Abstract
ATP-dependent nucleosome remodeling and core histone acetylation and deacetylation represent mechanisms to alter nucleosome structure. NuRD is a multisubunit complex containing nucleosome remodeling and histone deacetylase activities. The histone deacetylases HDAC1 and HDAC2 and the histone binding proteins RbAp48 and RbAp46 form a core complex shared between NuRD and Sin3-histone deacetylase complexes. The histone deacetylase activity of the core complex is severely compromised. A novel polypeptide highly related to the metastasis-associated protein 1, MTA2, and the methyl-CpG-binding domain-containing protein, MBD3, were found to be subunits of the NuRD complex. MTA2 modulates the enzymatic activity of the histone deacetylase core complex. MBD3 mediates the association of MTA2 with the core histone deacetylase complex. MBD3 does not directly bind methylated DNA but is highly related to MBD2, a polypeptide that binds to methylated DNA and has been reported to possess demethylase activity. MBD2 interacts with the NuRD complex and directs the complex to methylated DNA. NuRD may provide a means of gene silencing by DNA methylation.
Keywords: DNA methylation, histone deacetylase complex, nucleosome remodeling, gene silencing
Packaging of DNA into chromatin allows the cell to store its genetic information efficiently and has an important role in regulating gene expression (Workman and Kingston 1998). Dynamic changes in chromatin structure can facilitate or prevent the access of the transcription machinery to nucleosomal DNA, leading to transcription regulation. Recent studies have revealed two mechanisms by which chromatin structure can be altered. One mechanism involves multisubunit protein complexes that use the energy derived from ATP hydrolysis to alter the structure of, or ‘remodel’, nucleosomes (for review, see Tsukiyama and Wu 1997; Kadonaga 1998; Varga-Weisz and Becker 1998; Travers 1999). The other mechanism involves covalent modification of nucleosomes, in particular acetylation of the core histone tails and methylation of DNA (for review, see Grunstein 1997; Kuo and Allis 1998; Struhl 1998; Ng and Bird 1999).
Since the discovery of histone acetylation by Allfrey et al. (1964), a general correlation between histone acetylation and gene activity has been established (Hebbes et al. 1988). The enzymes that catalyze histone acetylation and deacetylation have been identified (Brownell et al. 1996; Taunton et al. 1996). Several transcriptional coactivators have histone acetyltransferase (HAT) activity, whereas several transcriptional corepressors have histone deacetylase activity (for review, see Grunstein 1997; Kuo and Allis 1998; Struhl 1998). In addition, mutagenesis studies with Gcn5 and Rpd3, the prototypical histone acetyltransferase and deacetylase, respectively, confirmed the long-speculated role of histone acetylation and deacetylation in transcription regulation (Kadosh and Struhl 1998a; Kuo et al. 1998; Wang et al. 1998). Moreover, Rpd3/Sin3-dependent repression has been shown to be directly associated with the deacetylation of lysine 5 of histone H4 in the promoters of UME6-regulated genes (Kadosh and Struhl 1998b; Rundlett et al. 1998). However, how core histone acetylation/deacetylation leads to transcriptional activation/repression remains to be elucidated.
Methylation of cytosine at CpG dinucleotides is a common feature of many higher eukaryotic genomes. Many studies have established a general correlation between DNA methylation and gene inactivation (Razin and Riggs 1980). However, the underlying molecular mechanism remained unknown until recently. It was found that MeCP2, a protein that specifically binds to methylated DNA, copurifies with the Sin3A/HDAC corepressor complex and that the histone deacetylase inhibitor TSA relieves MeCP2-mediated transcriptional repression (Jones et al. 1998; Nan et al. 1998). Recently, four mammalian proteins containing regions homologous to the MeCP2 methyl-CpG-binding domain, MBD1-4, were identified by searching the expressed sequence tag (EST) databases (Hendrich and Bird 1998). Interestingly, MBD2 was claimed to be a DNA demethylase, and MBD4 was shown to be an endonuclease potentially involved in DNA mismatch repair (Bellacosa et al. 1999; Bhattacharya et al. 1999). The functions of MBD1 and MBD3 are unknown.
To develop a mechanistic understanding of how core histone acetylation regulates transcription, we have studied the histone deacetylases HDAC1 and HDAC2 (Taunton et al. 1996; Yang et al. 1996). Using a combination of conventional and affinity chromatography, we previously purified and characterized two HDAC1/HDAC2-containing histone deacetylase complexes, the Sin3A/HDAC complex, and the NuRD complex (Zhang et al. 1997, 1998a,b). The two protein complexes share four polypeptides: HDAC1, HDAC2, RbAp46, and RbAp48. In addition, each complex contains three unique polypeptides (Sin3A, SAP30, and SAP18 in the Sin3 complex, and Mi2, p70, and p32 in the NuRD complex). Interestingly, the NuRD complex also possesses nucleosome remodeling activity, most likely because of the presence of Mi2, a member of the SWI2/SNF2 helicase/ATPase family (Tong et al. 1998; Xue et al. 1998; Zhang et al. 1998a).
To gain insight into the function of the NuRD complex, we have identified its p70 and p32 subunits. We demonstrate that these polypeptides have an important role in modulating the histone deacetylase activity of NuRD. Furthermore, we provide evidence linking NuRD function to methylated DNA.
Results
MTA2 and MBD3 are components of the NuRD complex
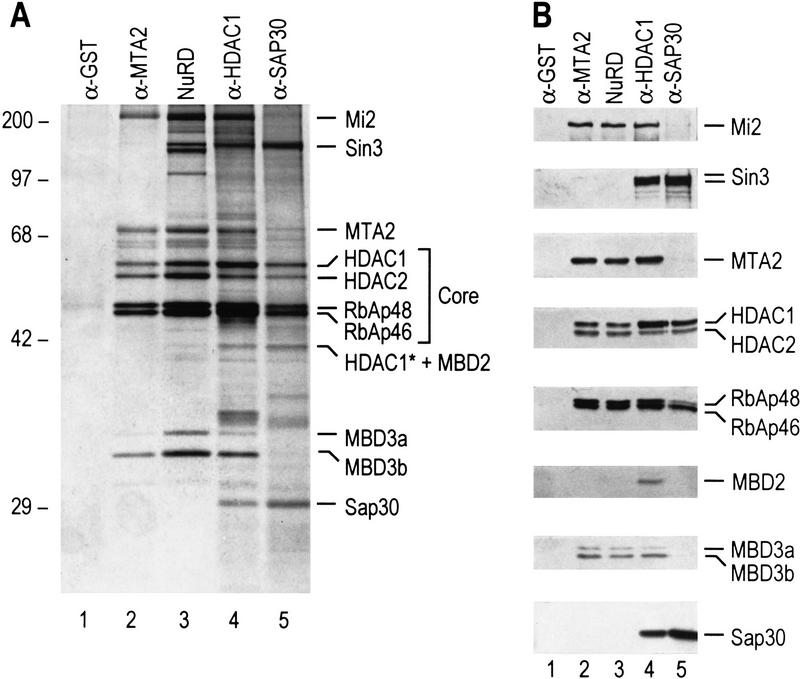
Previously, we reported the purification of NuRD, a multisubunit complex containing both histone deacetylase and nucleosome remodeling activities (Zhang et al. 1998a). Through extensive purification using conventional methods, combined with affinity purification using antibodies against Mi2, the largest subunit of the complex, we determined that this complex is composed of seven subunits, including the SWI2/SNF2 helicase/ATPase domain-containing Mi2 protein, the two histone deacetylases HDAC1 and HDAC2, the two histone-binding proteins RbAp46 and RbAp48, and polypeptides of 70 and 32 kD (Zhang et al. 1998a). The conventionally purified complex also contains minor contaminating polypeptides that did not copurify with NuRD activity (Fig. 1A, lane 3; see also Fig. 3B of Zhang et al. 1998a).
Figure 1.
MTA2 and MBD3 are components of the NuRD complex. (A) Silver staining of an SDS-polyacrylamide gel containing purified NuRD (lane 3) and samples derived from different antibody columns, as indicated at top. The identity of each polypeptide was defined by microsequencing and/or Western blot analysis and is indicated at right. The histone deacetylase core complex present in both the Sin3/SAP30 complex and the NuRD complex is indicated with a bracket and denoted as Core. Peptide sequences derived from the bands labeled as MTA2, MBD3a, and MBD3b are presented in Fig. 2. Sequencing of the band labeled as HDAC1* + MBD2 derived from HDAC1 antibody column suggest the presence of both HDAC1 and MBD2. Peptides specific for HDAC1 and MBD2 are VMTVSFHK and GLQGVGPGSNDETLLSAVASALHTSSAPITGQVSAAVEK, respectively. Mass-spectrometric analysis of the three polypeptides between Mi2 and MTA2 found in the conventionally purified NuRD fraction (lane 3) identified these contaminants as HCAP (GenBank accession no. AF020043), SA-1 (GenBank accession no. Z75330), and SB1/DXS423E (GenBank accession no. S78271). (B) Western blot analysis of the samples used in A. The proteins detected are indicated at right.
