacking leucine to ensure maintenance of the ACT1–CUP1 reporter plasmid. Cultures were then diluted to an OD600 of 0.5, and a frogger was used to stamp equivalent amounts of cells from each strain onto plates containing different concentrations of copper (Lesser and Guthrie 1993a). For assays of the effect of dominant-negative mutations in U6 A51 (Fig. 6A), cells were grown on medium and copper plates lacking histidine, to ensure maintenance of the pSE362 plasmid containing the dominant-negative U6 mutant.
Mapping of suppressor mutations
Because each mutagenized prp8 fragment was ∼2 kb, and contained multiple mutations, the location of mutations sufficient to confer suppression was mapped to a smaller region of PRP8. Fragments of the mutant prp8 were PCR amplified in nonmutagenic conditions with Pfu polymerase. The PCR fragments were then cotransformed with appropriately gapped plasmid. The fragments of mutant prp8 coding sequence that could reconstruct the suppressor phenotype for each mutant are shown in Table 3.
Table 3.
prp8 fragments that confer suppression
| Allele
|
Fragment PCR-amplified from mutant
|
Enzyme used to gap pJU225a
|
|---|---|---|
| C-133, C-134 | 4262–5161 | BstEII (w. EtBr) |
| D-135, D-136, D-143 | 5340–6147 | MscI |
| D-144 | 6017–7215 | StuI |
| B-131, B-132, B-141, B-142 | 1635–3322 | AflII (w. EtBr) |
| or | ||
| 2431–3735 | SpeI |
Oligonucleotides used to amplify each fragment ranged in size from 20 to 22 nucleotides.
For enzymes with two sites in pJU225, the plasmid was partially digested in the presence of 100 μg/ml ethidium bromide to enrich for singly cut plasmids (Umen and Guthrie 1996).
The region listed for each allele in Table 3 was then sequenced at the Biomolecular Resource Center DNA Sequencing facility at UCSF, with the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems). The program ALIGN (Myers and Miller 1988) was used at the GENESTREAM network server (http://www2.igh.cnrs.fr) to identify changes in the PRP8 sequence.
The three mutations in the sequenced region of D-144 were isolated from each other by standard cloning techniques, with the intervening StuI and SphI sites. Each isolate was confirmed by sequencing, and found to confer modest suppression of 5′SS U2A. Likewise, D-134 and D-143 mutations were separated from additional mutations by the MscI site. These additional mutations (not shown) were not required for the suppression phenotype (Table 3).
Primer extension analysis
RNA preparation and primer extension assays were performed as described previously (Frank and Guthrie 1992; Lesser and Guthrie 1993a). U14 snoRNA was primer extended for an internal control for the amount of total RNA in each lane (Noble and Guthrie 1996). Products were quantified by PhosphorImager analysis of duplicate or triplicate samples. The total splicing efficiency and the efficiency of the exon ligation step were estimated as derived previously (Pikielny and Rosbash 1985; Fouser and Friesen 1986), by calculating the ratio of levels of mature/precursor species for the total splicing efficiency, and mature/lariat for the efficiency of the exon ligation step.
Bead-beat extracts
For Figure 6C, strains described above were grown to late log phase in medium lacking uridine, to ensure maintenance of the dominant-negative U6 plasmid. A total of 100 mls of culture were pelleted and resuspended in 150 μl of Lysis Buffer (45 mm HEPES at pH 7.9, 400 mm NaCl, 0.5% NP-40 (vol/vol), 1 mm EDTA, 20% glycerol (vol/vol), DEPC-treated dH20, and protease inhibitors (1 mm PMSF, 1 μg/ml leupeptin, 1 mm benzamidine)). Cells were lysed by vortexing with 600 μl of acid-washed glass beads (size 0.5 mm), in 1-min bursts followed by a 1-min rest on ice, repeated six times. Insoluble material in the lysate was then removed by microfuging for 10 min at maximum speed. The supernatant lysate was diluted with an equal volume of dilution buffer (45 mm HEPES at pH 7.9, 1 mm EDTA, 20% glycerol, with a fresh addition of protease inhibitors). All steps were conducted at 4°C.
