Abstract
The phytochrome family of photoreceptors has a well-defined role in regulating gene expression in response to informational light signals. Little is known, however, of the early steps of phytochrome signal transduction. Here we describe a new Arabidopsis mutant, far1 (far-red-impaired response), which has reduced responsiveness to continuous far-red light, but responds normally to other light wavelengths. This phenotype implies a specific requirement for FAR1 in phyA signal transduction. The far1 locus maps to the south arm of chromosome 4, and is not allelic to photomorphogenic loci identified previously. All five far1 alleles isolated have single nucleotide substitutions that introduce stop codons in a single ORF. The FAR1 gene encodes a protein with no significant sequence similarity to any proteins of known function. The FAR1 protein contains a predicted nuclear localization signal and is targeted to the nucleus in transient transfection assays. This result supports an emerging view that early steps in phytochrome signaling may be centered in the nucleus. The FAR1 gene defines a new multigene family, which consists of at least four genes in Arabidopsis. This observation raises the possibility of redundancy in the phyA-signaling pathway, which could account for the incomplete block of phyA signaling observed in the far1 mutant.
Keywords: Signal transduction, phytochrome, far-red, nuclear, Arabidopsis
Just as animals modify their behavior in response to sensory information, so plants modify their growth and development according to information on light conditions from a system of photoreceptors (Kendrick and Kronenberg 1994). Among the best characterized of these photoreceptors are the phytochromes. The phytochromes are able to absorb red (R) and far-red (FR) light via a bilin chromophore covalently attached to the phytochrome polypeptide. The absorbed energy causes a photoreversible conformational change in the protein. Photoreceptor activation requires absorption of a photon that causes a phytochrome molecule to convert to the biologically active Pfr form. The Pfr form can then transfer information on the light environment to downstream-signaling pathway elements, leading to changes in gene expression. By this means, plants can optimize their growth and development to the prevailing light conditions (Kendrick and Kronenberg 1994).
In Arabidopsis, five phytochromes, designated phyA through phyE, have been characterized. They are all soluble chromoproteins of 120–130 kD encoded by single-copy members of the PHY gene family (Sharrock and Quail 1989; Clack et al. 1994). This family shows extensive amino acid sequence homology among its members, but differential transcriptional and post-translational regulation (Quail 1998). The transcription of the PHYA gene is strongly reduced in response to light, and phyA is specifically and rapidly degraded when in the active Pfr form. The other four phytochrome genes and proteins are less strongly regulated in response to light (Quail 1991; Clough and Vierstra 1997; Hirschfeld et al. 1998).
Differential roles of individual phytochromes in light perception have been revealed by the photoperception phenotypes of null mutants in the PHY genes. Responses to continuous FR (FRc) are eliminated in phyA null mutants (Dehesh et al. 1993; Whitelam et al. 1993) implying that phyA alone mediates the FRc responses. Mutations in the PHYB gene strongly reduce responses to continuous R (Rc) whereas the FRc responses are unaffected (Reed et al. 1993), thereby showing that phyB functions specifically in Rc perception.
Whereas the phytochromes are well characterized, the pathway by which they affect downstream processes such as gene expression is not. Research on the genetics and biochemistry of phytochrome signal transduction has been intensive, but our understanding remains fragmentary. The biochemical characterization of purified phytochrome preparations has revealed protein kinase activity (Ahmad et al. 1998; Yeh and Lagarias 1998). Farther downstream in the pathway, analysis of potential second-messenger involvement in phytochrome signaling has been addressed by microinjection and pharmacological techniques (e.g., Bowler et al. 1994). These techniques gave evidence for the involvement of cyclic GMP, G proteins and calcium/calmodulin in phytochrome signaling. However, genetic evidence for the involvement of these components has so far been lacking.
For a complete understanding of the phytochrome-signaling pathway, the genes encoding the proteins involved must be characterized. One approach is to screen expression libraries for phytochrome interaction partners. Using the yeast two-hybrid system, Ni et al. (1998) recently isolated PIF3, a phyA- and phyB-interacting protein with strong homology to basic helix–loop–helix transcription factors. Reverse genetic evidence indicates a requirement for PIF3 for normal phyA and phyB signal transduction in vivo (Ni et al. 1998).
Mutants with altered light perception provide another way to access the components of light-signaling pathways. Many well-characterized Arabidopsis mutants are available that show altered light responses, but are not allelic to photoreceptor mutations. These mutants can be divided into three main categories.
The first category of photosignal transduction mutants includes loci designated cop, det, and fus. These mutants grow in darkness as if they were subject to light stimuli (McNellis and Deng 1995; Chory et al. 1996; Wei and Deng 1996). A number of these loci have been cloned (e.g., Deng et al. 1992; Li et al. 1996). The phenotypes of these mutants indicate that these components are negative regulators of morphogenic signals, possibly shared by many signaling pathways (Mayer et al. 1996). These components appear to be part of one or more mechanisms of transcriptional repression, which are central to the control of plant gene expression. However, work on these components has so far not explained how signals are transmitted from the phytochromes to these repressors of photomorphogenesis.
A second category of mutants exhibit light-dependent phenotypes. They show no phenotype when grown in darkness, and aberrant light responses mediated by more than one photoreceptor. They are thus likely to be deficient in shared regulators specific to light signaling. Examples include hy5 and pef1 (Ahmad and Cashmore 1996; Oyama et al. 1997). The HY5 gene product, a bZIP protein, is now known to interact with COP1 (Ang et al. 1997). The pef1 locus, which shows a loss of Rc and FRc signaling, may also encode a positive regulator such as HY5. The psi2 mutant causes hypersensitivity to R and FR light but has no dark phenotype (Genoud et al. 1998). Currently, neither PEF1 nor PSI2 are characterized at the molecular level.
The third class of mutants shows a perturbation in signaling from a single photoreceptor, such as phyA or phyB. Mutants in integral components specific to the phyA-signaling pathway would be expected to show strong effects on FRc perception, but limited effects on Rc responses. Mutations in integral, phyB-specific signaling components would be expected to show strong phenotypes in Rc, but no phenotype under FRc. Because they are specific to a single photoreceptor, the components encoded by loci identified by mutations such as these are likely to mediate early steps in the signaling pathway.
Mutants that show enhanced phyA- or phyB-specific signaling are likely to encode early, negatively acting signaling components. Such mutants have been described in the poc1 mutant and the spa1 mutant (Hoecker et al. 1998; Halliday et al. 1999). The poc1 mutant is hypersensitive to Rc as a result of aberrant overexpression of the phytochrome-interacting protein PIF3 under Rc (Halliday et al. 1999). The spa1 mutant is a loss-of-function mutant specifically enhanced in phyA signaling (Hoecker et al. 1998). The SPA1 locus has been cloned, and encodes a WD-repeat protein with a specific role in the negative regulation of phyA signaling (Hoecker et al. 1999). Because of this specificity, SPA1 is likely to be involved in a signaling step close to the phyA photoreceptor (Hoecker et al 1998). However, because SPA1 is a suppressor of phyA signaling, its action is likely to be a modulating one on phyA itself, or on phyA-specific components of the pathway. For this reason, SPA1 may not be an integral component of the phyA-signaling pathway.
In the case of positively acting signaling components, the expected phenotype would be a loss of sensitivity to Rc or FRc specifically. Both classes of mutant have been reported, the Rc-specific class consisting of red1, (Wagner et al. 1997), pef2 and pef3 (Ahmad and Cashmore 1996), and the FRc-specific class of fhy1, fhy3 (Whitelam et al. 1993) and possibly fin2 (Soh et al. 1998). No molecular characterization of any of these loci has yet been reported.