We expanded our previous studies by identifying each of the polypeptides present in the conventionally purified fraction of NuRD. Protein sequencing of the doublet migrating around 32 kD revealed that it corresponds to two in-frame spliced forms of MBD3, a member of a protein family containing the methyl-CpG binding domain (Hendrich and Bird 1998). We named the two spliced forms MBD3a and MBD3b. The major form in the NuRD complex is MBD3b, which only contains a portion of the methyl-CpG binding domain (Fig. 2A,B). Interestingly, MBD3 is highly related to MBD2 (80% similar, 72% identical; see Fig. 2A), a protein recently claimed to have DNA demethylase activity (Bhattacharya et al. 1999).
Figure 2.
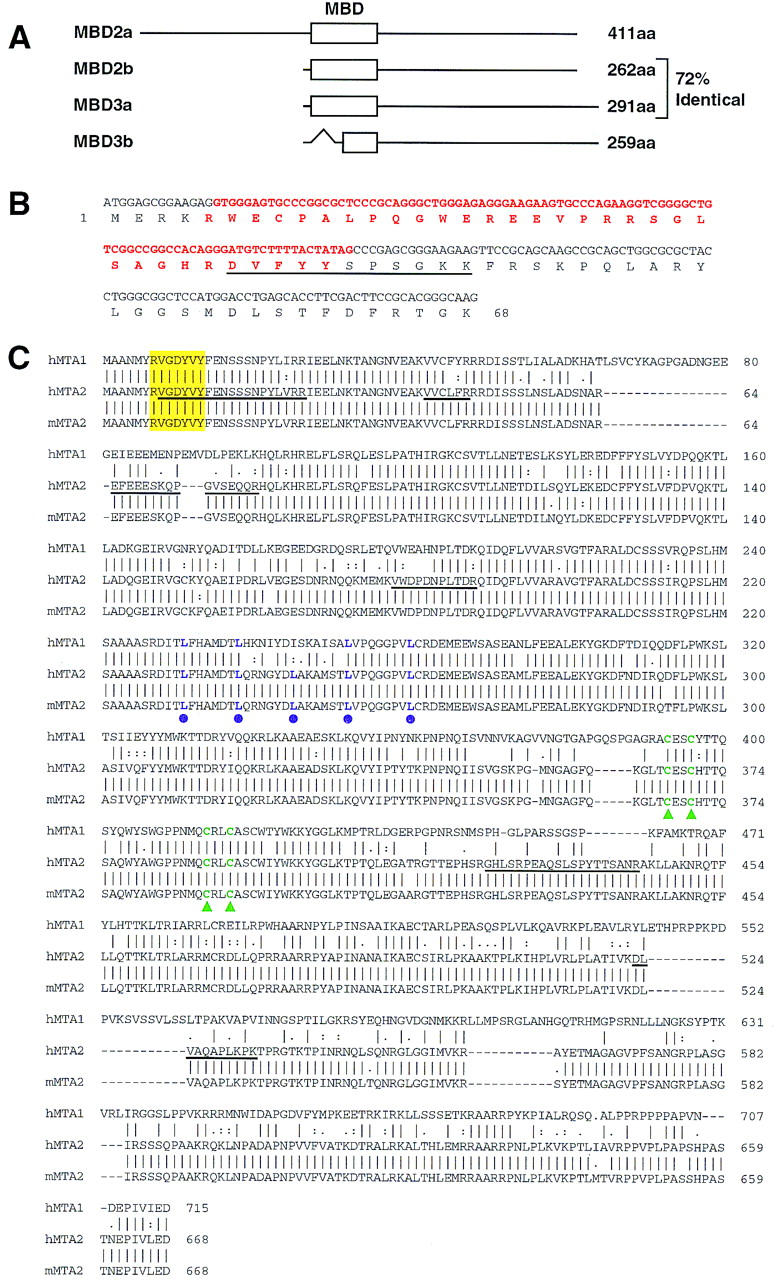
Sequence analysis of MBD3 and MTA2. (A) Schematic representation of the two forms of MBD2 and MBD3. The open box represent the methyl-CpG binding domain (MBD) initially identified in MeCP2 (also see B). The GenBank accession nos. for MBD2 and MBD3 are AF072242 and AF072247, respectively. The in-frame spliced form MBD3b only contains part of the MBD. (B) Sequences and splicing junctions of the MBD3a and MBD3b. The orange nucleotides that encode 5′ portion of MBD are spliced out in MBD3b. The peptide sequence derived from the band labeled as MBD3a in Fig. 1A is underlined. Other peptides derived from the MBD3 bands of Fig. 1A have perfect match to MBD3: KQEELVQQVR, TMDLPK, GKPDLNTALPVR, and NPGVWLNTTQPLCK. (C) MTA2 is related to the metastasis-associated protein MTA1. Amino acid sequence alignment of human MTA2 (GenBank accession no. AB016591), mouse MTA2 (GenBank accession no. AF159259) with human MTA1 (GenBank accession no. U35113). Sequence alignment was performed using Gap of the GCG program (University of Wisconsin, Madison). Peptide sequences obtained from microsequencing of the band labeled MTA2 in Fig. 1A are underlined. The putative zinc-finger is indicated by green triangles. The putative leucine zipper is indicated by purple dots. The yellow box indicates a potential tyrosine kinase phosphorylation site. Human and mouse MTA2 are 98% identical and are 65% and 63% identical to human MTA1, respectively.
Sequencing of the 70-kD polypeptide identified it as a novel protein highly related (65% identical) to a candidate metastasis-associated protein, MTA1. Therefore, we refer to this polypeptide as MTA2 (Fig. 2C). Interestingly, MTA1 was reported to be a component of the NURD complex based on four peptide sequences that happen to be common between MTA1 and MTA2 (Xue et al. 1998). Analysis of the amino acid sequences of MTA proteins identified one putative zinc-finger domain, one leucine zipper domain, and a potential tyrosine kinase phosphorylation site (Fig. 2C). The expression level of MTA1 was found to be elevated in metastatic breast cancer cell lines and metastatic cancer tissues, such as colorectal, gastric, and esophageal carcinomas (Toh et al. 1994, 1997, 1999). Similarly, we also found that the MTA2 expression level is elevated in cervical cancer tissue (data not shown). However, the cause-and-effect relationship between cancer metastasis and overexpression of MTA proteins is not known.
To further establish that MTA2 and MBD3 are integral components of the NuRD complex, antibodies against these polypeptides were produced. Western blot analysis using MTA2 antibodies demonstrated that MTA2 copurified with the NuRD complex and its associated nucleosome remodeling and histone deacetylase activities (Zhang et al. 1998a). Affinity purification using anti-MTA2 antibodies resulted in the isolation of a protein complex composed of seven polypeptides, all present as major polypeptides in the conventionally purified NuRD complex (Fig. 1A, cf. lanes 2 and 3). The identity of the polypeptides present in the anti-MTA2 affinity purified complex was verified by Western blot analyses as shown in Fig. 1B (lane 2). The other polypeptides present in the conventionally purified NuRD complex (lane 3) were absent in the complex isolated through affinity chromatography using anti-Mi2 (Zhang et al. 1998a) or anti-MTA2 (Fig. 1A, lane 2) antibodies. Moreover, these polypeptides did not coelute with subunits of the NuRD complex through conventional chromatography (see Fig. 3B of Zhang et al. 1998a). Thus, we conclude that these polypeptides are contaminants.
Antibodies against MBD3 were capable of immunoprecipitating recombinant MBD3 protein, however, these antibodies failed to immunoprecipitate the NuRD complex (data not shown), suggesting that MBD3 is not accessible in the NuRD complex (see below).
Immunopurification experiments using antibodies against HDAC1, which should result in the isolation of most of the polypeptides associated with HDAC1, resulted in a complex pattern of polypeptides (Fig. 1A, lane 4). Comparison of the polypeptides present in this fraction with those present in the NuRD and Sin3 complexes established that most polypeptides in the anti-HDAC1 purified sample were present in the NuRD or Sin3 complex (Fig. 1A,B). Interestingly, the reported demethylase MBD2 was also present in the anti-HDAC1 purified sample (Fig. 1A,B, lanes 4; see below). In addition, we found that four polypeptides (HDAC1, HDAC2, RbAp48, RbAp46) were common to the Sin3 and NuRD complexes (Fig. 1A), whereas the other polypeptides (Sin3, SAP30, Mi2, MTA2, MBD3) were specific to one of the two complexes (Fig. 1A,B, lanes 2,5).
The studies described above establish that both MTA2 and MBD3 are integral components of the NuRD complex and suggest the existence of a shared core histone deacetylase complex composed of HDAC1, HDAC2, RbAp48, and RbAp46. These results also suggest a possible connection between the NuRD complex and methyl-CpG binding proteins.
