Abstract
Yeast cells of different cell type exhibit distinct budding patterns that reflect the organization of the actin cytoskeleton. Bud1p (Rsr1p), a Ras-like GTPase, and Bud2p, a GTPase-activating protein for Bud1p, are essential for proper budding pattern. We show that Bud2p is localized at the presumptive bud site in G1 cells in all cell types and that this localization is independent of Bud1p. Bud2p subsequently localizes to the mother-bud neck after bud emergence; this localization depends on the integrity of the septins. These observations indicate that Bud2p becomes positioned in G1 cells by recognizing cell type-specific landmarks at the presumptive bud site.
Keywords: GTPase-activating protein, cell polarity, cell type, yeast
Oriented cell division and establishment of cell polarity are central to development of many organisms. Yeast cells exhibit simple, defined patterns of oriented cell divisions by choosing a specific bud site on their cell cortex. The site for bud formation is determined in a genetically programmed manner that depends on cell type. In a and α haploid cells, mother cells select a bud site immediately adjacent to their previous daughter, and daughter cells bud toward their mothers. This is the axial pattern. In a/α diploid cells, mother cells select bud sites adjacent to their daughter cells or on the opposite end of the cell, whereas daughter cells always choose a bud site directed away from their mother. This is the bipolar pattern (Freifelder 1960; Hicks et al. 1977; Chant and Herskowitz 1991). It is believed that cells respond to cortical cues that mark positions on the cell cortex to establish these cell type-specific budding patterns (Chant and Herskowitz 1991; Flescher et al. 1993).
Genetic screens have identified a large number of nonessential genes that are necessary for budding in the correct location. These genes have been termed BUD (bud-site selection) and can be divided into three distinct classes based on their requirement in the different cell types. The first class, which includes BUD3, BUD4, AXL1, and AXL2/BUD10, is required in a and α cells but not in a/α cells. Genetic analyses suggest that these gene products might provide the cortical marks for axial budding, as mutations in these genes specifically affect the axial pattern (Chant and Herskowitz 1991; Fujita et al. 1994; Halme et al. 1996; Roemer et al. 1996). The second class of genes, which includes SPA2, PEA2, and BUD6–BUD9, is required specifically in a/α cells. Mutations in any of these genes cause a random budding pattern in a/α cells but do not affect the axial budding pattern of a and α cells (Snyder 1989; Valtz and Herskowitz 1996; Zahner et al. 1996). These genes are hypothesized to determine the bipolar cortical cues. The third class of genes includes BUD1 (also known as RSR1), BUD2, and BUD5, which are required for bud-site selection in all cell types: mutants defective in any of these genes exhibit a random budding pattern in a, α, and a/α cells (Bender and Pringle 1989; Chant and Herskowitz 1991; Chant et al. 1991). It has been proposed that Bud1p, Bud2p, and Bud5p function as general bud-site selection machinery that brings proteins required for bud formation to the cell type-specific landmarks (Park et al. 1997). BUD1 encodes a protein with strong similarity to the Ras family of proteins. Subsequent studies demonstrated that it is a GTPase (Holden et al. 1991; Park et al. 1993). BUD2 encodes a GTPase-activating protein (GAP) for Bud1p: Bud2p has sequence similarity to the Ras GAP family of proteins and activates GTP hydrolysis by Bud1p (Park et al. 1993). BUD5 encodes a GDP–GTP exchange factor (GEF) for Bud1p (Zheng et al. 1995). Bud1p, Bud2p, and Bud5p thus constitute a functional GTPase module—a GTPase and its regulatory proteins—that is necessary for bud-site selection. A component of this module may interact with an axial- or bipolar-specific component to establish cell type-specific budding patterns.
We proposed previously that the Bud1 GTPase module directs bud-site assembly to occur at specific locations by recruiting components required for bud formation to that site. Subsequent work showed that Bud1p interacts with proteins necessary for bud-site assembly (Cdc24p, Bem1p, Cdc42p) in a guanine nucleotide-dependent manner (Zheng et al. 1995; Park et al. 1997; H.-O. Park and I. Herskowitz, unpubl.), which leads to the hypothesis that Bud1p brings these proteins to the correct site. Recruiting these proteins to the presumptive bud site is thought to direct the cytoskeleton and secretory apparatus toward the bud site, thereby restricting new growth to the bud.
To understand the mechanism of action of the Bud1 GTPase module, it is crucial to determine whether any of its components are localized at the presumptive bud site. Bud1p is not localized to the incipient budding site but rather is distributed uniformly around the plasma membrane (Michelitch and Chant 1996) and thus does not appear to be sufficient to direct positioning of bud-site assembly proteins. Localization of a regulator, Bud2p or Bud5p, may be crucial to direct bud-site assembly to the correct location. Previously, we hypothesized that Bud2 protein is localized to the presumptive bud site, where it stimulates conversion of Bud1–GTP to Bud1–GDP, ultimately leading to activation of Cdc42 at the bud site (Park et al. 1997). Here we show that Bud2 protein is localized at the presumptive bud site in the different cell types and is subsequently localized at the mother-bud neck.