To characterize potentially positively acting components of phyA signaling, we chose to examine FRc-specific, loss-of-signal-transmission mutants. As these mutants show a loss of positive signal transmission, they are likely to be deficient in components of the phyA pathway that participate directly in relaying the signal. Also, it is likely that previous mutant screens, such as those identifying fhy1 and fhy3 in this class (Whitelam et al. 1993), did not fully saturate the pathway. We therefore carried out a new screen to isolate more mutants of this type. Here we describe the characterization of far1, a phyA-specific, loss-of-signaling mutant of Arabidopsis. We also report positional cloning of the far1 locus and the molecular characterization of the FAR1 gene product.
Results
Mutant screening and isolation
Transgenic Arabidopsis seedlings (designated AOX) overexpressing the PHYA gene from Avena (Boylan and Quail 1991) were mutagenized with the ethyl ester of methanesulfonic acid (EMS). The M2 generation of mutagenized plants was screened for individuals displaying a long hypocotyl phenotype after 3 days of treatment with FRc of ∼10 μmoles/m2 per sec. Twelve potential mutants were isolated that showed a FRc-specific long hypocotyl phenotype in the next generation and that segregated independently from the transgene in the first backcross to the wild-type No-0 background. Once free of the AOX transgene, these mutant lines were further backcrossed twice before further analyses were performed. Genetic complementation analysis revealed that these mutants fell into two complementation groups, one of seven lines and one of five. Crosses to mutants described previously revealed that the former group was allelic to fhy3 (Whitelam et al. 1993). The latter group did not correspond to phyA, fhy1, or fhy3 (Whitelam et al. 1993). This group showed the degree of partial dominance characteristic of many photoreceptor loss-of-function mutations [such as phyA (Whitelam et al. 1993)]. This new locus was called far1 (far-red impaired response) and the five alleles were numbered far1-1 through far1-5. Because the far1-5 line also carries a clavata-type mutation, which was not removed by the first two backcrosses, it was not used for the physiological characterization of the mutant phenotype.
Responses to FRc, mediated by phyA, are specifically attenuated in far1 mutants
The phenotype of far1 seedlings grown under FRc of moderate-to-high fluence rate includes longer hypocotyls and reduced expansion of cotyledons relative to the wild type (Fig. 1A). When four alleles of far1 were treated with a range of FRc fluence rates for 3 days, the far1 alleles showed a very similar phenotype. When grown in the dark or under low fluence rates of FRc, far1 seedlings displayed a hypocotyl length similar to the isogenic wild type (No-0) (Fig. 1A). The response curve to FRc in the mutants is, however, shifted, with the mutants showing reduced inhibition of hypocotyl elongation at higher fluence rates. As a control, the phyA-101 null mutant is shown together with its isogenic wild type, the RLD ecotype. It can be seen that whereas elimination of the phyA photoreceptor causes complete loss of FRc responsivity, the far1 mutation reduces, but does not eliminate, sensitivity to FRc.
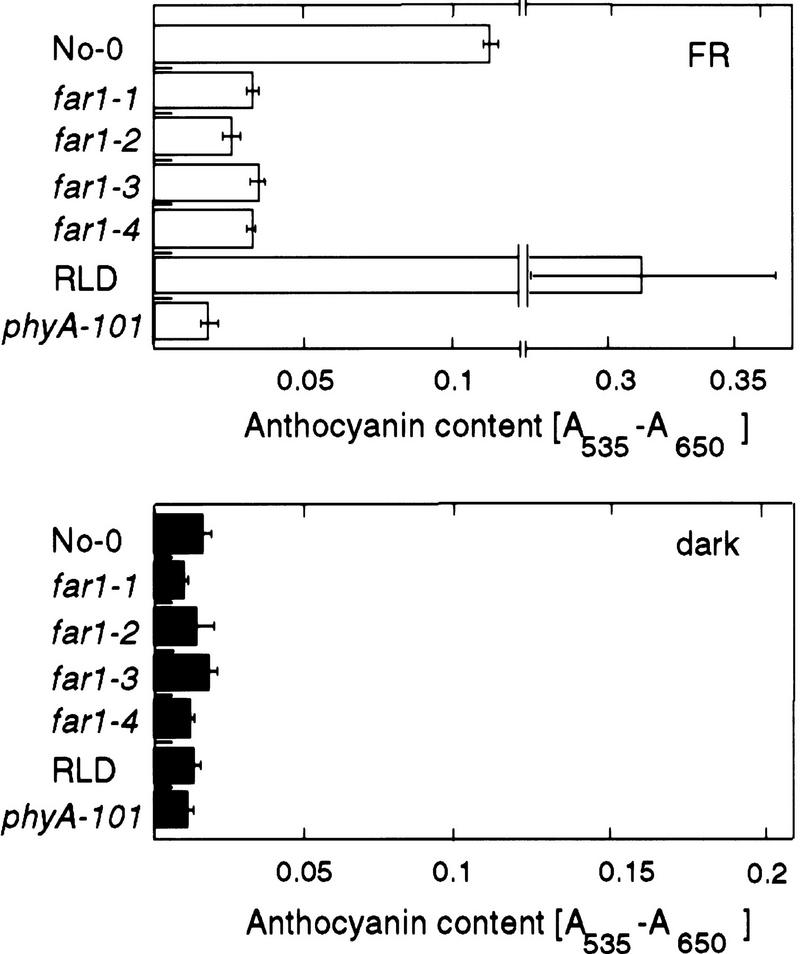
Figure 1.
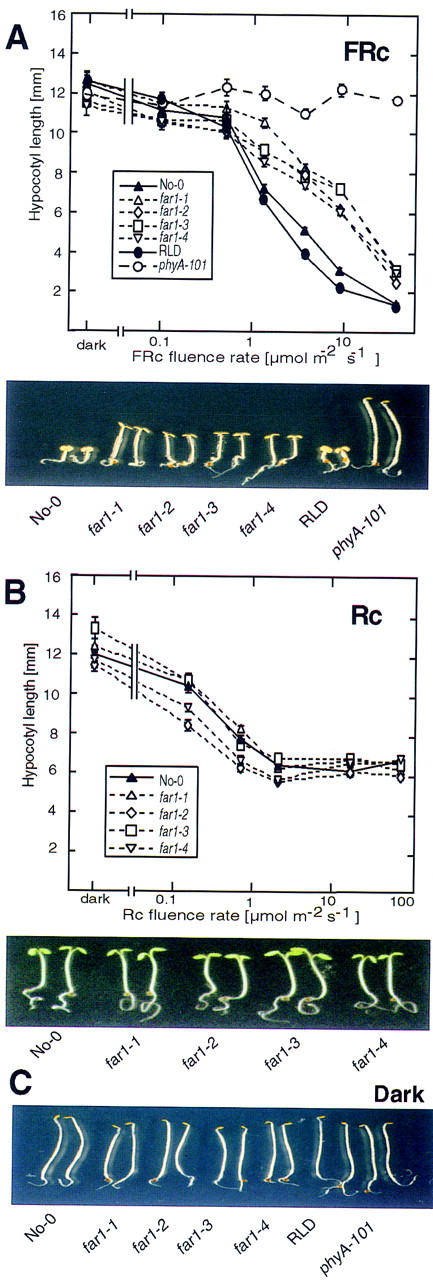
far1 seedlings are deficient in FRc-induced deetiolation. (A) (Top) Response of the hypocotyl length of seedlings grown under a logarithmic range of fluence rates of FRc. far1 seedlings (far1-1, far1-2, far1-3, and far1-4 alleles); wild-type No-0; a phyA null mutant phyA-101 and its wild-type RLD were grown for 3 days in the dark or under the indicated fluence rates of FRc. Error bars, s.e.m. (Bottom) The photograph shows seedlings grown for 3 days under 10 μmoles/m2 per sec FRc. (B) (Top) Fluence rate response curves for hypocotyl length of seedlings grown under Rc. far1 seedlings (far1-1, far1-2, far1-3, and far1-4 alleles) and the wild-type No-0 were grown for 3 days in the dark or under increasing fluence rates of Rc. Error bars, s.e.m. (Bottom) The photograph shows seedlings grown for 3 days under 80 μmoles/m2 per sec Rc. (C) Photograph showing seedlings as in A grown for 3 days in darkness.