MTA2 promotes the assembly of a catalytically active histone deacetylase complex
Having established that HDAC1, HDAC2, RbAp48, and RbAp46 are shared components of the Sin3 and NuRD complexes, we asked whether these four polypeptides can form a core protein complex. We began the studies by investigating whether a histone deacetylase core complex could be isolated from HeLa cells. These studies uncovered different complexes containing these four polypeptides (data not shown). However, we were unable to isolate a complex containing only these four polypeptides. A possible explanation is that the putative HDAC/RbAp core complex is limiting with respect to polypeptides that may associate with it.
We next asked whether an HDAC/RbAp core complex could be reconstituted from recombinant polypeptides. In agreement with our previous studies, which demonstrated that the HDAC and RbAp association requires cotranslation and/or the presence of a molecular chaperone (Zhang et al. 1998b), we failed to reconstitute the core complex using individually purified HDAC1 and HDAC2 from baculovirus-infected SF9 cells and RbAp46 and RbAp48 purified from Escherichia coli (data not shown). Thus, we performed coinfection experiments using recombinant baculoviruses expressing each of the four putative core subunits with Flag-tagged HDAC1. HDAC1 and its associated polypeptides were purified through multiple chromatographic steps, including ion exchange, immunoaffinity, gel filtration, and glycerol gradient sedimentation (Fig. 3A). Western blot analysis demonstrated cofractionation of the four polypeptides during each of the purification steps (data not shown). Silver staining of the sample derived from the last purification step revealed the presence of four major polypeptides (Fig. 3B). The identity of these polypeptides was confirmed by Western blot analysis (Fig. 3C). The contaminating protein of about 70 kD (Fig. 3B) does not react with MTA2 antibodies (Fig. 3C). These results suggest that a complex containing HDAC and RbAp polypeptides can be formed. However, the histone deacetylase activity of this core complex was severely compromised compared to that of native NuRD complex when equal Western blot units of HDACs or RbAps were used (Table 1; data not shown).
Figure 3.
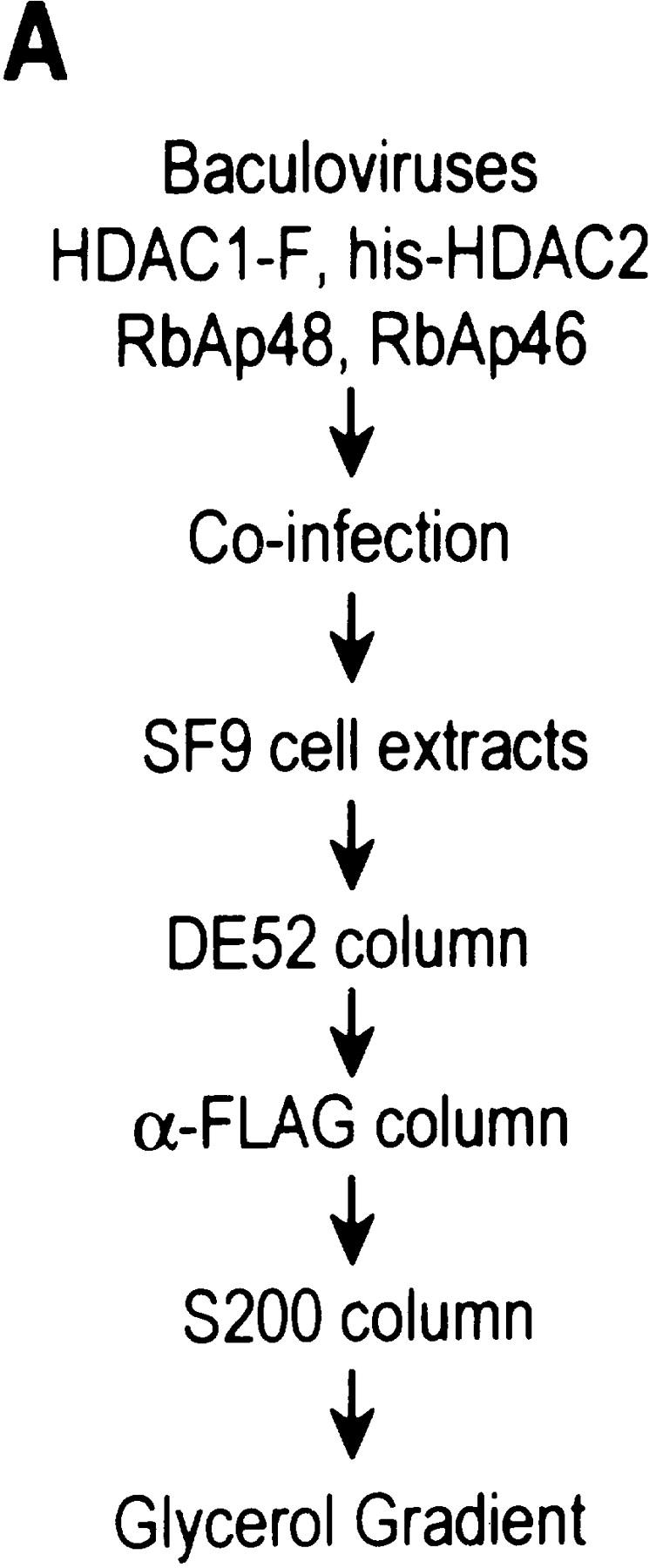
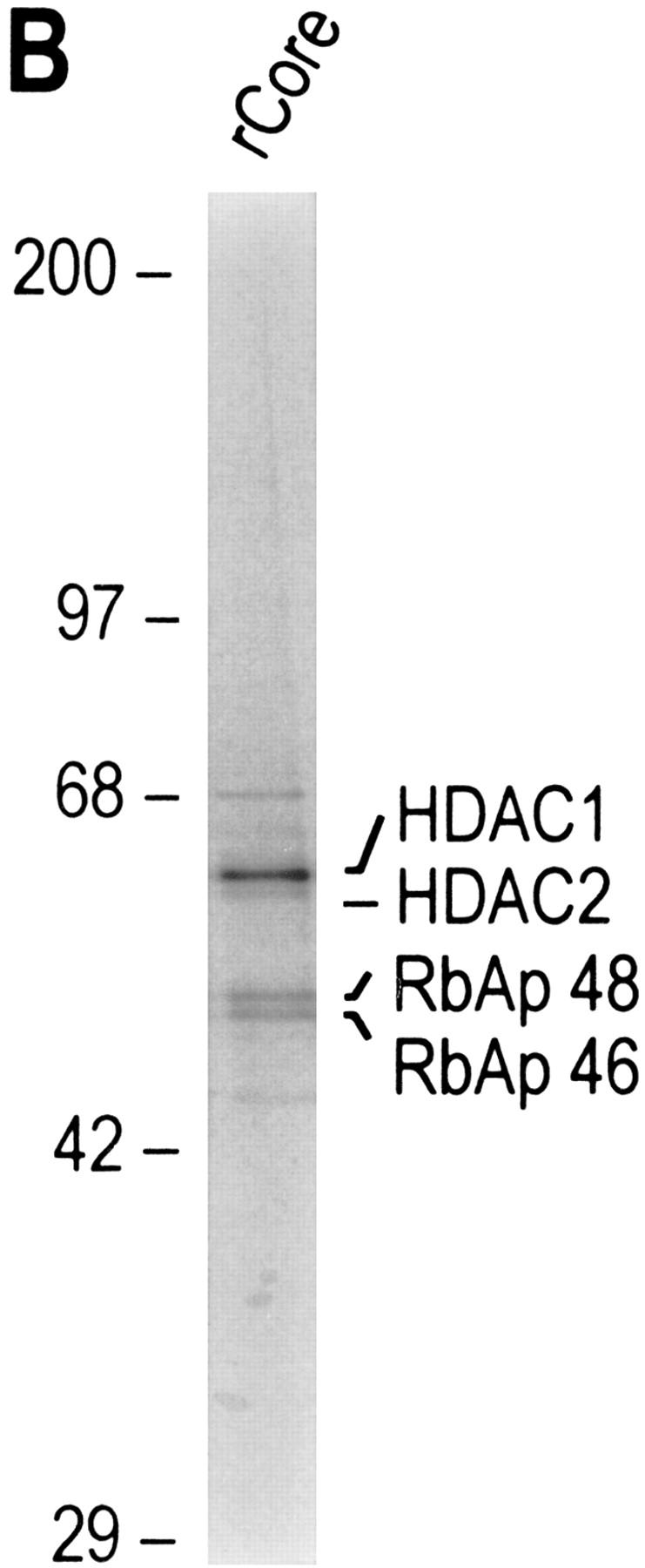
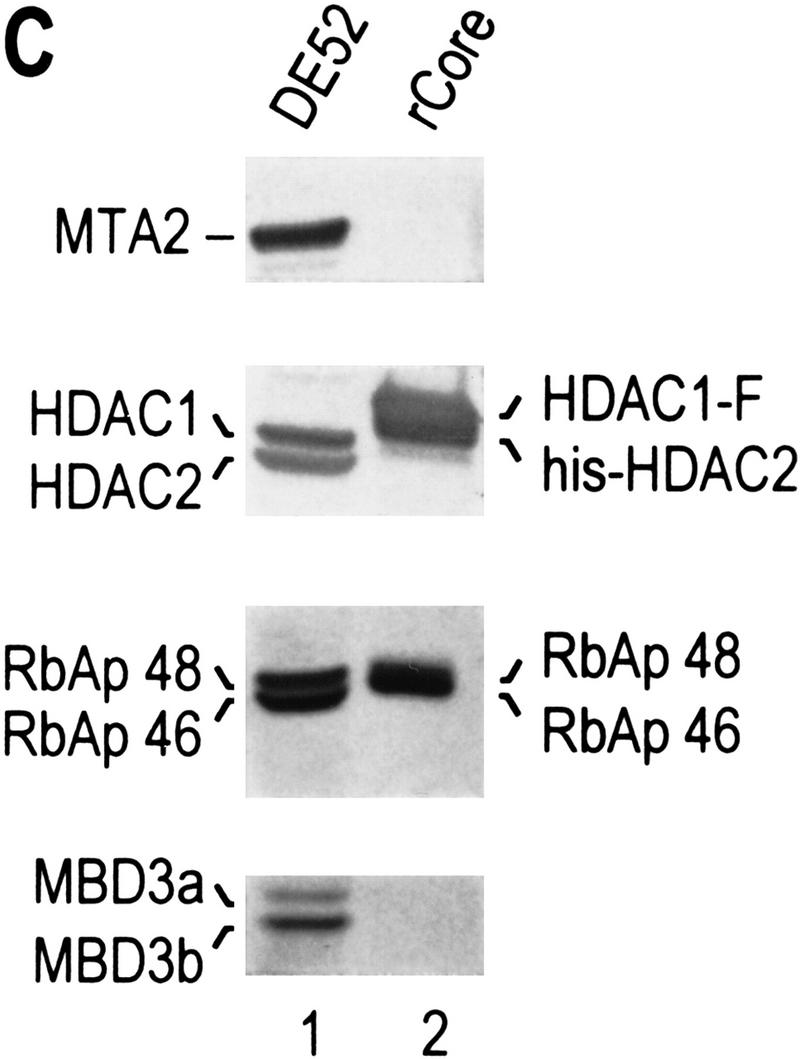
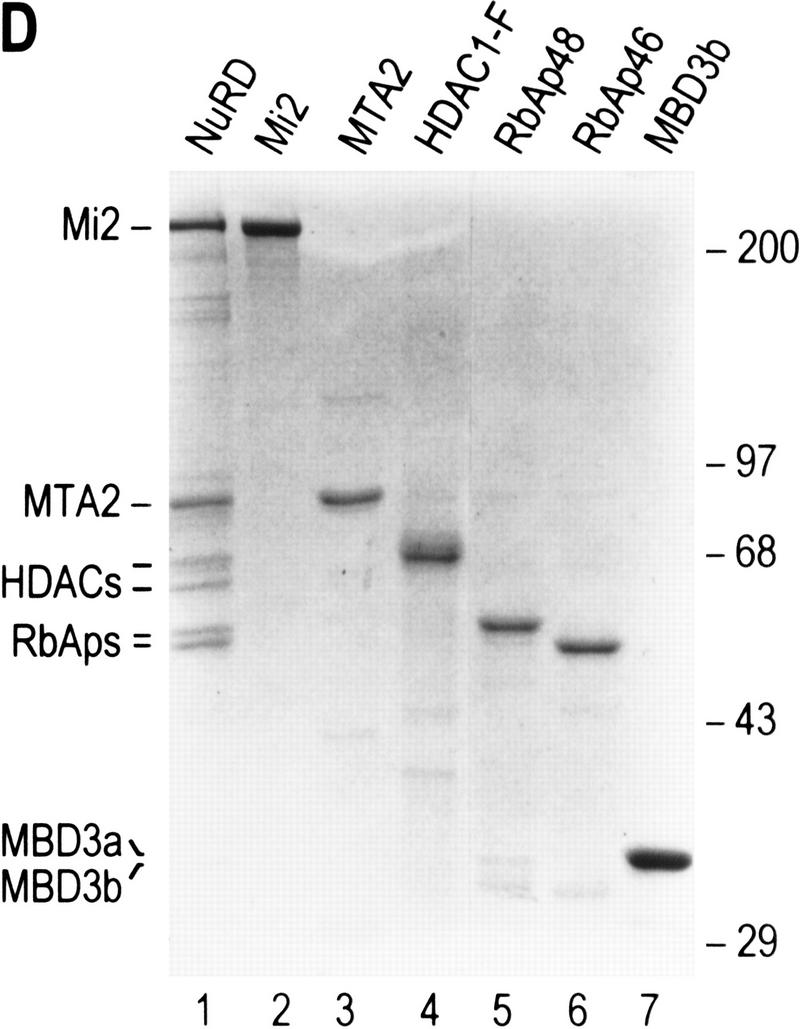
Purification of a recombinant histone deacetylase core complex. (A) Schematic representation of the steps used to purify the histone deacetylase core complex. (B) Silver staining of an SDS-polyacrylamide gel containing the purified core complex. The identities of the major polypeptides and protein size markers are indicated. (C) Western blot analysis of a partially purified HeLa nuclear extracts (lane 1) and the purified core complex (lane 2). The identities of the polypeptides are indicated. (D) Coomassie blue staining of an SDS-polyacrylamide gel containing the purified NuRD complex (lane 1) and individual recombinant NuRD components. Mi2, HDAC1, and MBD3b are purified from baculovirus-infected SF9 cells. MTA2 and RbAps were produced in Escherichia coli. Protein size makers are indicated.