Results
Subcellular localization of Bud2p in different cell types
The localization of Bud2p in haploid yeast cells was determined using a Bud2–GFP (green fluorescent protein) fusion. A plasmid (pHP726) expressing Bud2–GFP was constructed by inserting a 720-bp DNA fragment encoding GFP just after the start codon of BUD2 (see Materials and Methods). The plasmid was then inserted into the BUD2 locus of a bud2Δ strain (HPY24). The resulting strain (HPY252) budded in an axial manner (Table 1), indicating that the Bud2–GFP has Bud2p activity. About 20%–30% of the unbudded cells displayed localized Bud2–GFP in a patch at the presumptive bud site (Fig. 1A, a,c). More than 95% of cells with small- and medium-sized buds displayed a ring of Bud2–GFP encircling the mother-bud neck (Fig. 1A, a,b). Among those, 30%–40% of cells showed a double ring structure at the neck. Cells with a larger bud showed either very faint signal at the neck or no localized Bud2–GFP signal (data not shown). Bud2–GFP localized similarly in haploid α cells (data not shown). The previous bud site was marked by staining bud scars with Calcofluor, and the relative position of Bud2–GFP to the bud scar was ascertained. Bud2–GFP appeared immediately adjacent to the bud scar in 90%–95% of unbudded cells that had a localized Bud2–GFP signal (Fig. 1A, c), confirming that Bud2–GFP localizes at the axial position.
Table 1.
Budding patterns of yeast strains expressing Bud2–GFP
| Strainsa
|
Relevant genotype
|
Axial (%)
|
Bipolar (%)
|
Random (%)
|
|---|---|---|---|---|
| HPY22 | a BUD2 | 93 | 2 | 5 |
| HPY24 | a bud2Δ | 1 | 3 | 96 |
| HPY252 | a BUD2–GFP | 90 | 4 | 6 |
| HPY23 | α BUD2 | 90 | 3 | 7 |
| HPY293 | α BUD2–GFP | 89 | 3 | 8 |
| HPY19 | a/α BUD2/BUD2 | 6 | 70 | 24 |
| HPY40 | a/α bud2Δ/bud2Δ | 0 | 6 | 94 |
| HPY294 | a/α BUD2–GFP/ BUD2–GFP | 1 | 61 | 37 |
The isogenic strains derived from HPY22 (Park et al. 1993).
The budding patterns of log-phase cells was determined by Calcofluor staining as described in Pringle et al. (1989). Cells (200–300) were analyzed for each strain; budding pattern is given as percent of cells exhibiting each pattern.
Figure 1.
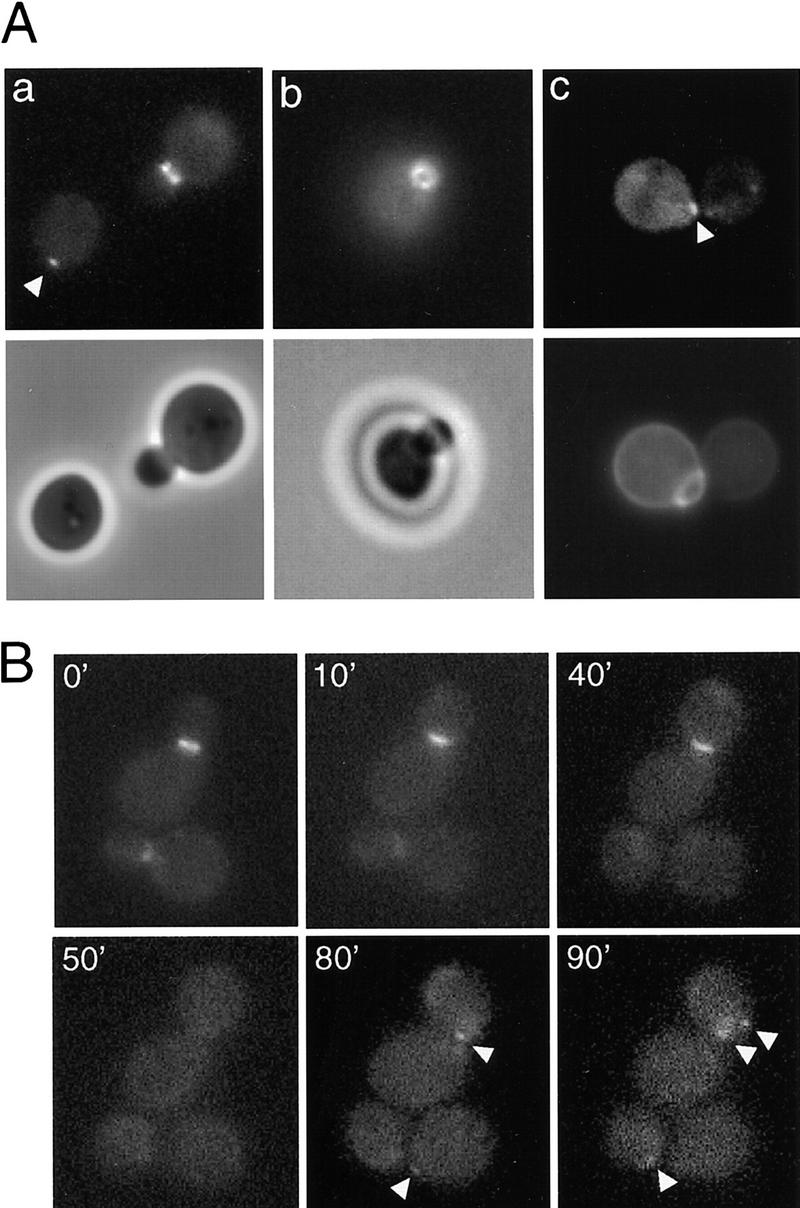
Localization of Bud2 protein in haploid a cells. (A) Localization of Bud2–GFP in a cells. Cells of wild-type haploid strain HPY252 (a BUD2–GFP) were grown to early log phase and visualized by fluorescence microscopy. (a,b, bottom) Phase-contrast images; (c, bottom) a fluorescent image of a bud scar stained with Calcofluor. Arrowheads indicate BUD2–GFP signal at incipient bud sites. (B) Time-lapse images of Bud2–GFP in mitotically growing haploid a cells. Cells of strain HPY252 were examined by time-lapse microscopy as described in Materials and Methods. Numbers indicate the time in minutes from the beginning of observation. Arrowheads indicate BUD2–GFP signal at the incipient bud sites.