The response of far1 alleles to a range of Rc fluence rates is shown in Figure 1B. Although there is no decrease in responsiveness to Rc in the mutants, some of the alleles show a marginal increase in sensitivity. Increased Rc sensitivity is also seen in the FRc response mutant fhy3 (Whitelam et al. 1993) and may be due to elevated levels of phyA (Fairchild and Quail 1998).
The phenotype of the far1 and wild-type seedlings grown in the dark is shown in Figure 1C. There is no effect of the mutation on any observable characteristic when the seedlings are grown in the absence of light, indicating that the mutant phenotype is light dependent.
Anthocyanin accumulation under FRc, as with other FRc-induced responses, is dependent on phyA (Kunkel et al. 1996). It thus provides a way to confirm that phyA signaling, rather than some process specific to hypocotyl elongation, is affected by the far1 mutation. The level of anthocyanin was thus measured in FRc-grown far1 seedlings. As shown in Figure 2A, far1 seedlings grown under FRc accumulated significantly less anthocyanin than the wild type. Seedlings of the phyA null mutant phyA-101 entirely failed to accumulate anthocyanin in response to FRc, compared with its RLD wild type. As expected, no difference between the far1 and wild-type phenotypes was observed in seedlings grown in the dark, confirming that the effect of far1 on anthocyanin levels is also light dependent.
Figure 2.
Response of anthocyanin levels to FRc in wild-type and mutant seedlings. The amount of anthocyanin accumulated in seedlings of the far1 alleles (far1-1, far1-2, far1-3, far1-4), wild-type No-0, the phyA-null mutant phyA-101, and the relevant wild-type RLD was measured. Seedlings were grown for 3 days under FRc (10 μmoles/m2 per sec) (Top) or in the dark (Bottom). Error bars, s.e.m.
The far1 phenotype is not caused by reduced levels of phyA
It has been shown that the levels of phytochromes have a significant and proportional effect on the sensitivity of seedlings to light, both from overexpression studies and the partial dominance of phyA and phyB mutations at intermediate fluence rates (Boylan and Quail 1991; Whitelam et al. 1993; Wester et al. 1995). For this reason it was necessary to determine whether the reduced sensitivity of far1 seedlings to FRc is due to lower levels of phyA. In dark-grown seedlings no difference in phyA levels between far1 and wild-type was detected (Fig. 3A). FRc-grown far1 seedlings showed slightly increased levels of phyA compared with the wild type (Fig. 3B). Thus, the far1 phenotype is not the result of lower levels of phyA.
Figure 3.
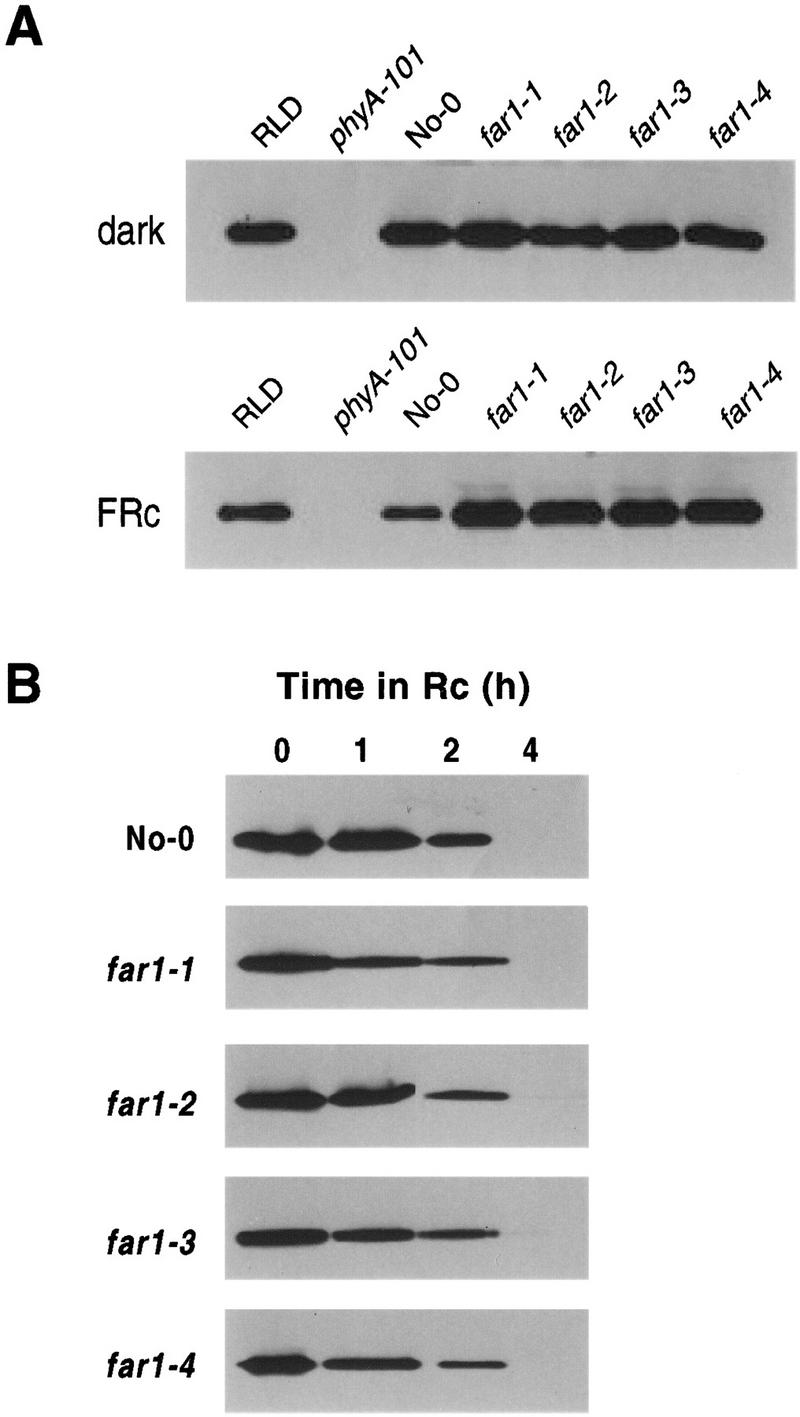
Phytochrome A levels in the far1 mutants. (A) Steady-state phyA levels measured by protein blot. The figure shows the bands detected by the phyA-specific monoclonal antibody 073D (Hirschfeld et al. 1998). Protein loading was equalized by Bradford assay, and is identical within each panel. Seedlings were grown for 3 days in the dark (top) or under FRc (10 μmoles/m2 per sec) (bottom). Bottom lanes contain 1.5× the protein loading of the top blot. (B) Time course of phyA loss in response to Rc. Seedlings of the four far1 mutant alleles and the wild type (No-0) were grown for 2 days in darkness, then subjected to 0, 1, 2 or 4 hr of RC (16 μmoles/m2 per sec) before protein extraction. PHYA protein was assayed as above.