Table 1.
Effect of NuRD subunits on histone deacetylase activity
| Complex/protein
|
3H-labeled acetate released (cpm)
|
|---|---|
| — | 32 ± 6 |
| NuRD | 6176 ± 213 |
| Core (SF9) | 301 ± 32 |
| Core (SF9) + MTA2 (E. coli) | 357 ± 17 |
| Core (SF9) + MBD3 (SF9) | 308 ± 19 |
| Core (SF9) + Mi2 (SF9) | 324 ± 7 |
| Core (SF9) + MTA2 (SF9) | 2466 ± 250 |
| MTA2 (SF9) | 1604 ± 106 |
Different subunits of the NuRD complex were analyzed for histone deacetylase activity and for their ability to regulate the histone deacetylase activity of the HDAC/RbAp core complex. The activity was compared to that of the NuRD complex using equal Western blot units of HDACs. Assays were performed as described in Materials and Methods. The numbers shown represent an average of at least three independent assays.
We next analyzed whether the addition of the other NuRD subunits (Mi2, MTA2, and MBD3) to the core complex could restore the activity to the level of the native NuRD complex. The addition of highly purified (Fig. 3D) recombinant Mi2 and MBD3b (or MBD3a, data not shown) purified from baculovirus infected-SF9 cells, or MTA2 produced in Escherichia coli, independently or in combination, did not affect the histone deacetylase activity (Table 1, and data not shown). In light of these negative results and to determine the identity of the subunits affecting enzymatic activity, we investigated the polypeptides of the NuRD complex that interact with subunits of the core HDAC/RbAp complex.
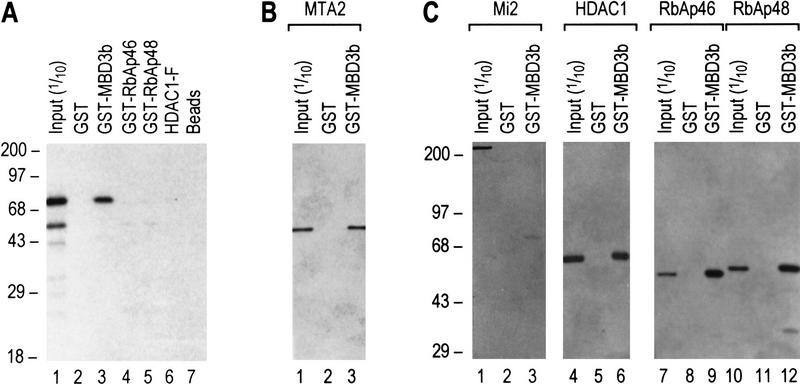
We began the study by asking whether subunits of the putative core complex, as GST-fusion or Flag-tagged proteins, could interact with MTA2 that was translated in vitro using the rabbit reticulolysate system. We found that none of the core subunits interacted with MTA2 in this assay (Fig. 4A). However, we observed an interaction between GST–MBD3b and MTA2 under the same conditions (Fig. 4A). We extended this finding by demonstrating a direct interaction using GST–MBD3b and MTA2 produced in Escherichia coli (Fig. 4B). We next analyzed whether MBD3b interacts with components of the core complex. This analysis demonstrated that MBD3b is able to interact with highly purified (Fig. 3D) HDAC1, RbAp48, and RbAp46 in a GST pull-down assay (Fig. 4C, lanes 6,9,12). These interactions appear to be specific because MBD3b failed to interact with Mi2 under the same conditions (Fig. 4C, lane 3). The finding that MBD3 engages in multiple interactions with subunits of the core complex is consistent with the result demonstrating that antibodies against MBD3 could immunoprecipitate recombinant MBD3, but failed to immunoprecipitate the MBD3-containing NuRD complex, as discussed above. Collectively, these findings suggest that MBD3 is embedded within the NuRD complex.
