Abstract
Dynamic changes in intracellular calcium concentration in response to various stimuli regulates many cellular processes such as proliferation, differentiation, and apoptosis1. During apoptosis, calcium accumulation in mitochondria promotes the release of pro-apoptotic factors from the mitochondria into the cytosol2. It is therefore of interest to directly measure mitochondrial calcium in living cells in situ during apoptosis. High-resolution fluorescent imaging of cells loaded with dual-excitation ratiometric and non-ratiometric synthetic calcium indicator dyes has been proven to be a reliable and versatile tool to study various aspects of intracellular calcium signaling. Measuring cytosolic calcium fluxes using these techniques is relatively straightforward. However, measuring intramitochondrial calcium levels in intact cells using synthetic calcium indicators such as rhod-2 and rhod-FF is more challenging. Synthetic indicators targeted to mitochondria have blunted responses to repetitive increases in mitochondrial calcium, and disrupt mitochondrial morphology3. Additionally, synthetic indicators tend to leak out of mitochondria over several hours which makes them unsuitable for long-term experiments. Thus, genetically encoded calcium indicators based upon green fluorescent protein (GFP)4 or aequorin5 targeted to mitochondria have greatly facilitated measurement of mitochondrial calcium dynamics. Here, we describe a simple method for real-time measurement of mitochondrial calcium fluxes in response to different stimuli. The method is based on fluorescence microscopy of 'ratiometric-pericam' which is selectively targeted to mitochondria. Ratiometric pericam is a calcium indicator based on a fusion of circularly permuted yellow fluorescent protein and calmodulin4. Binding of calcium to ratiometric pericam causes a shift of its excitation peak from 415 nm to 494 nm, while the emission spectrum, which peaks around 515 nm, remains unchanged. Ratiometric pericam binds a single calcium ion with a dissociation constant in vitro of ~1.7 μM4. These properties of ratiometric pericam allow the quantification of rapid and long-term changes in mitochondrial calcium concentration. Furthermore, we describe adaptation of this methodology to a standard wide-field calcium imaging microscope with commonly available filter sets. Using two distinct agonists, the purinergic agonist ATP and apoptosis-inducing drug staurosporine, we demonstrate that this method is appropriate for monitoring changes in mitochondrial calcium concentration with a temporal resolution of seconds to hours. Furthermore, we also demonstrate that ratiometric pericam is also useful for measuring mitochondrial fission/fragmentation during apoptosis. Thus, ratiometric pericam is particularly well suited for continuous long-term measurement of mitochondrial calcium dynamics during apoptosis.
Keywords: Cellular Biology, Issue 50, Ratiometric pericam, mitochondria, calcium, apoptosis, staurosporine, live cell imaging
Protocol
1. Pericam-mt Transfection and Cell Preparation.
Plate HeLa cells the night before transfection on sterile glass coverslips placed in a standard 6-well culture plate.
Prepare DNA/Lipofectamine 2000 complexes as suggested by the manufacturer (Invitrogen). We use 4 μg of ratiometric-pericam-mt expression vector and 10 μL of Lipofectamine 2000 diluted in 0.5 mL of Opti-MEM for each well. The cells should be ~70% confluent before transfection.
Replace HeLa culture media (Dulbecco's modified eagles medium, 10% FBS, penicillin/streptomycin) in each well with 1.5 mL of the same media without antibiotics.
Add the DNA/Lipofectamine complexes to each well to be transfected. Mix gently with rocking and incubate for 4 hours in a cell culture incubator at 37oC, 5 % CO2.
Replace antibiotic-free media with culture media with antibiotics. Allow cells to express ratiometric pericam for 1-2 days before imaging.
2. Microscope Setup and Image Acquisition
Place a coverslip with HeLa cells into an appropriate imaging chamber with imaging solution: 107mM NaCl, 7.2mM KCl, 1.2mM MgCl2, 1mM CaCl2,11.5mM glucose, 0.1% bovine serum albumin, and 20mM HEPES 7.2. Our laboratory uses the Attofluor cell chamber from Invitrogen. Mount the imaging chamber in an inverted microscope stage.