The possibility was raised by this result that the phyA detected by immunoblot analysis was in some way less photoactive in the mutant than in the wild type. This would result in altered degradation kinetics of phyA in the far1 mutant, because phyA must perceive FRc and convert to the Pfr form before it is degraded. Consequently, phyA degradation time courses under Rc were measured. As shown in Figure 3B, no substantial difference in the degradation rate of phyA is visible between far1 and the wild type. This result demonstrates that the phyA detected was photoactive, and that its capacity for photoperception was not noticeably impaired by the far1 mutation.
Mapping and molecular identification of the far1 locus
Mapping populations were established by crossing far1 to the ecotypes Landsberg erecta (La-er) and Columbia (Col-0). Using simple sequence length polymorphism (SSLP, Bell and Ecker 1994) and cleaved amplified polymorphic sequence (CAPS; Konieczny and Ausubel 1993) markers, we mapped the far1 locus to a region of chromosome 4 between the SSLP marker nga8 and the CAPS marker AG (Fig. 4). Analysis of recombination frequencies indicated that the far1 mutation lay 14.4 cm south of nga8 and 7.0 cm north of AG. We then narrowed down the possible physical position of the far1 lesion to within the sequenced ESSA1 contig (Bevan et al. 1998), close to the photomorphogenic loci PHYD, PHYE, and COP9 (Fig. 4). Knowing the genomic sequence of the entire 1.9-Mb contiguous region, we were able to design CAPS markers by synthesizing primers to amplify predicted intergenic regions, which showed a high degree of polymorphism. The PCR products from the ecotypes Col-0, La-er, and No-0 were sequenced. The sequence was analyzed for restriction polymorphisms between the ecotypes and the initial primers could then be used to generate CAPS markers if a suitable polymorphism was found. Many such markers were developed; those that were critical in the eventual positional cloning are shown in Figure 4. These markers will be made available to the compilers of CAPS marker databases.
Figure 4.
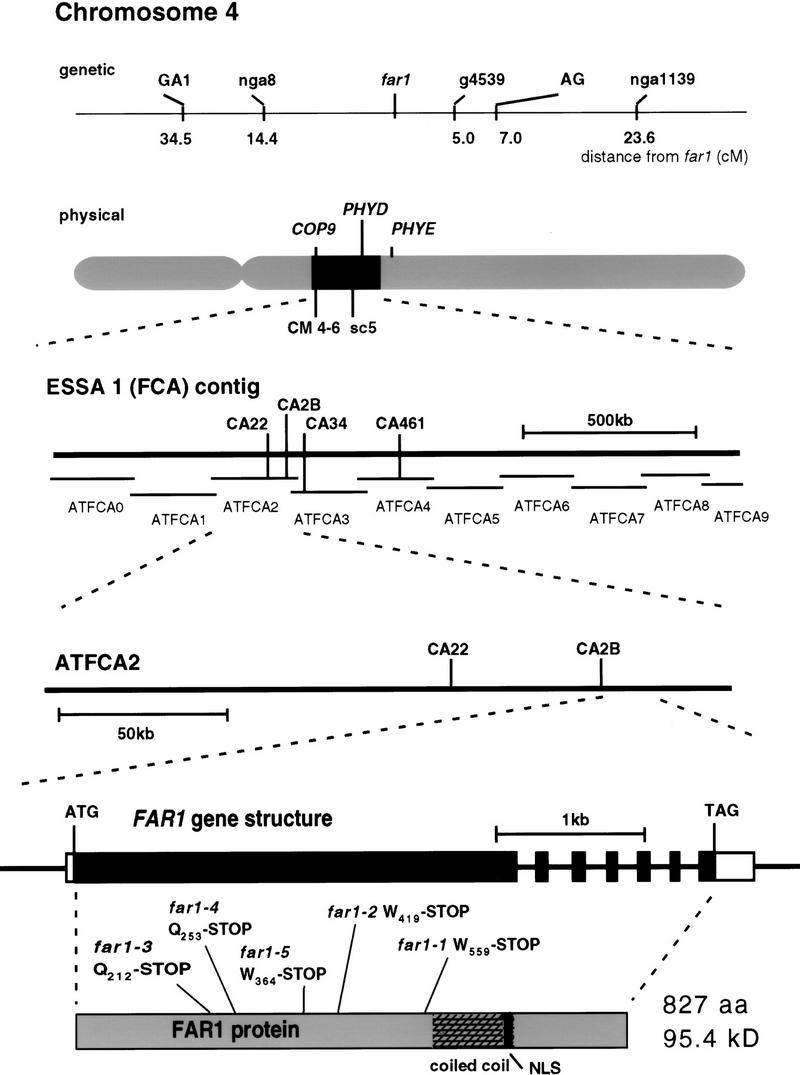
Positional cloning of the far1 mutant locus. The position of the far1 locus is shown on the genetic map; recombination frequencies were calculated from a population of 88 plants. We chose the markers shown on the basis of the RI map of Lister and Dean (http://genome-www.stanford.edu/Arabidopsis/). We narrowed the physical location of far1 to the highlighted region of chromosome 4, close to the photomorphogenic loci shown, using the publicly available markers CM 4-6 and sc5. Bevan et al (1998) described the sequence of this region. A number of markers throughout this region (part of the ESSA1 contig) were generated (only key markers mapping within 1 cm of far1 are shown). Using these markers, we located far1 between CA22 and CA34, with 0 recombination at CA2B. By directly sequencing the 65 kb between CA22 and CA34 from mutant and wild-type DNA, we discovered the indicated mutations in the gene shown, all of which cause the introduction of stop codons in the same large ORF. The intron structure derived from cDNA clones is shown along with the regions encoding the predicted coiled–coil domain and nuclear localization signal.
Because polymorphisms were much more abundant between Col-0 and No-0 (ecotype Nossen) than between La-er and No-0 in the region of interest, fine mapping was accomplished with a far1 × Col-0 F2 population. Using DNA preparations from a mapping population of 700 plants, two recombinants from the northern side and one from the southern side were identified at markers CA22 and CA34, respectively. No recombinants were found at CA2B. The far1 locus was hence genetically located to 65 kb between CA22 and CA34. ORFs and their flanking regions that we judged likely to represent candidate genes were amplified from the mutants and No-0 wild type by PCR and sequenced. Six such putative genes were located between markers CA22 and CA3. The G → A/C → T substitution mutations expected from EMS mutagenesis were located by comparison with PCR products from genomic DNA isolated from the wild type (No-0) and the genomic sequence already available (Col-0). All five far1 alleles were found to have G → A or complementary C → T mutations within a single ORF, located between position 179,800 and 182,000 on the subcontig ATFCA2 (GenBank accession no. Z97337, Bevan et al. 1998), within 2 kb of the marker CA2B (Fig. 4). The marker CA2B hence provides a CAPS marker for the FAR1 locus. No other mutations were detected in a total of >50 kb of genomic DNA, which was sequenced in the No-0 wild type and at least two far1 alleles.