Figure 4.
MBD3 interacts with MTA2 and components of the histone deacetylase core complex. (A) GST pull-down assays show that in vitro-translated MTA2 only interacts with MBD3. Equal amounts (5 μl) of in vitro-translated and labeled MTA2 was incubated with 10 μl of glutathione–agarose beads or anti-Flag beads coated with 2 μg of proteins as indicated at the top. After extensive wash, bound proteins were eluted, resolved by SDS-PAGE, and visualized by autoradiography. Ten percent of input was loaded on lane 1. (B) MTA2 directly interacts with MBD3. An experimental procedure similar to A was used except that MTA2 was purified from Escherichia coli (Fig. 3D, lane 3). Bound proteins were eluted, resolved by SDS-PAGE, and visualized by Western blot. (C) MBD3 interacts with components of the core complex in a GST pull-down assay. Assays were performed as in B. Input proteins are indicated at top and are the same as those used in Fig. 3D.
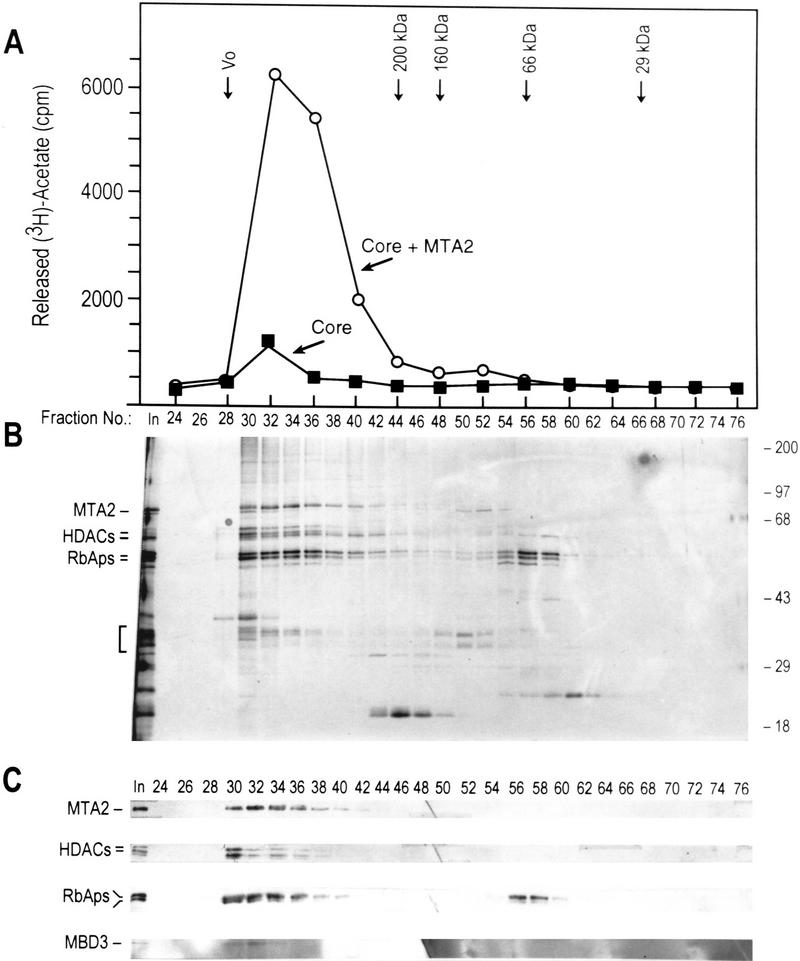
To verify these in vitro protein–protein interaction studies, we coinfected SF9 cells with five baculoviruses each expressing one of the four core subunits and MTA2. The complex was purified as described above (Fig. 3A). In light of the protein–protein interaction results described above, we expected that during affinity purification and gel-filtration chromatography the four subunits of the core complex would copurify, but would be separated from MTA2. However, to our surprise we observed copurification of MTA2 with the core HDAC/RbAp complex during affinity purification and gel-filtration chromatography (Fig. 5; data not shown). More importantly, we found that this complex is active in deacetylating core histones (Fig. 5A; Table 1). The activity was similar (within 2.5-fold) to that of the native NuRD complex (Table 1). Our interpretation of these results is that either the association of MTA2 with the core complex requires cotranslation or, alternatively, that mammalian MTA2 can associate with the endogenous insect MBD3 that mediates the association between MTA2 and core. Silver staining (Fig. 5B) and Western blot (Fig. 5C) analyses of the fractions derived from the gel-filtration column indicate the presence of MTA2 and the HDAC/RbAp core polypeptides coeluting with HDAC activity. Moreover, the silver-staining analysis revealed polypeptides in the 30-kD range coeluting with histone deacetylase activity (Fig. 5B). Western blot analysis revealed that antibodies directed against the human MBD3 protein reacted, although weakly, with a polypeptide of ∼30 kD that coeluted with the histone deacetylase activity. We concluded that the MBD3-immunoreactive 30-kD polypeptide is likely the SF9 cell-derived MBD3 (Fig. 5C). The silver-staining analysis demonstrates that the complex is highly pure and suggests that the coelution of HDACs, RbAps, MTA2, and insect-derived MBD3 is a functional association and not the result of protein aggregation.
Figure 5.
MTA2 is required for the formation of a functional histone deacetylase complex. (A) Histone deacetylase activities of the fractions derived from gel filtration S200 columns. Histone deacetylase core complex as well as core plus MTA2 were purified using the procedure described in Fig. 3A (see Materials and Methods for details). In the histone deacetylation assays using core- and core + MTA2-derived fractions approximately equal Western blot units of HDACs were used. The elution profiles of different size markers are indicated. (B) Silver staining of an SDS-polyacrylamide gel containing the core + MTA2 fractions derived from the S200 gel filtration column analyzed in A. The identities of the polypeptides are indicated and were confirmed by Western blot analysis which is shown in C. (C) Western blot analysis of the fractions shown in B.
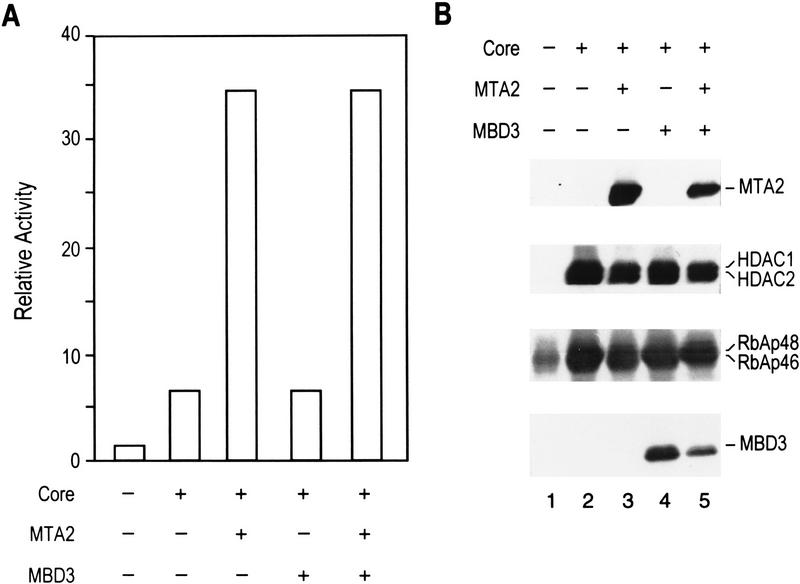
To further analyze the specificity of MTA2 in directing the formation of an enzymatically active histone deacetylase complex, we performed coinfections as above, but coinfected a baculovirus-expressing MBD3 with components of the HDAC core complex in the presence and absence of MTA2. Following affinity purification, the purified complexes were divided and analyzed for histone deacetylase activity (Fig. 6A) and protein composition by western blots (Fig. 6B). As above, the core complex was severely compromised in its enzymatic activity (Fig. 6A). The presence of MBD3 was without effect. However, coinfection with MTA2 in the presence or absence of MBD3 resulted in the recovery of a complex that was enzymatically active (Fig. 6A). Consistent with our finding that MTA2 can direct the formation of an enzymatically active histone deacetylase complex, we found that infection of SF9 cells with a recombinant baculovirus-expressing Flag-tagged MTA2 followed by purification on a column containing antibodies against the Flag tag resulted in the isolation of an MTA2-containing complex that was enzymatically active (Table 1). Silver staining and Western blot analysis revealed that in addition to MTA2, polypeptides corresponding to the insect HDAC and RbAp were present in the affinity-purified sample (data not shown). Collectively, these studies demonstrate that MTA2 directs the assembly of an active histone deacetylase complex and that the association of MTA2 with the core HDAC/RbAp complex requires MBD3.