Next, we examined the localization of Bud2–GFP in mitotically growing a cells using time-lapse microscopy. As shown in Figure 1B, in cells with small- and medium-sized buds, Bud2–GFP localized to the mother-bud neck region. The Bud2–GFP signal at the neck became much fainter or disappeared in cells with large buds, generating unbudded cells with no Bud2–GFP signal (at 50 min). In unbudded cells, Bud2–GFP appeared to be localized in a small patch adjacent to the previous cell division site (see arrowheads at 80 and 90 min).
We also used HA epitope-tagged Bud2p, which functions as the wild-type protein, to determine its localization by indirect immunofluorescence. The localization pattern of Bud2p by indirect immunofluorescence was similar to that observed using Bud2–GFP throughout the cell cycle (data not shown), which also confirmed that the localization of Bud2p was no longer apparent in cells after mitosis (as judged by DAPI staining; data not shown).
Taken together, it appears that Bud2p localizes to the presumptive bud site (in an axial position) in unbudded a and α cells. After bud emergence, Bud2p localizes to the mother-bud neck region in cells with a small- or medium-sized bud. The neck staining of Bud2p becomes much fainter or disappears in cells with a large bud. Such cells then divide to give rise to unbudded cells without localized Bud2p.
Because a/α cells exhibit bipolar budding, whereas a or α cells exhibit axial budding, we determined the localization of Bud2p in a/α cells. The Bud2–GFP fusion restored bipolar budding to an a/α bud2Δ/bud2Δ strain (HPY294; Table 1), indicating that the fusion protein is functional in a/α cells. Twenty percent to 30% of unbudded a/α cells exhibited Bud2–GFP localization in a patch at the presumptive bud site at the proximal pole (Fig. 2a) or distal pole (Fig. 2b) of mother cells, and at the distal pole of daughter cells (Fig. 2c). Bud2–GFP also localized at the mother-bud neck in most (>95%) of cells with small- or medium-sized buds (Fig. 2d). As seen in a or α cells, ∼30% of these cells showed a double ring structure at the neck. No such signal was observed in cells expressing native Bud2p (Fig. 2e,f). Cells with a large-sized bud show either a very faint signal at the neck or no obvious Bud2–GFP signal.
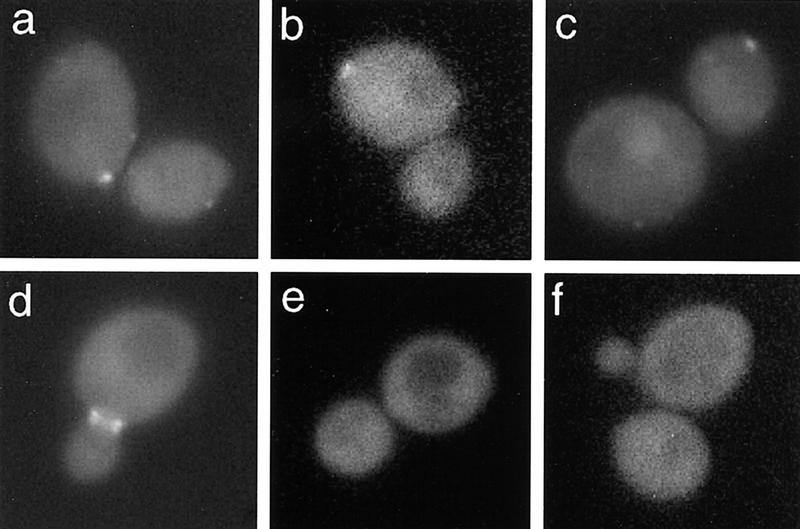
Figure 2.