Analysis of the genomic sequences produced indicated a discrepancy between the genome project sequence and the No-0 genome, extending the ORF in a 5′ direction from the gene predicted by the annotators (Bevan et al. 1998). The discrepancy was the T residue at 180,101 of ATFCA2, which we found to be inserted in the database relative to our genomic sequence. We believe this difference is due to an error in the Genome Project sequence in GenBank (GenBank accession no. Z97337) as we have also sequenced this region from Col-0 and find it to be identical to our FAR1 sequence from No-0.
Isolation and analysis of cDNA sequence
By use of a PCR product spanning the ORF in which the far1 mutations were located, a cDNA clone was isolated from a λ cDNA library derived from dark-grown seedlings (Kieber et al. 1993). One positive clone was identified from >300,000 phage, indicating a low abundance of the transcript. Northern blot analysis, with RNA from wild-type Arabidopsis, revealed a rare transcript of ∼3.1 kb. We have found no significant effects of light treatment, tissue type, or developmental stage on the abundance of this transcript (data not shown). Comparison of the cDNA to the genomic sequence revealed a single large exon spanning the ORF containing the far1 mutations, followed by six smaller exons interspersed by introns of 200 bp or less (Fig. 4).
RT–PCR techniques were used to extend the cDNA in a 5′ direction and check the original λ clone for mutations (one of which was found). The corrected and extended sequence of the cDNA has been submitted to GenBank (accession no. AF159587). When the genomic mutations are introduced to the reading frame of the cDNA sequence, they all produce stop codons. The locations of the truncations are as follows: far1-3, Q212–Stop; far1-4, Q253–Stop; far1-5, W364–Stop; far1-2, W419–Stop; and far1-1, W559–Stop. The far1-2 mutant also has a secondary mutation causing the substitution G413–E.
The FAR1 gene is a member of a multigene family in Arabidopsis, and homologous transcripts exist in other plant species
Searches of sequences submitted previously to GenBank with BLAST2 (Altschul et al 1997) revealed that the conceptual translation of the FAR1 cDNA gives a protein sequence containing no significant homology to currently recognized proteins of known function (Fig. 5). Use of the web-based program coils (http://dot.imgen.bcm.tmc.edu:9331/seq-search/struc-predict.html) predicts that residues in the 600- to 700-amino-acid region may form a coiled–coil structure (maximum probability 0.6) (Figs. 4 and 5). The FAR1 sequence also contains a basic region, which could act as a nuclear localization signal (NLS) (Figs. 4 and 5).
Figure 5.
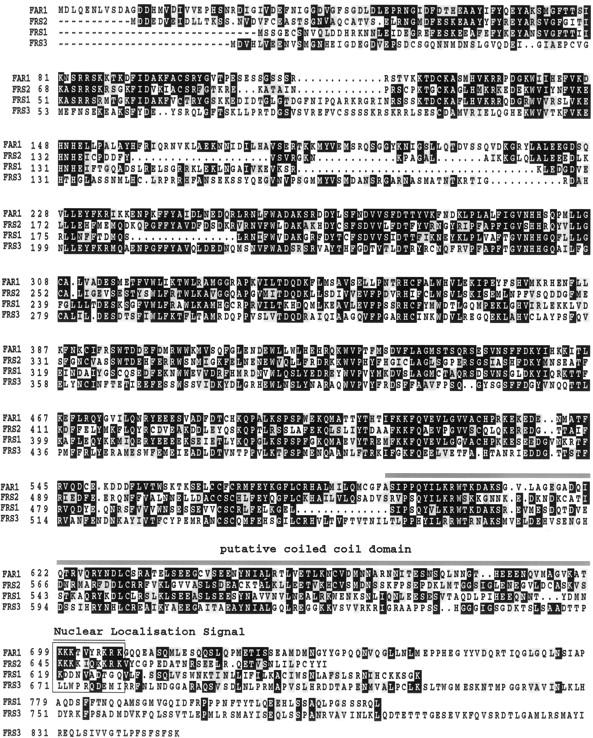
Alignment of the FAR1 amino acid sequence with homologs from the gene family in Arabidopsis. The conceptual translation of the FAR1 cDNA is presented, aligned to amino acid sequences predicted from genomic sequence from the Arabidopsis genome project. Residues identical to those in FAR1 are highlighted in black; those in gray are conserved substitutions. The putative coiled–coil and nuclear localization signal sequences of FAR1 are indicated.
BLAST searches revealed sequence from the Arabidopsis genome project that contained previously undescribed genes with striking amino-acid-level similarity to FAR1 (Fig. 5). To date, three such genes have been identified, one on the ESSA II contig south of FAR1 on chromosome 4 (BAC F18F4) and two on chromosome 2, located on BACs T32F6 and T20P8. We identify these genes as FAR1-Related Sequences FRS1, FRS2, and FRS3 in the order of their submission to the database. The shared motifs between the conceptual translations of these sequences can be clearly seen in Figure 5. Note that the putative NLS in FAR1 is conserved in FRS2, but not in FRS1 or FRS3, which we predict to be cytoplasmic proteins. Interestingly, although FRS2 has the highest amino acid similarity score to FAR1, FRS1 is predicted to be its closest evolutionary relative by use of the neighbor-joining method. FRS3 is the least similar to FAR1 by all analyses used (data not shown).
EST sequences in cotton and rice, the conceptual translations of which share substantial amino acid sequence similarity to FAR1, are present in the dbEST database [accession nos. AI054610, AI055286 (cotton), C72410, D43616 (rice)]. This observation provides evidence for the presence of conserved, expressed FAR1-like sequences in other plant species including monocotyledons. The absence of Arabidopsis ESTs, not only for FAR1 itself but also for FRS1 and FRS2, may be due to low expression in the tissues used to create the libraries. On the other hand, FRS3 has two Arabidopsis ESTs, GenBank accession nos. N37149 and T14215. The ESTs with GenBank accession nos. T20465 and T14014 encode sequences homologous to, but not identical with, FAR 1 and FRS 1, FRS 2, and FRS 3, implying a still larger gene family is present in Arabidopsis.
FAR1 has a functional nuclear localization signal
The FAR1 amino acid sequence contains the monopartite NLS RKRK, which, together with other basic residues nearby, could contribute to nuclear localization (Fig. 5). To investigate whether FAR1 could be targeted to the nucleus, the complete coding region of the FAR1 cDNA was isolated from first-strand cDNA by RT–PCR. The resulting sequence was fused to the 3′ end of the GUS reporter gene in a modified version of the vector pTEX2, as described by Hoecker et al. (1999). The vector containing the GUS–FAR1 fusion protein was introduced with a particle gun into onion epidermal cells as described earlier (Ni et al. 1998). After incubation in white light and treatment with the X-gluc substrate, GUS staining in transformed cells due to the GUS–FAR1 fusion was found to be localized to the nucleus, whereas the GUS-only control was mainly seen in the cytoplasm of the onion cells (Fig. 6). The position of nuclei was confirmed by DAPI staining (Fig. 6).
Figure 6.