Polyclonal α-PRP8 antibodies
The LacZ–PRP8 fusion plasmid, pFP8.4, was provided by Jean Beggs (University of Edinburgh, Edinburgh, Scotland), and expression of the fusion protein was heat induced in the Escherichia coli strain pop2136 as described previously (Lossky et al. 1987). The insoluble pellet, which contained the majority of the fusion protein, was prepared from lysed cells, and purified according to Harlow (1988). The ∼170-kD fusion protein was further purified by subjecting the sample to SDS-polyacrylamide gel electrophoresis and removing the band after staining with 0.05% Coomassie blue in distilled water. Gel slices, containing a total of ∼0.5 mg of fusion protein, were solubilized in Freund’s adjuvant and used to immunize rabbits. The rabbits were boosted in 3-week intervals with 0.25 mg of gel-purified fusion protein. Preimmune sera were collected prior to immunization, and antisera were obtained ∼10 days after the fourth boost. All rabbit work was done by Berkeley Antibody Company (BAbCO) in Richmond, CA.
The ability of the antibodies to immunoprecipitate Prp8 was confirmed with extracts from strains that contain hemagglutinin (HA) epitope-tagged PRP8 (Umen and Guthrie 1995a), and subjecting the pellets and supernatants to Western blot analysis with a monoclonal antibody specific for the HA epitope.
Immunoprecipitations
For immunoprecipitation of snRNAs, IgG was prebound to 30 μl of protein A–Sepharose (Sigma) in NTN buffer [150 mm NaCl, 50 mm Tris-HCl at pH 7.4, 0.1% NP-40 (vol/vol)], and washed five times with 600 μl of the same buffer at 4°C. For each experiment, 200 μl of bead-beat extract was added to 15 μl of prebound serum, and incubated at 4°C, with protease inhibitors (above), for 2.5 hr. The antibody complexes were centrifuged, and washed four times with 600 μl of NTN buffer. RNA was then prepared from the pellets, (and 10 μl of total extract), and subjected to Northern analysis, with a 32P-end-labeled oligonucleotide probe against U6 snRNA (Noble and Guthrie 1996).
Acknowledgments
We thank Andreas Kuhn and Dave Brow for communicating unpublished results; Magda Konarska, Mirka Siatecka, and José Reyes for communicating results and for extensive collaborative discussions; Jim Umen, Jon Staley, Stephen Rader, and David McPheeters for helpful discussions; Stephen Rader, Amy Kistler, Kent Duncan, Maki Inada, Jon Staley, James Wilhelm, Alan Frankel, and John Abelson for critical comments on this manuscript; and Lucita Esperas and Carol Pudlow for excellent technical assistance. C.C. is a predoctoral student in the Biophysics Graduate Group at UCSF. C.G. is an American Cancer Society Research Professor of Molecular Genetics. This work was supported by a grant from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL guthrie@cgl.ucsf.edu; FAX (415) 502-5306.
References
- Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M, Hornig H, Weissmann C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Bone R, Silen JL, Agard DA. Structural plasticity broadens the specificity of an engineered protease. Nature. 1989;339:191–195. doi: 10.1038/339191a0. [DOI] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Cech TR. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell. 1986;44:207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Legrain P, Dujon B, Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3′ splice site selection in S. cerevisiae. Nucleic Acids Res. 1994;22:1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K, Reed R. Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes & Dev. 1999;13:841–850. doi: 10.1101/gad.13.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- Deirdre A, Scadden J, Smith CW. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- Dix I, Russell CS, O’Keefe RT, Newman AJ, Beggs JD. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;250:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- Fouser LA, Friesen JD. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- ————— Effects on mRNA splicing of mutations in the 3′ region of the Saccharomyces cerevisiae actin intron. Mol Cell Biol. 1987;7:225–230. doi: 10.1128/mcb.7.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes & Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Gutierrez A, Yoshimura T, Fuchikami Y, Soda K, Esaki N. A mutant D-amino acid aminotransferase with broad substrate specificity: Construction by replacement of the interdomain loop Pro119- Arg120-Pro121 by Gly-Gly-Gly. Protein Eng. 1998;11:53–58. doi: 10.1093/protein/11.1.53. [DOI] [PubMed] [Google Scholar]
- Harlow E. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hodges PE, Jackson SP, Brown JD, Beggs JD. Extraordinary sequence conservation of the PRP8 splicing factor. Yeast. 1995;11:337–342. doi: 10.1002/yea.320110406. [DOI] [PubMed] [Google Scholar]