Localization of Bud2–GFP in diploid a/α cells. Wild-type diploid a/α cells (HPY294) expressing Bud2–GFP were grown to early log phase and visualized by fluorescence microscopy. A mother cell with Bud2–GFP signal at the proximal pole (a) and the distal pole (b); (c) a daughter cell with Bud2–GFP signal at the distal pole; (d) a cell with Bud2–GFP signal as a double ring at the mother-bud neck; (e,f) cells expressing native Bud2.
These results suggest that Bud2p localizes to the presumptive bud site at late G1 in a/α cells, which is likely to contribute to the bipolar budding pattern of these cells. Mother a/α cells bud at their proximal or distal pole, whereas daughter cells bud preferentially at the distal pole. After bud emergence, Bud2–GFP localizes to the mother-bud neck and delocalizes later in the cell cycle.
Because localization of Bud2p changes over the cell cycle, we asked whether its level changes during the cell cycle. Bud2p levels were determined by immunoblotting extracts from yeast cells arrested at specific points of the cell cycle using drugs and also from synchronized cell cultures using α-factor. The level of Bud2p was approximately equal in both a and a/α cells and did not change noticeably over the cell cycle (data not shown).
Localization of Bud2p in the absence of Bud1p
Previously, we proposed that Bud1p guides bud-site assembly proteins to the presumptive bud site where Bud2p is located (Park et al. 1997). Based on this model, we expected that localization of Bud2p to the presumptive bud site is independent of Bud1p.
To determine whether localization of Bud2p depends on Bud1p, we examined localization of Bud2–GFP in an a bud1Δ strain (HPY289) in which the only functional Bud2p was Bud2–GFP (see Materials and Methods). Bud2–GFP localized in a patch in 20%–25% of unbudded haploid a bud1Δ cells. Among those, 50%–60% cells showed localization near the previous bud site (in an axial position) (Fig. 3A, a) and the rest at a random position (Fig. 3A, b). Interestingly, Bud2–GFP appeared to be localized in two patches in ∼10% of G1 cells—often one strong and another relatively weak signal that was not adjacent to the other. Bud2–GFP localized as a ring at the mother-bud neck in most of the bud1Δ cells (>95%) (Fig. 3A, c), all of which budded at a random site (as judged by staining with Calcofluor; data not shown). We interpret these observations to indicate that Bud2p initially localizes at the site adjacent to the previous bud site (the correct position for the axial budding pattern) and then relocalizes to a random location at bud emergence in the absence of Bud1p. The G1 cells with two patches of Bud2–GFP may be at intermediate stages when the initial patch of Bud2–GFP delocalizes at one site and a new patch develops at another site.
Figure 3.
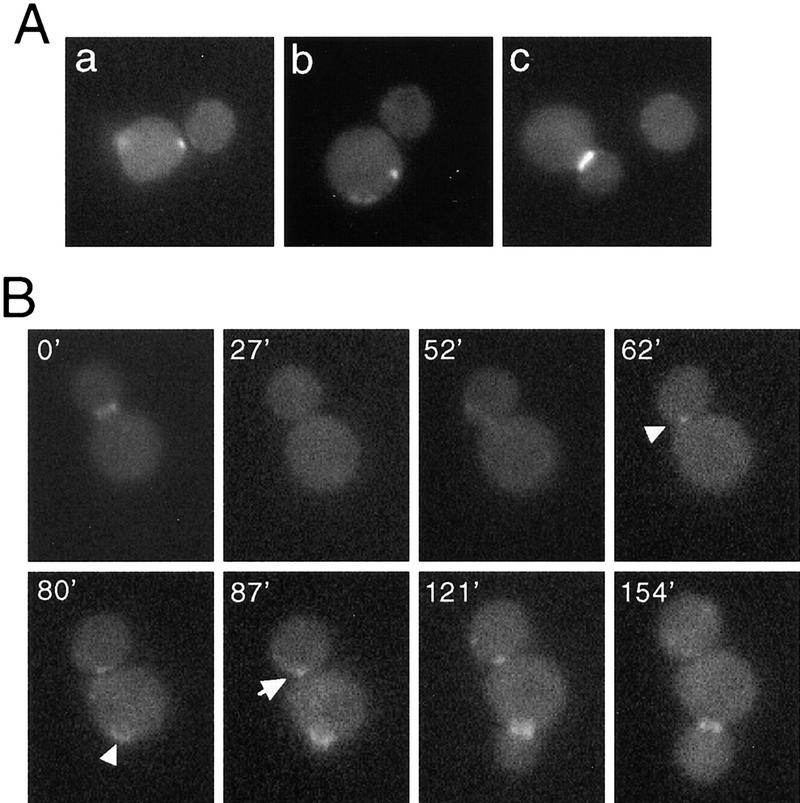
(A) Localization of Bud2–GFP in a bud1 deletion mutant. Cells of a bud1Δ bud2Δ strain (HPY289) expressing Bud2–GFP were grown to early log phase and examined by fluorescence microscopy. (a) A mother cell with Bud2–GFP signal at the axial position; (b) a mother cell with Bud2–GFP signal at a random position; (c) budded cell with Bud2–GFP signal at the mother-bud neck. (B) Time-lapse images of Bud2–GFP in a mitotically growing a bud1 deletion mutant. Cells of strain HPY289 were examined by time-lapse microscopy as described in Materials and Methods. Numbers indicate the time in minutes from the beginning of observation. Arrowheads indicate changes of Bud2–GFP signal from the axial position in G1 (62 min) to a random location (80 min) at bud emergence; the arrow indicates Bud2–GFP signal at the axial position of a daughter cell in G1 (87 min), which then disappears at the later time point (154 min).