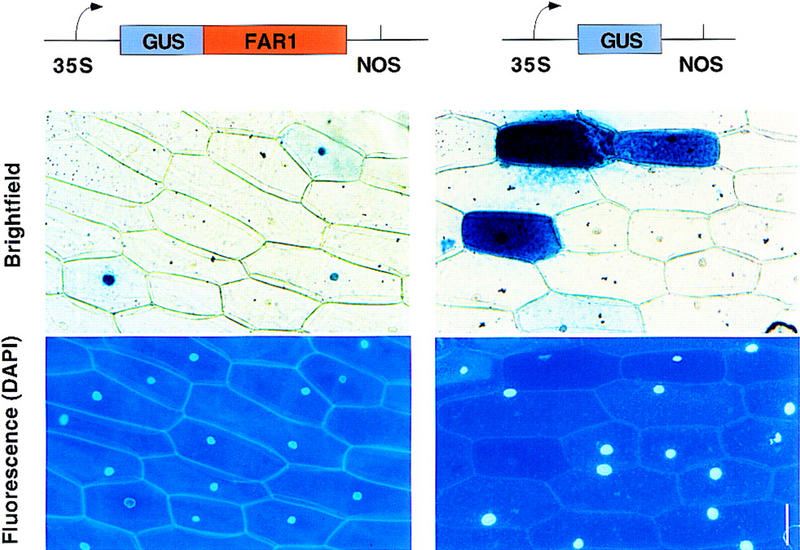
Subcellular location of a GUS–FAR1 fusion protein in transiently transfected onion cells. Shown is the GUS staining pattern of onion epidermal cells transfected by particle bombardment. The GUS–FAR1 fusion transfection is shown at left, together with a DAPI-stained fluorescence micrograph to demonstrate the location of the nuclei. A control GUS transfection is shown at right. Bar, 50 μm.
Discussion
The availability of Arabidopsis mutants selectively impaired in responsiveness to either Rc or FRc indicates the existence of components that act in separate signaling pathways, specific to either phyB or phyA, respectively (Quail 1998). The far1 mutant described here represents such a mutant, with a phenotype specific to phyA signaling. Prior to the recent cloning of SPA1, a negative regulator of the phyA pathway (Hoecker et al. 1999), no pathway-specific phytochrome-signaling components had been molecularly identified. The identification of the FAR1 gene sequence here provides the first insight into the molecular nature and function of a positively acting component specific to a single phytochrome-signaling pathway.
The evidence that FAR1 is a component specific to phyA signaling is as follows. The absence of a phenotype in dark-grown far1 seedlings establishes that the effect of the mutation is light conditional. This observation indicates a specific requirement for the locus in the transmission of light signals. The FRc specificity of the light-conditional phenotype establishes that the requirement for this locus is specific to phyA signaling. The FRc specificity is unlikely to be due to FRc-induced changes in FAR1 transcript levels, as RNA blot analysis indicates that the expression of FAR1 is constitutive (data not shown). The FRc specificity could also have been explained by reduced levels of active phyA, but the data in Figure 3 demonstrate that phyA is present in at least normal levels in the far1 mutant alleles and that it is capable of actively perceiving light. Any deleterious effect on chromophore biosynthesis would be expected to reduce all phytochrome responses, such as phyB responses to Rc, as well as phyA responses to FRc. No such decrease in Rc responsiveness is observed in far1 (Fig. 1). As there is also no visible increased elongation of the white-light-grown adult far1 mutant, compared with the wild type (data not shown), the phenotype is consistent with a specific effect of the loss of FAR1 on phyA signaling, leaving other phytochrome-signaling pathways unaffected.
The marginal increase in Rc sensitivity exhibited by some of the mutant alleles could be the consequence of elevated phyA levels in the mutant (Fig. 3). Because phyA is responsible for the perception of very low fluence rates of Rc, higher phyA levels can enhance this response (Boylan et al. 1991). Higher levels of phyA under FRc, and increased Rc sensitivity, have been reported for the FRc hyposensitive fhy3 mutant (Whitelam et al. 1993; Fairchild and Quail 1998). This observation implies that elevated phyA levels and increased Rc sensitivity may be a general consequence of reduced phyA signal-transduction activity. The elevated phyA levels could be a consequence of decreased feedback down-regulation of the PHYA promoter (F. Canton and P.H. Quail, unpubl.) or a result of a link between phyA degradation and signal transduction.
Regardless of the mechanism, observed overexpression of phyA in fhy3 and far1 might be expected to partially compensate for the reduced signaling, thereby tending to reduce the severity of their phenotypes. The partial block of FRc sensitivity in the far1 mutant must therefore be viewed in the context of a plant that has higher than normal levels of phyA, and thus far1 may have a more significant effect on phyA signaling than is apparent from its phenotype in FRc.
The genetic analysis performed here indicates that FAR1 is a new locus, not allelic to photomorphogenic mutants reported previously. The genetic evidence also indicates that the far1 mutations are partially dominant (data not shown), and that they are due to the introduction of stop codons in an ORF of the FAR1 genomic coding sequence. The far1 mutant phenotype is therefore due to loss of function of the FAR1 protein, and the partial dominance of the mutation is almost certainly due to haploinsufficiency in the heterozygote. Because loss of FAR1 function causes a loss of phyA signaling, FAR1 must either be an integral component of the transduction pathway or a positive regulator of it.
The loss of an essential integral pathway component would be expected to completely stop signal transmission in a single phyA-signaling pathway. The far1 mutant still has substantial phyA-signaling activity. FAR1 may therefore be a positive regulator of one or more phyA-specific pathways, or it may be an essential component of only one of multiple, partially redundant phyA-signaling pathways. Alternatively, because FAR1 is encoded by a member of a multigene family, there may also be genetic redundancy between FAR1 and one of the other members, FRS1, FRS2, FRS3, and perhaps more proteins. Therefore, these homologous gene products may transmit a reduced phyA signal in the absence of FAR1.
The components of the phyA and phyB signal transduction pathways in which mutants have been isolated may be placed in a tentative order, largely on the basis of interpretation of the mutant phenotypes. Molecular and reverse-genetic evidence indicates that a shared signaling component, PIF3, binds phyA and phyB directly. This result, combined with the genetic evidence for components specific to either phyA or phyB, gives rise to the proposal that there may be multiple signaling pathways from phyA and phyB, some of which are specific to phyA or phyB and some of which are shared (Ni et al. 1998). In this case, FAR1 would act in one or more of the phyA-specific pathways, as represented in Figure 7A (i). Alternatively, a single phyA-pathway model can be postulated, with some components being specific to phyA and others being shared with phyB. Because far1 appears to be specifically blocked in phyA signaling, its action in a single-pathway model could therefore be placed upstream of that of components shared between phyA and phyB signaling (Fig. 7A, ii). The reality is likely to be more complex and could include elements from both models.
Figure 7.