Figure 6.
MTA2 directs the formation of an enzymatically active histone deacetylase complex. (A) Relative histone deacetylase activities of different coinfections. SF9 cells were infected with different combinations of viruses. The complexes were affinity purified through Flag-tagged HDAC1 using antibodies against Flag. The purified complexes were divided for deacetylase assay shown in A and Western blot analysis shown in B. (+) Presence of the viruses in the coinfection. (B) Western blot analysis of the samples used in histone deacetylation assays shown in A. Antibodies against RbAps cross-reacted with bleached heavy chain in lane 1.
The NuRD complex interacts with MBD2 and is tethered to methylated DNA
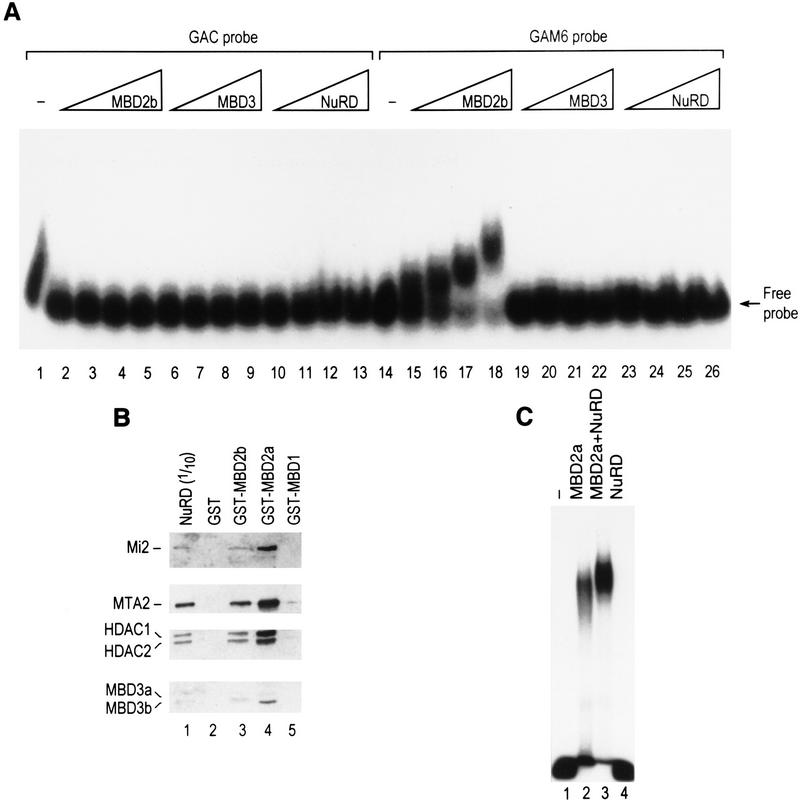
The finding that MBD3 is a component of the NuRD complex suggests a possible connection between the NuRD complex and methylated DNA. Therefore, we investigated whether the NuRD complex could specifically bind to methyl-CpG-containing DNA using a gel mobility-shift assay. We found that NuRD and MBD3 failed to bind to DNA. However, under the same conditions, MBD2 bound specifically to methylated DNA (Fig. 7A, cf. lanes 2–6 with 15–18). This is in agreement with previous studies (Hendrich and Bird 1998). Moreover, because MBD3 is likely embedded in the NuRD complex, and the major form of MBD3 present in the complex is MBD3b, which only contains part of the methyl-CpG-binding domain (Fig. 2A), it is not surprising that NuRD failed to directly bind to methylated DNA.
Figure 7.
The NuRD complex can be targeted to methylated DNA by MBD2. (A) Gel mobility shift assay shown that the NuRD complex does not bind to methylated DNA directly. The GAM6 probe contains six methyl-CpG sites and was previously described (Nan et al. 1993). Binding reactions were resolved on a 2% agarose gel as described in Materials and Methods. (B) GST pull-down assays demonstrates an interaction between MBD2 and the NuRD complex. The assays were performed as described in Fig. 4B using different GST fusion proteins as indicated at the top. (C) Gel mobility-shift assay shown that the NuRD complex is able to super-shift the DNA–MBD2 binary complex (cf. lanes 2 and 3). The probe used is MeCG11, which contains 27 methyl-CpG sites and was described previously (Ng et al. 1999). Binding reactions were resolved on a 1% agarose gel as described in Materials and Methods.
Because MBD3 is highly related to MBD2 and we found that MBD3 is able to interact with each component of the histone deacetylase core complex (Fig. 4C), we investigated whether MBD2 associates with histone deacetylase complexes. Immunoprecipitation experiments using antibodies against HDAC1 resulted in the immunoprecipitation of MBD2 (Fig. 1B, lane 4). However, as described above, MBD2 is absent from the NuRD complex isolated through conventional (Fig. 1B, lane 3) or affinity (Fig. 1B, lane 2) purification procedures. Nonetheless, the similarity between MBD2 and MBD3 proteins prompted us to analyze whether MBD2 could physically interact with the NuRD complex. In this experiment, full-length and a truncated form of MBD2 (Hendrich and Bird 1998) were analyzed. The repression domain of MBD1 (H-H. Ng and A. Bird, unpubl.), a protein related to MBD2 and MBD3 through the methyl-CpG DNA-binding domain (Hendrich and Bird 1998), was used as a control. Different GST–MBD fusion proteins were independently incubated with purified NuRD complex and possible interactions were analyzed by GST pull-down assays followed by Western blot analyses using antibodies against different components of the NuRD complex. Whereas GST–MBD1 failed to interact with the NuRD complex, both forms of MBD2 interacted with the NuRD complex, and the full-length form of MBD2 interacted more efficiently (Fig. 7B). Thus, we conclude that MBD2 is able to interact directly with components of the NuRD complex.
The interaction between MBD2 and the NuRD complex prompted us to analyze whether MBD2 could tether the NuRD complex to methylated DNA. We analyzed this possibility using the gel mobility-shift assay with methylated DNA as the probe. Under these conditions, and in agreement with the studies presented above, NuRD failed to bind to DNA, but MBD2 bound to the methylated DNA specifically (Fig. 7C; data not shown). Importantly, the addition of NuRD to a DNA-binding assay containing MBD2 resulted in the production of a new DNA–protein complex that migrates slower than the MBD2–DNA protein complex. Additionally the amount of DNA in the NuRD–MBD2 ternary complex was greater than the amount of DNA in the binary MBD2-containing complex. The observed supershift is specific because addition of 3 μg of BSA (10 × the amount of NuRD used) or other proteins was without affect (data not shown). Therefore, we conclude that NuRD is tethered to methylated DNA via MBD2 and that the interaction of NuRD with MBD2 stabilizes the MBD2–DNA protein complex.
Discussion
In this study we have identified MTA2 and MBD3 as two subunits of the NuRD complex. MTA2 is related to the metastasis-associated protein MTA1, whereas MBD3 is related to MBD2, a methyl-CpG-binding domain-containing protein recently claimed to possess demethylase activity. In addition, we provide evidence that MTA2, through an interaction with MBD3, has an important role in modulating the enzymatic activity of the histone deacetylase core complex composed of HDAC1 and HDAC2 and RbAp48 and RbAp46. Furthermore, we demonstrate that the methyl-CpG binding protein MBD2 is able to tether the NuRD complex to CpG-methylated DNA.
The connection between NuRD and cellular proliferation
The nucleosome remodeling and histone deacetylase NuRD complex contains seven subunits. In addition to the histone deacetylase core HDAC1, HDAC2, RbAp46, and RbAp48, it contains Mi2, MTA2, and MBD3. It is interesting to note that MTA2 is 65% identical to the metastasis-associated protein MTA1, whose expression level was found to be elevated in different types of cancer tissue as well as metastatic breast cancer cell lines (Toh et al. 1994, 1997, 1999). Similarly, we found that MTA2 is highly expressed in rapidly dividing cells at both the RNA and protein level (data not shown) indicating a correlation between MTA2 expression and cellular proliferation. Another component that links the NuRD complex to cellular transformation is MBD3. This protein was initially identified because it contains a domain related to the methyl-CpG-binding domain of MeCP2 (Hendrich and Bird 1998). However, in agreement with a previous report (Hendrich and Bird 1998), MBD3 is not able to bind to methyl-CpG specifically (Fig. 7A). Moreover, two alternately spliced forms of MBD3 were identified. The predominant form present in the NuRD complex is a spliced variant deleted of the methylated CpG-binding domain. Interestingly, MBD3 is highly related to MBD2, which was also identified as a colon cancer antigen (Scanlan et al. 1998). The fact that two components of the NuRD complex are linked to malignancy, in conjunction with the fact that patients with dermatomyositis, who produce antibodies against Mi2 (Seelig et al. 1996), have an increased risk to malignancy (Airio et al. 1995), suggests a link between the NuRD complex and malignancy. Indeed the accumulative evidence strongly suggest that histone deacetylases, in particular HDAC1 and HDAC2, are associated with polypeptides that regulate cellular proliferation. The first mammalian HDAC complex to be described was Sin3, which associates with the Max–Mad heterodimer required for cellular differentiation (for review, see Amati and Land 1994; Pazin and Kadonaga 1997). Additionally, Sin3 associates with the corepressor NcoR/SMRT, which negatively regulates transcription of genes targeted by nuclear hormone receptors and has been linked with acute promyelocytic leukemia (Grignani et al. 1998; Lin et al. 1998). Furthermore, HDAC1 was found to be associated with the tumor suppressor Rb protein (Brehm et al. 1998; Luo et al. 1998; Magnaghi-Jaulin et al. 1998). Moreover, our recent studies have uncovered that SAP30, a component of the Sin3 complex (Laherty et al. 1998; Zhang et al. 1998b), associates with the Rb-binding protein RBP1 as well as with the p53-binding protein p33ING1 (Y. Zhang and D. Reinberg unpubl.).