To confirm that Bud2–GFP relocalizes at bud emergence in bud1 mutants, we used time-lapse fluorescence microscopy. Examination of Bud2–GFP in mitotically growing bud1Δ cells confirmed the interpretation from the static pictures. As shown in Figure 3B, Bud2–GFP was initially localized in a patch adjacent to the previous mother-bud junction, but the signal soon disappeared and relocalized at the mother-bud neck around bud emergence (in >90% of cells), which is no longer at the axial position (see arrowheads in Fig. 3B). These results are also consistent with the fact that cells of bud1Δ mutant bud in a random position (Bender and Pringle 1989; Chant and Herskowitz 1991).
We also examined Bud2p localization in a/α bud1Δ/bud1Δ cells (HPY297) in which Bud2–GFP was the only functional Bud2p. In G1, Bud2–GFP localized similarly in a/α bud1Δ/bud1Δ cells as in wild-type a/α cells: Bud2–GFP localized to the distal pole of daughter cells and either the proximal or distal pole of mother cells after cytokinesis. Over 90% of a/α bud1Δ/bud1Δ cells with small- or medium-sized buds had Bud2–GFP signal localized at the mother-bud neck, which was in a random location (data not shown).
In summary, in the absence of Bud1p, Bud2p initially localizes to axial or bipolar positions in G1 cells and appears to relocalize around bud emergence to a random position, finally becoming localized to the mother-bud neck until M phase. Thus, it appears that initial localization of Bud2p to the presumptive bud site in G1 cells is independent of Bud1p but that maintenance of the correct localization requires Bud1p.
Localization of Bud2p is dependent on integrity of the septins
As described above, Bud2p is localized at the mother-bud neck in all cell types and also in bud1Δ mutants. This localization is similar to that of septins, which assemble as a small ring on the cell surface prior to bud emergence and then are present at the mother-bud neck throughout the rest of the cell cycle (Byers and Goetsch 1976; Haarer and Pringle 1987; Ford and Pringle 1991; Kim et al. 1991). Although Bud2p becomes delocalized at later stages of the cell cycle and the septins remain until cytokinesis, the apparent colocalization of Bud2p and septins during the early stage of the cell cycle suggests that Bud2p might assemble on septins.
To test this hypothesis, we examined Bud2p localization in a temperature-sensitive cdc12 mutant, in which the neck filaments and associated proteins disassemble rapidly after shift from permissive (23°C) to restrictive (37°C) temperature. Bud2–GFP localized to the mother-bud neck in the cdc12-6 mutant during growth at 23°C (Fig. 4A, a), although the signal was generally weaker than the signal in the isogenic wild-type cells. Bud2–GFP was rapidly delocalized in the cdc12-6 mutant shifted to 37°C for 10 min (Fig. 4A, b). In contrast, Bud2–GFP was properly localized in the wild-type cells at both temperatures (Fig. 4A, c,d).
Figure 4.