Find an area of interest containing one or more cells expressing ratiometric pericam using a 40x (or higher) oil immersion objective. We have found that ratiometric pericam is less resistant to photobleaching at 380 nm, and thus we use this wavelength to identify suitable cells. For the images acquired in Figure 1, a Nikon Superfluor 40X objective with a 1.3 numerical aperture was used. Higher magnification objectives would also be appropriate (60-100x). Excitation filters used were Chroma Technology filters D380/30x (380 +/- 30 nm) and D495/10 (495 +/- 5 nm), beamsplitter was 505DCXR (505nm longpass), and emission filter was HQ535/50m (535 +/- 25nm). Our microscope used to acquire the images in Figure 1 consists of a Nikon TE2000 inverted microscope equipped with a Roper Scientific Coolsnap HQ cooled CCD camera, Ludl MAC6000 rapid filter wheel and changer, and a MetaFluor data acquisition and analysis software. This protocol can be adapted to any standard wide-field microscope with the appropriate filters and software capable of capturing and processing ratiometric images.
Set up the software for dual excitation using the 495 and 380 nm filters. Calcium binding to ratiometric pericam increases absorbance at 495 nm, thus the ratio should be set up to be 495 nm/380 nm. Ratiometric pericam has a calcium-free excitation peak at 415 nm4. In this protocol we suggest using a 380 nm filter only because it is ubiquitous in most calcium imaging laboratories. If a 410 nm filter is available, it is preferable to the 380 nm filter.
Acquire images sequentially by alternatively exciting at 495 and 380 nm. Acquire images of baseline calcium levels for at least 30 s before application of agonist via a perfusion apparatus. Mark the addition time for each agonist. Exposure times and acquisition intervals should be optimized to prevent photobleaching while still allowing sufficient temporal resolution.For example, for semi-rapid calcium dynamics monitored in response to agonist stimulation, images were acquired every two seconds in Figures 1B and C. Much faster acquisition rates are also possible6. Long-term imaging after staurosporine administration was acquired with a longer interval (30s) between acquisitions (Figures 1 D and E).
3. Image Processing and Analysis
Once the experiment is complete, obtained images can be analyzed offline. Define regions of interest (ROI) in which you want to monitor changes in calcium levels. Use a ROI selected within an empty area of the field to subtract background if necessary. It should be noted that mitochondria are quite motile and dynamic, and extrapolating events in a single mitochondrion over extended periods is not always possible. Thus, considering the high motility of this organelle in some cell types, selection of ROI should be carefully monitored. Furthermore, as evidenced in Figure 1, mitochondria response heterogeneously to various stimuli. It is therefore important to analyze data offline and monitor RO1 placement on a frame-by-frame basis. A detailed description of measuring mitochondrial calcium in highly motile mitochondria has been described elsewhere7.
Obtained ratio measurements from the ROI can be imported into Excel or similar graphingsoftware. Data can be presented as a trace representing changes in mitochondrial calcium levels in a single cell or single mitochondrion, or averaged fluorescence signals from several ROI. We have also used this technique to measure average mitochondrial calcium levels in a population of lymphocytes undergoing apoptosis induced by Fas ligand8.
4. Representative Results:
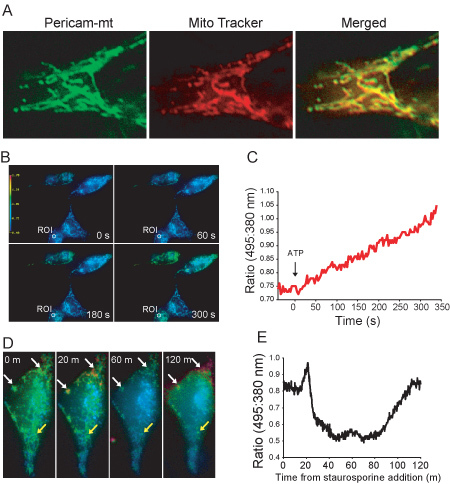 Figure 1. (A) Subcellular localization of ratiometric-pericam-mt. This first image shows live HeLa cell expressing ratiometric-pericam-mt. The second image shows fluorescent staining with mitochondrion-selective dye MitoTracker Red CMXRos. The yellow fluorescence in the merged image demonstrates co-localization of ratiometric-pericam-mt and MitoTracker fluorescence. (B) A series of pseudocolor ratio (495:380 nm) images of 4 HeLa cells expressing mt-ratiometric pericam treated with 10 mM ATP. (C) Quantification of changes in mitochondrial calcium levels in the region of interest (ROI) indicated in (B) treated with 10 mM ATP. (D) Series of pseudocolor ratio (495:380 nm) images of a HeLa cell expressing mt-ratiometric pericam treated with 0.5 μM staurosporine. Heterogeneity in the calcium response in individual mitochondria is evident, as well as significant fragmentation of mitochondria by 60 minutes. Some mitochondria have oscillatory increases in calcium (white arrow head), whereas others do not show significant changes in calcium level (yellow arrow head).(E) Quantification of increases in global mitochondrial calcium levels in a single HeLa cell after induction of apoptosis with 0.5 μM staurosporine.