- Kandels LS, Séraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- Konforti BB, Konarska MM. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes & Dev. 1994;8:1962–1973. doi: 10.1101/gad.8.16.1962. [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Li Z, Brow DA. Splicing factor Prp8 governs unwinding during activation of the spliceosome. Mol Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993a;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science. 1993b;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- Lossky M, Anderson GJ, Jackson SP, Beggs JD. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell. 1987;51:1019–1026. doi: 10.1016/0092-8674(87)90588-5. [DOI] [PubMed] [Google Scholar]
- Lucke S, Klockner T, Palfi Z, Boshart M, Bindereif A. Trans mRNA splicing in trypanosomes: Cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 1997;16:4433–4440. doi: 10.1093/emboj/16.14.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Moreau GA, Levin N, Moore MJ. The human Prp8 protein a common component U2- and U12-dependent spliceosomes. RNA. 1999;5:893–908. doi: 10.1017/s1355838299990520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Séraphin B. Construction of an in vivo-regulated U6 snRNA transcription unit as a tool to study U6 function. RNA. 1998a;4:231–238. [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Séraphin B. Genetic interaction between U6 snRNA and the first intron nucleotide in Saccharomyces cerevisiae. RNA. 1998b;4:167–180. [PMC free article] [PubMed] [Google Scholar]
- Mace JE, Wilk BJ, Agard DA. Functional linkage between the active site of alpha-lytic protease and distant regions of structure: Scanning alanine mutagenesis of a surface loop affects activity and substrate specificity. J Mol Biol. 1995;251:116–134. doi: 10.1006/jmbi.1995.0420. [DOI] [PubMed] [Google Scholar]
- MacMillan AM, Query CC, Allerson CR, Chen S, Verdine GL, Sharp PA. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Bordonne R, Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes & Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Morton A, Matthews BW. Specificity of ligand binding in a buried nonpolar cavity of T4 lysozyme: Linkage of dynamics and structural plasticity. Biochemistry. 1995;34:8576–8588. doi: 10.1021/bi00027a007. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Newman A. RNA splicing. Activity in the spliceosome. Curr Biol. 1994;4:462–464. doi: 10.1016/s0960-9822(00)00104-4. [DOI] [PubMed] [Google Scholar]
- ————— The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- ————— U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. RNA-RNA interactions in the spliceosome: Unraveling the ties that bind. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- O’Keefe RT, Newman AJ. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano PG. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Kois P, Konforti BB, Konarska MM. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA. 1996;2:213–225. [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Gustafson HE, Luo HR, Moore MJ, Konarska MM. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis BL, Kivens WJ, Siliciano PG. The interaction between the first and last intron nucleotides in the second step of pre-mRNA splicing is independent of other conserved intron nucleotides. Nucleic Acids Res. 1994;22:5190–5195. doi: 10.1093/nar/22.24.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B, Green MR. Role of the 3′ splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985;317:732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on Prp16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA. Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA. 1999;5:525–538. doi: 10.1017/s1355838299982213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka, M., J.L. Reyes, and M.M. Konarska. 1999. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Sun S, Piccirilli JA. Metal ion catalysis during splicing of premessenger RNA. Nature. 1997;388:801–805. doi: 10.1038/42068. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S, Newman AJ, Beggs JD. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 1995a;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- ————— The second catalytic step of pre-mRNA splicing. RNA. 1995b;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- ————— Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Weeks KM, Cech TR. Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry. 1995a;34:7728–7738. doi: 10.1021/bi00023a020. [DOI] [PubMed] [Google Scholar]
- ————— Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell. 1995b;82:221–230. doi: 10.1016/0092-8674(95)90309-7. [DOI] [PubMed] [Google Scholar]
- ————— Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science. 1996;271:345–348. doi: 10.1126/science.271.5247.345. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: The structure of the L11- RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- Wyatt JR, Sontheimer EJ, Steitz JA. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]