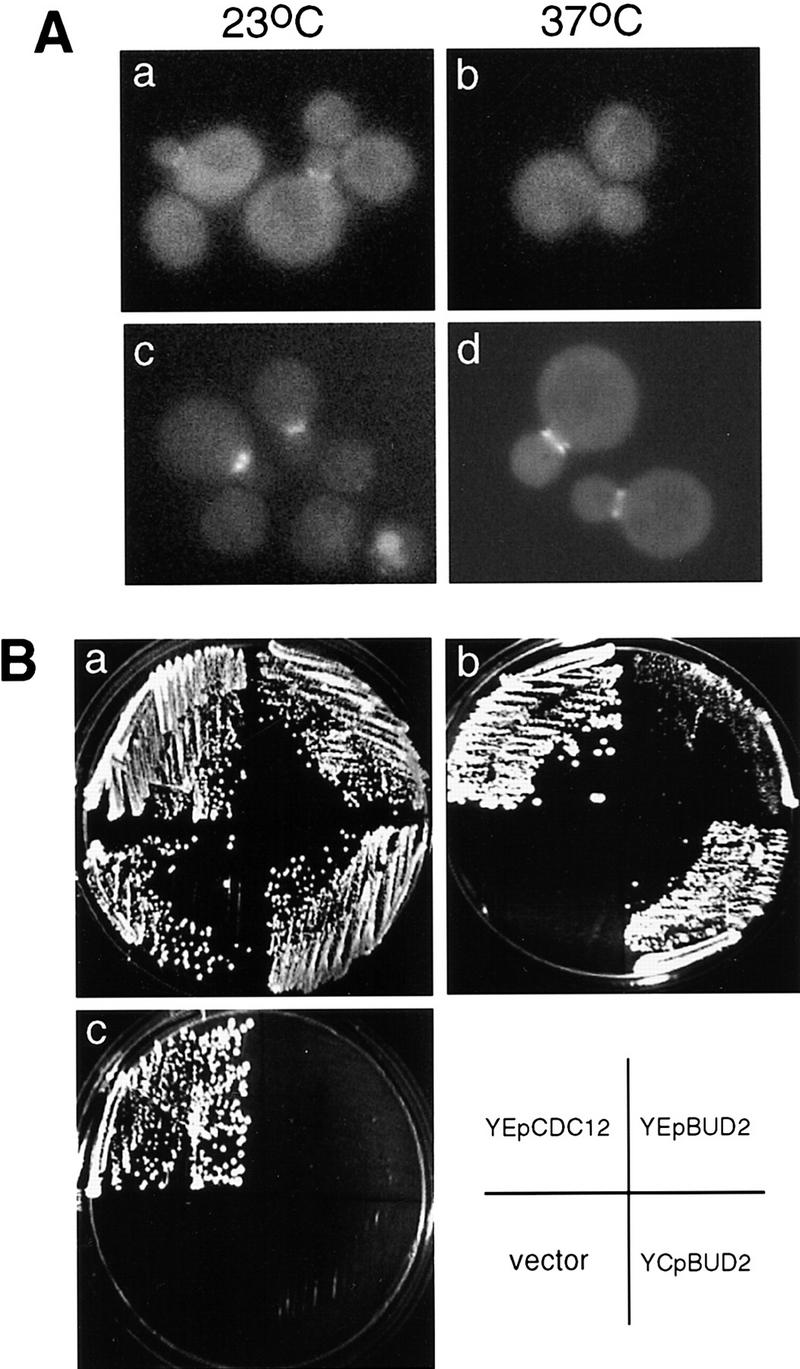
Localization of Bud2p is dependent on integrity of septins. (A) Bud2–GFP in a cdc12-6 mutant. Cells of haploid a cdc12-6 bud2Δ strain (HPY279) carrying a plasmid expressing Bud2–GFP (pHP725) (a,b) and the parental strain a bud2Δ (HPY24) carrying pHP725 (c,d) were grown at 23°C and shifted to 37°C for 10 min and examined by fluorescence microscopy. Over 80% of the cdc12-6 cells with small- or medium-sized bud showed Bud2–GFP at the neck at 23°C, but none of the mutant cells showed localized Bud2–GFP signal at 37°C. In contrast, >90% of wild-type cells with small- or medium-sized bud showed Bud2–GFP signal at the neck at both temperatures. (B) BUD2 can partially suppress temperature-sensitive growth of a cdc12-6 mutant. cdc12-6 mutants carrying YEpCDC12, YEpBUD2, YCpBUD2, or the vector control (YCp50) were streaked at 23°C (a), 30.5°C (b), and 37°C (c).
BUD2 also exhibited genetic interactions with CDC12. Temperature-sensitive growth of the cdc12-6 mutant was partially suppressed by extra copies of the BUD2 gene. The cdc12-6 mutant carrying BUD2 on a low copy plasmid (YCpBUD2) or a multicopy plasmid (YEpBUD2) grew at 30.5°C but not at 37°C, whereas the cdc12-6 mutant carrying the vector control (YCp50) could not grow at either temperature (Fig. 4B). The low copy BUD2 plasmid restored growth to cdc12-6 mutants at 30.5°C better than the multicopy plasmid did. Taken together, these observations are consistent with the idea that Bud2p and the septin proteins physically interact.