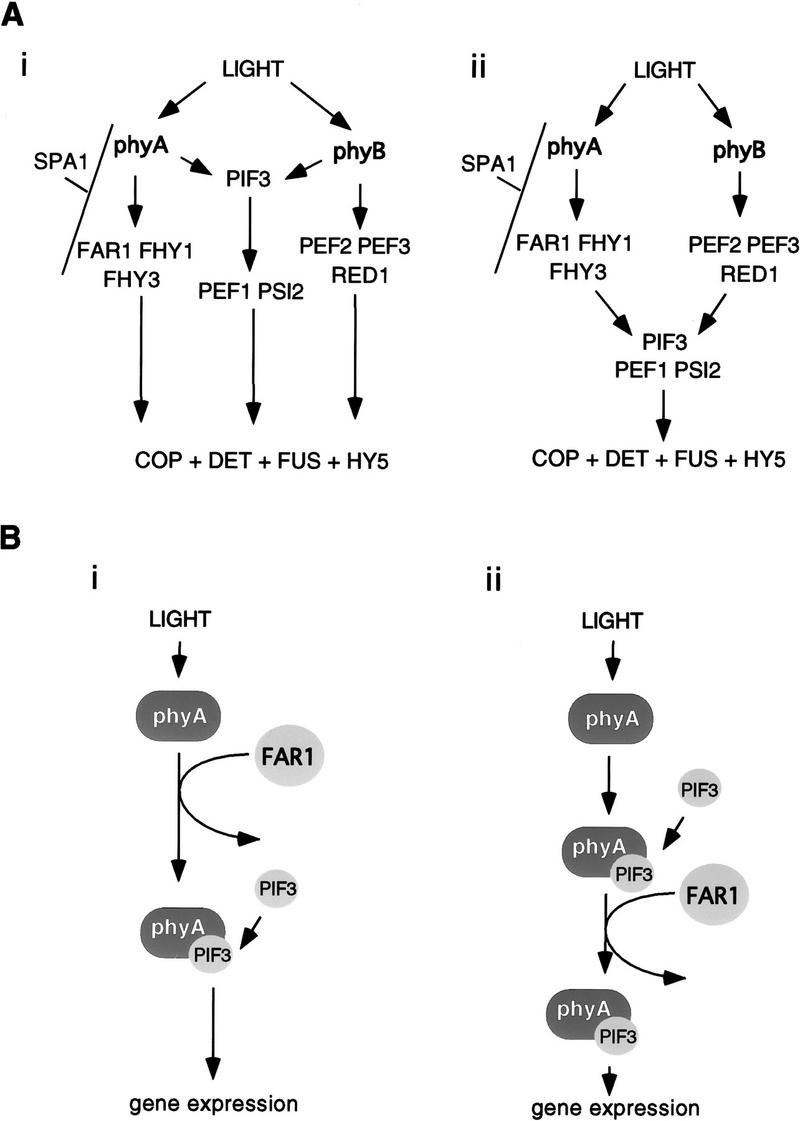
Models for the possible action of FAR1. (A) Genetic models of the pathway of phytochrome signal transduction. In the multiple-pathway model (i) specific and shared signaling components can transduce signals independently of one another. In the alternative single-pathway model shown (ii), loci specific to a single phytochrome-signaling pathway (e.g., FAR1) are shown upstream of those components shared by multiple pathways (e.g., PIF3). (B) Speculative molecular modes of action for FAR1. Although we speculate in A that FAR1 is upstream of PIF3 on the basis of genetic data, its biochemical action may lie before that of PIF3 (i) or after it (ii). Either could produce a specific effect on phyA signaling.
It is not easy to reconcile an action of FAR1 upstream of PIF3 with the direct interaction of PIF3 with phyA. A phyA-specific component such as FAR1 could, however, still act upstream of PIF3 binding phyA (Fig. 7B, i), for example, by being involved in the transport of phyA from cytoplasm to nucleus. In darkness, phyA appears to be confined to the cytosol (Kircher et al. 1999) and to interact with PIF3 [thought likely to be constitutively nuclear (Ni et al. 1998)], it presumably must be imported into the nucleus in response to light. If FAR1, and also possibly FHY1 and FHY3, were involved in nuclear transport of phyA, they would be positive facilitators of phyA signaling rather than direct participants in signal transfer.
Action of FAR1 after PIF3 binding could also result in a specific effect on phyA signaling, however. An example of how this could occur is in a regulatory complex of phyA, PIF3, and FAR1, which are all required for full transmission of the phyA signal (Fig. 7B, ii). FAR1 could act after PIF3, binding and yet still be specific to phyA.
It is intriguing that the majority of likely phytochrome signal-transduction components that have been characterized at the molecular level are nuclear localized. This has been known for some time of the components likely to act later in the pathway [such as HY5 (Oyama et al. 1997) and COP1 (Deng et al. 1993)]. The transcriptional repressor COP1 is not constitutively nuclear, but its nuclear abundance is regulated by light, and its site of action is thought to lie in the nucleus, in which it interacts with HY5 (von Arnim et al. 1994; Ang et al. 1998). However, less predictable was the fact that the upstream components characterized recently, such as PIF3 [which directly binds phytochromes (Ni et al. 1998) and SPA1 which is phyA specific (Hoecker et al. 1999)] would also be nuclear localized. It appears, therefore, that the nucleus is emerging as an important venue for events both early and later on in phytochrome-signal transduction.
This notion is supported here by the finding that the FAR1 protein is also targeted to the nucleus, suggesting a role in phyA-regulated gene expression. The roles FAR1 might play are highly speculative, however, given our lack of knowledge of its structure or biochemistry. The one structural feature indicated by the FAR1 amino acid sequence is the putative coiled–coil structure (Fig. 4 and 5). Similar structures are involved in mediating specific protein–protein interactions, such as homo- or heterodimerization or oligomerization, in other signaling systems (Kohn et al. 1997). Many other light-signal transduction components have such a predicted structure, for example, COP1 (Deng et al. 1993) and SPA1 (Hoecker et al. 1999). FAR1 could interact with other signaling components such as these by this coiled–coil structure.
PIF3 shares strong homology to the DNA-binding domains of basic helix–loop–helix transcription factors, suggesting a direct link between phytochromes and transcriptional regulation. In contrast, the FAR1 and SPA1 sequences lack canonical DNA-binding motifs. Although FAR1 lacks obvious homology to DNA-binding proteins, it could still bind DNA via a previously undescribed structure. It is perhaps more likely, however, that FAR1 does not bind DNA itself but regulates transcription by interacting with DNA-binding complexes, in the manner of a coactivator. If this is the case, then the FAR1 gene family may represent a new class of transcriptional regulators.
Material and methods
Growth of seedlings
Seeds dried for 10 days under anhydrous CaSO4 were surface sterilized with 20% bleach (1.05% sodium hypochlorite), 0.03% Triton X-100 for 10 min, washed five times with sterile water, and sown on growth medium (GM) (Valvekens et al. 1988) without sucrose, containing 1% agar. After stratification for 5 days at 4°C, induction of germination by 3 hr irradiation with white light, and subsequent storage for 21 hr in the dark at room temperature, the plates were transferred for 3 days to appropriate light conditions at 21°C. The light sources used are described elsewhere (Wagner et al. 1991). The fluence rates of light were measured with a spectroradiometer (model LI-1800, Li-Cor, Lincoln, NE).
Plant material and EMS mutagenesis
A transgenic Arabidopsis line, No-0, containing one copy of an oat phyA overexpression construct, was used for the EMS mutagenesis. Boylan and Quail (1991) described the transgenic line (AOX). Fifty thousand seeds were mutagenized for 12 hr with 0.2% (EMS) and sown on soil. These M1 plants were grown up in 50 families of 1000 individual plants each and the M2 seeds of each family were pooled. About 3000 M2 seeds of each family were then sown as described above on agar plates containing GM medium (1% sucrose) under FRc for 3 days and seedlings displaying a long hypocotyl phenotype were selected. These seedlings were recovered on sucrose-containing medium from the FRc treatment and grown up in the greenhouse.
Genetic analysis
The selected M2 seedlings were crossed with No-0 wild-type plants. The resulting F1 seedlings were heterozygous for the AOX transgene as well as the mutation causing the phenotype. F1 seedlings were propagated to the F2 generation, which segregated for the AOX transgene as well as the mutation. One hundred F2 seeds were sown on GM without sucrose, and the seedlings exhibiting the longest hypocotyl under FRc were selected, recovered from the FR treatment, and grown up in the greenhouse. Seeds of the next generation were grown on GM containing 80 mg/ml kanamycin as selectable marker to test for sensitivity to the antibiotic, that is, the absence of the AOX transgene, and on nonselective medium under FRc to confirm the mutant phenotype.