NuRD is targeted to methylated DNA
Since the discovery of the first nucleosome remodeling factor, the SWI/SNF complex, about a dozen different chromatin remodeling factors, from different organisms, have been described (for review, see Bjorklund et al. 1999; Travers 1999). However, it is not known whether these remodeling factors function on a genome-wide basis or whether they are targeted to specific genes. Genetic studies indicate that transcription of only a fraction of genes is affected by the SWI/SNF complex (Holstege et al. 1998), but until recently it was not clear how the SWI/SNF complex is recruited to promoters of SWI/SNF-regulated genes. The first evidence suggesting that the SWI/SNF complex can be targeted to specific genes came from studies on the human β-globin promoter. It was demonstrated that a SWI/SNF-related chromatin remodeling factor E-RC1 is required for the erythroid Krüppel-like factor, ELKF, to activate transcription in vitro using a chromatin-assembled template (Armstrong et al. 1998). Recently, Nasmyth and coworkers demonstrated that the SWI/SNF complex is recruited to the HO promoter by SWI5 in vivo in a cell cycle-dependent manner (Cosma et al. 1999). Therefore, targeted nucleosome remodeling is at least one mechanism by which nucleosome remodeling complexes can function to activate transcription. Similarly, histone deacetylase complexes may also be targeted to specific promoters. For example, the histone deacetylase HDAC2 interacts with the transcription factor YY1, and thus could be recruited to promoters containing YY1-binding sites (Yang et al. 1996). Moreover, the Sin3/HDAC complex can be recruited by Mad–Max, Ume6, and unliganded nuclear hormone receptors to specific promoters (for review, see Pazin and Kadonaga 1997). Additionally, the Sin3–histone deacetylase complex can also be recruited via the methyl-CpG binding protein MeCP2 to methylated regions of the genome (Jones et al. 1998; Nan et al. 1998). Therefore, targeted deacetylation represents at least one mechanism by which histone deacetylase complexes can be recruited.
Previously, we and others have shown that the NuRD complex has both nucleosome-remodeling and histone deacetylase activities (Tong et al. 1998; Xue et al. 1998; Zhang et al. 1998a). It was also shown that recruitment of the NuRD complex to a promoter results in transcription repression (Kehle et al. 1998; Xue et al. 1998). We proposed that the NuRD complex is targeted to specific genes by transcription factors (Zhang et al. 1998a).The NuRD-subunit MTA2 contains a zinc-finger (CX2CX17CX2C) belonging to the type found in transcription factors that bind to the GATA sequence involved in hematopoiesis and heart development (Orkin 1992; Lyons 1996). These observations prompted us to analyze whether MTA2 or the NuRD complex could bind to the human β-globin gene promoter using the gel mobility-shift assay. This assay failed to detect any DNA binding activity (data not shown). This negative result can be explained because several amino acids involved in GATA sequence recognition are not conserved in the MTA2 zinc-finger domain (Omichinski et al. 1993).
Similar to previous studies demonstrating that Mad, a protein that interacts with the Sin3–histone deacetylase corepressor complex, but is not an integral component of the complex, can recruit the corepressor complex to specific promoters (for review, see Pazin and Kadonaga 1997), we found that the methyl-CpG binding protein MBD2, although not an integral component of the NuRD complex (Fig. 1B), has the ability to directly interact with the NuRD complex and recruit it to methylated DNA (Fig. 7). The implication of this result is that the NuRD complex may play a role in methylation-associated gene silencing. In support of this hypothesis are recent findings demonstrating that artificial recruitment of MBD2 to promoters results in repression of transcription, which was reversed by the histone deacetylase inhibitor Trichostatin A (Ng et al. 1999). All of the evidence discussed above suggests that MBD2 functions as a transcriptional repressor. However, it is worth noting that MBD2 was recently reported to have demethylase activity (Bhattacharya et al. 1999). Because DNA methylation causes gene silencing, a demethylase is likely to function as a transcriptional activator. It remains to be determined how MBD2 activity can be converted from a repressor to an activator. Given the similarity between MBD2 and MBD3 (72% identical), we tested the recombinant MBD3 protein (produced in Escherichia coli or SF9 cells) and the native NuRD complex for demethylase activity and failed to detect such an activity (data not shown).
Can NuRD also be recruited to promoters by gene-specific DNA-binding protein? It has been shown that the Drosophila gap gene hunchback can interact with Drosophila Mi2 and potentially recruit the NuRD complex to repress hunchback-regulated HOX genes (Kehle et al. 1998). Recently, it was also reported that Ikaros, a zinc finger DNA-binding protein essential for lymphocyte lineage determination, through interaction with Mi2, recruits the NuRD complex to heterochromatin regions (Kim et al. 1999). It is therefore likely that DNA-binding proteins, which in principle have a restricted specificity as compared to MBD2, can also target the NuRD complex to specific genes. The interplay between sequence-specific DNA-binding proteins and MBD2 in recruiting the NuRD complex to specific genes may represent another pathway for regulation of the NuRD complex.
The function of the NuRD complex
The NuRD complex was purified based on its histone deacetylase and nucleosome remodeling activities (Tong et al. 1998; Xue et al. 1998; Zhang et al. 1998a). The biochemical association of the histone deacetylase and nucleosome remodeling activities suggest that these two enzymatic activities are functionally related. It is conceivable that nucleosome remodeling may be required for nucleosomal histone deacetylation in vivo. Therefore, the presence of these two activities in one protein complex may represent an efficient way to facilitate dynamic changes in nucleosome structure. Based on the finding that a similar protein complex is also present in Xenopus eggs (Wade et al. 1998), we believe that the function of the NuRD complex is rather general. Several reports have already shed light on its potential biological function. Studies in Drosophila implicated NuRD in repression of homeotic (HOX) genes and Polycomb-group genes (Kehle et al. 1998). In addition, NuRD has been shown to be associated with the zinc-finger DNA-binding protein Ikaros in toroidal structures presumed to be associated with centromeric heterochromatin in the G1 and S phase of the T-lymphocyte cell cycle (Brown et al. 1997; Kim et al. 1999). This suggests that NuRD is involved in centromeric silencing, consistent with the requirement of histone deacetylase activity for the maintenance of the underacetylated state of centromeric histones (Ekwall et al. 1997). It is interesting to note that Ikaros is required for the differentiation of all three lymphoid lineage (Georgopoulos et al. 1994). Because Ikaros can recruit NuRD, it is possible that NuRD may play an important role in lymphocyte development. We provide evidence indicating that NuRD is potentially involved in transcriptional repression of methyl-CpG through an interaction with the methyl-CpG-binding protein MBD2. Therefore, the two best-characterized histone deacetylase complexes, Sin3 and NuRD, are both involved in transcriptional repression of methylated DNA. MBD2 was recently shown to be a component of the MeCP1 complex (Meehan et al. 1989; Ng et al. 1999). Because MeCP2 binds methylated DNA much tighter than MeCP1 (Meehan et al. 1989; Lewis et al. 1992), it is possible that the Sin3/HDAC complex may be involved in long-term silencing of methylated DNA sequences, whereas the NuRD complex may be involved in transient silencing of some methylated genes.
To maintain epigenetic silencing of genes through multiple cell divisions, newly deposited histones that are acetylated in the cytoplasm must be deacetylated (Jeppesen 1997). The identity of the histone deacetylase complex responsible for this function is unknown. It is intriguing that Ikaros forms a higher-order toroidal structure with NuRD that colocalizes with components of the DNA replication machinery upon T-cell activation (Avitahl et al. 1999; Kim et al. 1999). Therefore, the nucleosome-remodeling and histone deacetylase activities of the NuRD complex are ideal candidates for deacetylating the deposition-related acetylated lysine residues during chromatin maturation.