Discussion
The Bud1 GTPase module is required for proper bud site selection in all cell types. It has been proposed that some components of this module recognize cell type-specific spatial signals, which then direct bud-site assembly to occur at that site (Chant and Herskowitz 1991; Chant and Pringle 1995; Park et al. 1997). Bud1p is not localized to the incipient budding site but, rather, is distributed uniformly around the plasma membrane (Michelitch and Chant 1996). Here we show that Bud2p is initially localized to the presumptive bud site and that its localization changes over the cell cycle. Bud5p is also initially localized at the presumptive bud site (H.-O. Park, unpubl.; J. Chant, pers. comm.). Although we have not shown directly that localization of Bud2p to the presumptive bud site is required for the proper budding pattern, its distinct localization in a, α, and a/α cells in G1 correlates with the budding pattern of these cell types. Bud2p may recognize an axial-specific component such as Bud3p or Bud4p in haploid a and α cells during G1 and then direct localization of bud-site assembly proteins and septins to this site. It remains to be tested whether localization of Bud2p in G1 cells is dependent on Bud3p and Bud4p. Likewise, Bud2p might be localized to the distal or proximal poles of a/α cells by interacting with proposed bipolar-specific landmarks in G1 (Zahner et al. 1996). Our observations indicate that Bud2p and perhaps Bud5p as well may link the bud-site landmark to the polarity establishment proteins.
The localization, but not the level, of Bud2p changes during the cell cycle. Bud2p is initially localized to a small patch at the presumptive bud site in G1 and then to the mother-bud neck region in cells with small- and medium-sized buds. Cells with large-sized buds do not exhibit localized Bud2p and generate new cells that lack localized Bud2p. Although it has been reported previously that BUD2 mRNA levels fluctuate about two- to threefold over the cell cycle (Cvrčková and Nasmyth 1993; Cho et al. 1998), Spellman et al. (1998) find little variation of the BUD2 mRNA levels over the cell cycle, and we detected no significant change in the level of Bud2p over the cell cycle (data not shown). We conclude that the loss of Bud2p localization at mitosis is not due to decreased levels of the protein but, rather, to delocalization. Perhaps Bud2p undergoes a cell cycle-dependent modification, which alters its interaction with other proteins at the neck.
Bud2p is localized to the mother-bud neck just after bud emergence in all cells tested: wild-type a, α, a/α, and a bud1 and a/α bud1/bud1 mutants. Several observations indicate that localization of Bud2p at the neck might be due to its interaction with septins. First, localization of Bud2p at this position requires the septin Cdc12p. Second, extra copies of BUD2 partially suppress a mutation in one of the septin genes (cdc12-6). Finally, overexpression of BUD2 causes a partial defect in cytokinesis (H.-O. Park, unpubl.). It is not clear yet what functions, if any, Bud2p has at the neck after bud emergence. Genetic interactions between bud2 and cln1 cln2 have suggested that Bud2p might have an additional role in bud emergence other than determining its position (Benton et al. 1993; Cvrčková and Nasmyth 1993). This role may simply be to switch Bud1–GTP to Bud1–GDP, thus allowing targeted release of proteins such as Cdc24p to the bud site (Benton et al. 1993; Park et al. 1993, 1997). Several proteins involved in bud-site selection, such as Bud3p, Bud4p, and Axl2p/Bud10p, are localized to the mother-bud neck (Chant et al. 1995; Halme et al. 1996; Roemer et al. 1996; Sanders and Herskowitz 1996). It is unlikely that localization of Bud2p to the neck results from interaction with Bud3p or Bud4p, as these proteins are not at this position during S and G2.
Localization of Bud2p does not require Bud1p. In the absence of Bud1p, Bud2p initially localizes to a patch at an axial position in haploid unbudded cells, as in wild-type cells. Bud2p then relocalizes to the mother-bud neck, which in bud1 mutants occurs in a random position relative to the initial patch of Bud2p. We suggest that the initial position of Bud2p during G1 is not influenced by Bud1p but, rather is determined by association with, for example, Bud3p/Bud4p. Bud2p then recruits Bud1p and associated proteins to the bud site, which leads to bud emergence at this position.
Our findings support the view that Bud2p itself is able to recognize the cell type-specific spatial information and then directs polarity establishment through the Bud1 GTPase. We showed previously that Bud2p is a GAP for Bud1p and that its GAP activity is essential for proper bud-site selection (Park et al. 1993). The findings reported here indicate that Bud2p has two important roles in bud-site selection—a structural role to identify the presumptive bud site and a regulatory role as a GAP for Bud1p, thus allowing activation of bud-site assembly at the proper bud site.