For the complementation analysis with the mutants fhy1 and fhy3, the new mutant lines, once free of the AOX transgene, were crossed with these mutants. Control crosses were made with their respective wild-type ecotypes (La-er and Col-0, respectively). The F1 seeds were grown for 3 days under FRc and the hypocotyl length was measured. To confirm the presence of genetically distinct loci, F1 seedlings were grown to the F2 generation, when segregation of the mutant phenotype was confirmed by the presence of seedlings displaying a wild-type phenotype. In all cases, the analysis of the F2 confirmed the result of the allelism test of the F1 generation.
Mapping of the far1 locus
F2 plants of the different far1 alleles were grown in the greenhouse under continuous white light and were crossed with plants of the ecotypes La-er and Col-0. The F1 seeds of these crosses were germinated on GM without sucrose under white light as described below, transferred to soil, and grown to seed in the greenhouse. The F2 seeds were sown on GM without sucrose and grown for 3 days under FRc (10 μmoles/m2 per sec). The seedlings showing the elongated hypocotyl phenotype of the homozygous far1 mutant were recovered on sucrose-containing medium. After greening, the plants were transferred to soil and grown up in the greenhouse. At this stage, leaf material was used to extract genomic DNA (Edwards et al. 1991), which was subsequently used to map the far1 locus with SSLP and CAPS markers. The F3 seeds of the selected plants were collected and tested for their mutant phenotype. The results from any F2 individuals giving segregating F3 populations were discarded. DNA sequencing to determine polymorphisms, mutations, and gene structure was from PCR products and λ clones with ABI chemistry and a 373 DNA sequencer (Perkin Elmer, Foster City, CA).
Analysis of seedling anthocyanin levels
Seeds were grown on GM containing 2% sucrose under FRc (10 μmoles/m2 per sec) or in the dark. Subsequently, anthocyanins were extracted from the seedlings under green safelight and anthocyanin content was spectrophotometrically determined as described by Schmidt and Mohr (1981). Thirty seeds were used per extraction and each data point represents the mean of two duplicate experiments.
phyA extraction and immunoblot detection
For phyA extraction, 200 seeds of each line were sown on GM medium as described above and grown for 3 days under FRc (10 μmoles/m2 per sec) or in the dark. Subsequently, seedlings were collected under green safelight and frozen in liquid nitrogen. After grinding the plant material with a mortar and pestle, proteins were extracted as described by Wagner et al. (1991) with the exception that, for protection from proteolytic degradation, a proteinase inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN) supplemented with 2 mm PMSF was used. Transfer of the proteins onto a PVDF membrane after separation on a 7% SDS-PAGE under denaturing conditions was performed as described by Wagner et al. (1991). Blocking of the membrane was done overnight with 1× TBS, 0.1% Tween 20 followed by 2 hr in 1× TBS, 0.1% Tween 20; 5% fat-free milk powder. Antibody incubations and washing were done in 1× TBST, 0.5% fat-free milk powder with the phyA-specific monoclonal antibody mAb O73D (Hirschfeld et al. 1998) and a anti-mouse antibody (Promega) suitable for detection with a chemiluminescent system (SuperSignal; Pierce, Rockford, IL).
Transient transfection of GUS fusions into onion cells
The full-length coding region of the FAR1 gene was isolated from first-strand c-DNA by proofreading RT–PCR and checked for mutations against genomic sequence. The sequence thus generated was fused to GUS in the transient expression vector TEX2 with a modified polylinker, as used by Ni et al. (1998). The transfection, staining and microscopy were all performed as in Ni et al. (1998).
Acknowledgments
We thank Yurah Kang and Sharon Moran for excellent technical assistance, Jim Tepperman for invaluable help with the transient expression experiments, Dr. Craig Fairchild for his intellectual input and FR-grown seedling-rescue method, Dr. Ute Hoecker for helpful discussions and Drs. Enamul Huq, Elena Monte, Jaime Martinez, and Craig Fairchild for critical reading of the manuscript. C. Ringli was the recipient of a Swiss National Science Foundation grant. This research was supported by National Institutes of Health grant GM47475 and U.S. Department of Agriculture CRIS grant 5335-21000-0010-00D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL quail@nature.berkeley.edu; FAX (510) 559-5678.
References
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng X-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Boylan MT, Quail PH. Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc Natl Acad Sci. 1991;88:10806–10810. doi: 10.1073/pnas.88.23.10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Chatterjee M, Cook RK, Elich T, Fankhauser C, Li J, Neff M, Pepper A, Poole D, Reed J, Vitart V. From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc Natl Acad Sci. 1996;93:12066–12071. doi: 10.1073/pnas.93.22.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes - The sequences and expression of phyD and phyE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant Cell Envir. 1997;20:713–721. [Google Scholar]
- Dehesh K, Franci C, Parks BM, Seeley KA, Short TW, Tepperman JM, Quail PH. Arabidopsis HY8 locus encodes phytochrome A. Plant Cell. 1993;5:1081–1088. doi: 10.1105/tpc.5.9.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Quail PH. Control of plant development: Genes and signals. Cambridge, UK: The Company of Biologists Ltd; 1998. The phytochromes: Photosensory perception and signal transduction; pp. 85–92. [PubMed] [Google Scholar]
- Genoud T, Millar AJ, Nishizawa N, Kay SA, Schäfer E, Nagatani A, Chua NH. An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M-M, Quail PH. poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics. 1998;149:523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in plants. 2nd ed. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rugner A, Schafer E, Harter K. Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J Cell Biol. 1999;144:201–211. doi: 10.1083/jcb.144.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn WD, Mant CT, Hodges RS. Alpha-helical protein assembly motifs. J Biol Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T, Neuhaus G, Batschauer A, Chua N-H, Schafer E. Functional analysis of yeast-derived phytochrome A and B phycocyanobilin adducts. Plant J. 1996;10:625–636. doi: 10.1046/j.1365-313x.1996.10040625.x. [DOI] [PubMed] [Google Scholar]
- Li JM, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Mayer R, Raventos D, Chua N-H. det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell. 1996;8:1951–1959. doi: 10.1105/tpc.8.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, Deng X-W. Light control of seedling morphogenetic pattern. Plant Cell. 1995;7:1749–1761. doi: 10.1105/tpc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome. A light-activated molecular switch that regulates plant gene expression. Annu Rev Genet. 1991;25:389–409. doi: 10.1146/annurev.ge.25.120191.002133. [DOI] [PubMed] [Google Scholar]
- ————— The phytochrome family: Dissection of functional roles and signalling pathways among family members. Phil Trans R Soc Lond. 1998;353:1399–1403. doi: 10.1098/rstb.1998.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Mohr H. Time-dependent changes in the responsiveness to light of phytochrome-mediated anthocyanin synthesis. Plant Cell Environ. 1981;4:433–437. [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes & Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Wagner D, Tepperman JM, Quail PH. Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell. 1991;3:1275–1288. doi: 10.1105/tpc.3.12.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Hoecker U, Quail PH. Red1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of the COP/DET/FUS genes in light control of Arabidopsis seedlings development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester L, Somers DE, Clack T, Sharrock RA. Transgenic complementation of the hy3 phytochrome B mutation and response to PHYB gene copy number in Arabidopsis. Plant J. 1994;5:261–272. doi: 10.1046/j.1365-313x.1994.05020261.x. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci. 1998;10:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]