Figure 1. (A) Subcellular localization of ratiometric-pericam-mt. This first image shows live HeLa cell expressing ratiometric-pericam-mt. The second image shows fluorescent staining with mitochondrion-selective dye MitoTracker Red CMXRos. The yellow fluorescence in the merged image demonstrates co-localization of ratiometric-pericam-mt and MitoTracker fluorescence. (B) A series of pseudocolor ratio (495:380 nm) images of 4 HeLa cells expressing mt-ratiometric pericam treated with 10 mM ATP. (C) Quantification of changes in mitochondrial calcium levels in the region of interest (ROI) indicated in (B) treated with 10 mM ATP. (D) Series of pseudocolor ratio (495:380 nm) images of a HeLa cell expressing mt-ratiometric pericam treated with 0.5 μM staurosporine. Heterogeneity in the calcium response in individual mitochondria is evident, as well as significant fragmentation of mitochondria by 60 minutes. Some mitochondria have oscillatory increases in calcium (white arrow head), whereas others do not show significant changes in calcium level (yellow arrow head).(E) Quantification of increases in global mitochondrial calcium levels in a single HeLa cell after induction of apoptosis with 0.5 μM staurosporine.
Discussion
Here we present a very simple method for measuring mitochondrial calcium using mitochondrial-targeted ratiometric pericam. As shown in Figure 1A, using standard widefield optics with no deconvolution it is possible to easily view individual mitochondria in HeLa cells with acceptable signal-to-noise ratio. This is because HeLa cells, like most cells in culture, flatten out significantly when adherent obviating the need for confocal microscopy or other specialized equipment. We have found similar results in Jurkat cells adhered to poly-lysine coated coverslips8. In contrast to methodologies using dyes, it is also possible to non-invasively monitor mitochondrial calcium levels for hours using genetically encoded calcium indicators such as ratiometric pericam (Figure 1E). This is especially important when analyzing mitochondrial calcium during cell death. Furthermore, it is also possible to visualize fission/fragmentation of mitochondria during the apoptotic process. As shown in Figures 1D and E, staurosporine treatment causes a slow increase in calcium in select subpopulations of mitochondria which peaks at 20 minutes. Calcium levels go down again concomitant with mitochondrial fragmentation before going up again 2 hours after treatment. This is consistent with the slow waves in cytosolic calcium induced by staurosporine treatment measured using Fura-29. Thus, important kinetic information can be obtained which is not possible with methods employing static measurements. Although we have presented ratios and not calcium concentrations in Figure 1, it is possible to calibrate the sensor in situ to calculate absolute calcium levels4, 10. One important caveat to consider is that ratiometric pericam is sensitive to pH4, 10. As both cytosolic and mitochondrial pH levels can change dramatically during apoptosis11, this is an important consideration. Titration of pH in isolated mitochondria expressing pericam demonstrate that increasing [H+] increases emission of pericam when excited at 495 nm, with little effect on emission stimulated by excitation at 410 nm, a property which has been exploited to simultaneously measure both mitochondrial calcium and pH12. Thus, selectively monitoring emission with 410 nm excitation provides a means to non-ratiometrically monitor mitochondrial calcium with reduced concern for pH. Finally, the protocol presented here only requires access to a standard epifluorescent microscope with widely available filters, thus making this technique accessible to most laboratories.
Disclosures
No conflicts of interest declared.
Acknowledgments
We would like to thank Atsushi Miyawaki for providing us with the ratiometric-pericam-mt expression construct. This work was supported by grant GM081685 from the National Institutes of Health (DB).
References
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G. Mitochondrial calcium signalling and cell death: approaches for assessing the role of mitochondrial Ca2+ uptake in apoptosis. Cell Calcium. 2006;40:553–560. doi: 10.1016/j.ceca.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2. Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Simpson AW, Brini M, Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- Miyawaki A. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Shimozono S. Confocal imaging of subcellular Ca2+ concentrations using a dual-excitation ratiometric indicator based on green fluorescent protein. Sci STKE. 2002. pp. pl4–pl4. [DOI] [PubMed]
- Wozniak AL. Requirement of biphasic calcium release from the endoplasmic reticulum for Fas-mediated apoptosis. J Cell Biol. 2006;175:709–714. doi: 10.1083/jcb.200608035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehning D. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 2003;5:1051–1061. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
- Csordas G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–147. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]