Materials and methods
Plasmids
To create a plasmid expressing Bud2p fused to GFP, first, a NotI site was introduced just after the start codon (ATG) of BUD2 using PCR, resulting in pHP724. A 720-bp fragment coding for GFP and the flanking NotI recognition sites was amplified by PCR using two primers, GFP-N and GFP-C, and the plasmid pKS-GFP2+ (a kind gift of E. O’Shea, University of California, San Francisco). The sequence of primer GFP-N, 5′-ATAAGAATGCGGCCGCATGAGTAAAGGAGAAGAA-3′, includes the start codon and codons for the first five amino acids of GFP. The sequence of primer GFP-C, 5′-ATAAGAATGCGGCCGCATTTGTATAGTTCATCCAT-3′, includes codons for the six carboxy-terminal amino acids of GFP. The resulting PCR product carrying GFP (F64L, S65T) was cloned into the NotI site of plasmid pHP724, generating pHP725. To create an integrating plasmid expressing Bud2–GFP, a 5-kb HindIII–SalI fragment from pHP725 was cloned into pRS304 (Sikorski and Hieter 1989), resulting in pHP726.
Yeast strains and growth conditions
Standard methods were used for growth, transformation, and genetic manipulations of Saccharomyces cerevisiae, as described in Rose et al. (1990).
Haploid a (HPY252) and α (HPY293) strains expressing Bud2–GFP were constructed by integration of a linearized plasmid (pHP726) into the BUD2 locus of a bud2Δ strain (HPY24) (Park et al. 1993). A diploid a/α strain expressing Bud2–GFP (HPY294) was constructed by mating HPY252 and the isogenic α strain (HPY293).
To avoid potential competition between wild-type Bud2p and Bud2–GFP fusion protein, we constructed a bud1Δ bud2Δ strain (HPY289) expressing Bud2–GFP by one-step gene disruption (Rothstein 1991) of HPY263 using plasmid pHP529 (Park et al. 1993), following targeted integration of the plasmid pHP726 to the BUD2 locus. A diploid a/α bud1Δ/bud1Δ bud2Δ/bud2Δ strain (HPY297) expressing Bud2–GFP was constructed by mating HPY289 to the isogenic α strain, HPY296.
Microscopy
Epifluorescence microscopy was carried out with a Nikon Eclipse microscope fitted with a 100× immersion objective (N.A. = 1.30) and standard filter cube sets. Images were collected using a Micromax digital camera (Princeton Instruments) for each time point. To minimize the drift of yeast cells over time, a drop of yeast cells was usually placed on top of agarose containing an appropriate medium. To minimize photo bleaching over time, an ND4 filter was used. Images were analyzed with IPLab software (Signal Analytics Corporation, Vienna, VA), and the figures were created with ClarisWorks. Manipulations of the images were confined to adjusting contrast and brightness and the addition of markers.
Acknowledgments
We thank R. Tabtiang and A. Sil for comments on the manuscript; J. Pringle and the members of the Herskowitz laboratory for valuable discussions; and R. Li, J. Pringle, E. O’Shea, and D. Kellogg for yeast strains, plasmids, and antibodies. This work was supported by a grant from the National Institutes of Health (GM48052 to I.H.), a Basil O’Connor Starter Scholar Research Award (5-FY98-0697) from the March of Dimes Birth Defects Foundation (to H.-O.P.), and an award from the American Cancer Society, Ohio Division (to H.-O.P.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL park.294@osu.edu; FAX (614) 292-4466.
References
- Bender A, Pringle JR. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc Natl Acad Sci. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B, Tinkelenberg AH, Jean D, Plump SD, Cross FR. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–5275. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Corrado K, Pringle JR, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cvrckova F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford SK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: Localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Bud position in Saccharomyces cerevisiae. J Bacteriol. 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature. 1994;372:567–570. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Pringle JR. Immunofluorescence Localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10 nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Michelitch M, Mitchell EL, Chant J. Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr Biol. 1996;6:570–579. doi: 10.1016/s0960-9822(02)00543-2. [DOI] [PubMed] [Google Scholar]
- Hicks JB, Strathern JN, Herskowitz I. Interconversion of yeast mating types. III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics. 1977;85:373–393. doi: 10.1093/genetics/85.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden JL, Nur-E-Kamal MSA, Fabri L, Nice E, Hammacher A, Maruta H. Rsr1 and Rap1 GTPases are activated by the same GTPase-activating protein and require threonine 65 for their activation. J Biol Chem. 1991;266:16992–16995. [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitch M, Chant J. A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr Biol. 1996;6:446–454. doi: 10.1016/s0960-9822(02)00512-2. [DOI] [PubMed] [Google Scholar]
- Park H-O, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- Park H-O, Bi E, Pringle J, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Roemer T, Madden K, Chang J, Snyder M. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes & Dev. 1996;10:777–793. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. pp. 1–123. [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sanders S, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner J, Harkins HI, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Bender A, Cerione RA. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J Biol Chem. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]