Materials and methods
Purification of the NuRD complex and cloning of MTA2 and MBD3
The procedure for conventional purification of the NuRD complex has been described (Zhang et al. 1998a). The purified NuRD complex derived from Mono S column was concentrated using a centricon concentrator and was resolved on a 10% SDS-polyacrylamide gel. After Coomassie blue staining, protein bands were excised and subjected to in-gel tryptic digestion. The previously identified polypeptides were verified by mass-spectrometric analysis (Erdjument-Bromage et al. 1998; Geromanos et al. 1998). Protein bands corresponding to p70 and p32 were sequenced as described (Zhang et al. 1997). The peptide sequences obtained were used to search the EST database. Multiple EST clones were identified and were used to construct the full-length cDNA clones encoding p70 and p32. Clones encoding alternative spliced forms of p32 (MBD3a and MBD3b) were identified in human and mouse by sequencing EST clones (see text). Affinity purification using anti-MTA2, anti-HDAC1, and anti-SAP30 was based on a previously published procedure (Zhang et al. 1998b) using HeLa nuclear extracts fractionated on phosphocellulose and DEAE-52 columns.
Baculoviruses, recombinant core complex, and antibodies
Baculoviruses expressing RbAp46, RbAp48, and HDAC1 have been described (Hassig et al. 1997; Verreault et al. 1998). Baculovirus-expressing HDAC2 was constructed using the pFastBac HTb vector (GIBCO BRL) and was a gift from Dr. Ed Seto (Moffitt Cancer Center, Tampa, FL). cDNAs encoding His-tagged Mi2 and MBD3 and Flag-tagged MTA2 were constructed using the pVL1392 vector and the recombinant viruses were generated using BaculoGold DNA (Pharmigen). Histone deacetylase core complex and core plus MTA2 were purified using the procedure shown in Figure 3A. Extracts derived from SF9-infected cells (11 grams) were loaded onto a 35-ml DEAE–cellulose column and were eluted with six column volumes using a linear gradient of buffer C (20 mm Tris-HCl at pH 7.9, 0.2 mm EDTA, 10 mm β-ME, 0.2 mm PMSF, 10% glycerol) from 50 mm to 400 mm of KCl (BC50–BC400). The fractions containing the peak of the recombinant core complex (160–269 mm KCl) were pooled and incubated with 0.5 ml of anti-Flag M2 affinity gel (Eastman Kodak) at 4°C for 2 hr. Proteins were eluted with 0.5 ml of BC400 containing 0.1 mg of Flag peptide. The affinity purified core complex was further purified through a gel filtration Sephadex-200 (10/30) column. The complex eluted with an apparent mass of ∼450 kD. Finally, the core complex was purified by sedimentation through a 15%-50% glycerol gradient. Recombinant HDAC1-Flag and MBD3-His proteins were purified using anti-Flag M2 affinity gel (Eastman Kodak) and Ni2+-NTA agarose (Qiagen), respectively. Recombinant RbAp48 and RbAp46 were generated by cleaving the GST fusion proteins produced in Escherichia coli with thrombin. Antibodies against Mi2, Sin3, HDAC1, HDAC2, RbAp48, RbAp46, SAP30, and MBD2 were described previously (Zhang et al. 1998a; Ng et al. 1999). Antibodies against MTA2 and MBD3 were generated by injecting recombinant proteins into rabbits.
Histone deacetylase assays
Core histone octamers were purified from HeLa cells as described (Zhang et al. 1998b) and acetylated with yeast Hat1p in buffer containing 50 mm HEPES (pH 7.8), 50 mm KCl, 0.1 mM EDTA, 1 mm DTT, 1 mm PMSF, 10 mm sodium butyrate, 10% glycerol, 5 μm [3H]-acetyl coenzyme A, and 0.5 mg/ml core histones. Acetylated core histones were purified on a phosphocellulose column. Histone deacetylase assays were performed as described (Zhang et al. 1998b) with 1 μg of labeled core histones.
Gel mobility shift assays
Gel mobility-shift assays were performed with modifications from a published procedure (Meehan et al. 1989; Nan et al. 1993). Data presented in Figure 7A used GAM6 probe and the binding reactions were carried out in 20 mm HEPES buffer at pH 7.9, 1 mm EDTA, 3 mm MgCl2, 10 mm 2-mercaptoethanol, 4% glycerol, 0.1% Triton X-100, and 500 ng of poly [d(G-C)] at 25°C for 30 min. Each reaction contains 0.1 ng of probe and 10–80 ng of recombinant MBDs or 50–300 ng of the NuRD complex. The reactions were loaded onto a 2% agarose gel and resolved with 0.5× TBE containing 5 mm magnesium acetate and 3% glycerol. The supershift assay shown in Figure 7C used the MeCG11 probe in a reaction similar to the above except it also contained 100 mm NaCl, 0.1 mg/ml BSA, and 300 ng of Escherichia coli DNA as competitor. Approximately 200 ng of GST–MBD2a and 300 ng of NuRD were used. Reactions were incubated on ice for 2 hr before electrophoresis on a 1% agarose gel.
GST pull-down assays
Approximately 2–3 μg of GST fusion proteins were bound to glutathione–Sepharose beads and incubated with in vitro translated, or purified recombinant proteins, or purified NuRD complex at 4°C for at least 4 hr. Beads were then washed three times with buffer containing 300 mm KCl and 0.05% NP40 for three times before loading onto SDS-PAGE. The bound proteins were revealed by fluorography or Western blot analysis.
Acknowledgments
We are grateful to Drs. E. Seto and S.C. Tsai for baculovirus expressing HDAC2, to S. Schreiber for baculovirus expressing HDAC1, to A. Verreault and B. Stillman for baculoviruses expressing RbAp46 and RbAp48, and to D. Gottschling for the plasmid encoding yeast Hat 1. We thank Dr. George Orphanides for critical reading of the manuscript. We also thank members of the Reinberg laboratory for stimulating discussions during the course of this work. Y.Z. is a recipient of a National Institutes of Health (NIH) fellowship (1F32GM19515-01). H.H.N. holds a Darwin Trust Scholarship. D.R. is supported by a grant from NIH (GM-48518) and from the HHMI. A.B. is supported by grants from the Wellcome Trust. P.T. is supported by grants from the National Science Foundation and the National Cancer Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL reinbedf@umdnj.edu; FAX (732) 235-5294.
References
- Airio A, Pukkala E, Isomaki H. Elevated cancer incidence in patients with dermatomyositis: A population based study. J Rheumatol. 1995;22:1300–1303. [PubMed] [Google Scholar]
- Allfrey V, Faulkner RM, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Land H. Myc-Max-Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity. 1999;10:333–343. doi: 10.1016/s1074-7613(00)80033-3. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, Golemis EA, Genuardi M, Neri G. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA [see comments] Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bjorklund S, Almouzni G, Davidson I, Nightingale KP, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. doi: 10.1016/s0092-8674(00)80586-3. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, McNulty DE, Carr SA, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Geromanos S, Philip J, Freckleton G, Tempst P. InJection adaptable fine ionization source (′JaFIS′) for continuous flow nano-electrospray. Rapid Commun Mass Spectrom. 1998;12:551–556. doi: 10.1002/(SICI)1097-0231(19980515)12:9<551::AID-RCM198>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Lazar MA, Minucci S, Pelicci PG. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Jeppesen P. Histone acetylation: A possible mechanism for the inheritance of cell memory at mitosis. BioEssays. 1997;19:67–74. doi: 10.1002/bies.950190111. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes & Dev. 1998a;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998b;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes & Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, Rosenfeld MG, Ayer DE, Eisenman RN. SAP30, a component of the mSin3 corepressor complex involved in N-CoR- mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Lyons GE. Vertebrate heart development. Curr Opin Genet Dev. 1996;6:454–460. doi: 10.1016/s0959-437x(96)80067-0. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Ng, H.H., Y. Zhang, B. Hendrich, C.A. Johnson, B.M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. (in press). [DOI] [PubMed]
- Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Tureci O, Sahin U, Pfreundschuh M, Old LJ. Characterization of human colon cancer antigens recognized by autologous antibodies. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Seelig HP, Renz M, Targoff IN, Ge Q, Frank MB. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 1996;39:1769–1771. doi: 10.1002/art.1780391029. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- Toh Y, Oki E, Oda S, Tokunaga E, Ohno S, Maehara Y, Nicolson GL, Sugimachi K. Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. Int J Cancer. 1997;74:459–463. doi: 10.1002/(sici)1097-0215(19970822)74:4<459::aid-ijc18>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Toh Y, Kuwano H, Mori M, Nicolson GL, Sugimachi K. Overexpression of metastasis-associated MTA1 mRNA in invasive oesophageal carcinomas. Br J Cancer. 1999;79:1723–1726. doi: 10.1038/sj.bjc.6690274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Travers A. An engine for nucleosome remodeling. Cell. 1999;96:311–314. doi: 10.1016/s0092-8674(00)80543-7. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes & Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998a;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998b;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]