Abstract
The diagnosis of viral causes of many infectious diseases is difficult due to the inherent sequence diversity of viruses as well as the ongoing emergence of novel viral pathogens, such as SARS coronavirus and 2009 pandemic H1N1 influenza virus, that are not detectable by traditional methods. To address these challenges, we have previously developed and validated a pan-viral microarray platform called the Virochip with the capacity to detect all known viruses as well as novel variants on the basis of conserved sequence homology1. Using the Virochip, we have identified the full spectrum of viruses associated with respiratory infections, including cases of unexplained critical illness in hospitalized patients, with a sensitivity equivalent to or superior to conventional clinical testing2-5. The Virochip has also been used to identify novel viruses, including the SARS coronavirus6,7, a novel rhinovirus clade5, XMRV (a retrovirus linked to prostate cancer)8, avian bornavirus (the cause of a wasting disease in parrots)9, and a novel cardiovirus in children with respiratory and diarrheal illness10. The current version of the Virochip has been ported to an Agilent microarray platform and consists of ~36,000 probes derived from over ~1,500 viruses in GenBank as of December of 2009. Here we demonstrate the steps involved in processing a Virochip assay from start to finish (~24 hour turnaround time), including sample nucleic acid extraction, PCR amplification using random primers, fluorescent dye incorporation, and microarray hybridization, scanning, and analysis.
Keywords: Immunology, Issue 50, virus, microarray, Virochip, viral detection, genomics, clinical diagnostics, viral discovery, metagenomics, novel pathogen discovery
Protocol
The steps of the Virochip assay include (1) nucleic acid extraction from clinical samples, (2) reverse transcription of extracted RNA and 2nd-strand cDNA synthesis, (3) PCR amplification of randomly primed cDNA, (3) Cy3 fluorescent dye incorporation, (4) hybridization of labeled material to the Virochip microarray, and (5) scanning and analysis (Figure 1). The protocol shown below will demonstrate the use of the Virochip assay on a nasal swab sample from a child with an influenza-like illness.
Primer Sequences
Primer A 5'-GTTTCCCAGTCACGATA-(N9)-3'
Primer B 5'-GTTTCCCAGTCACGATA-3'
1. Nucleic Acid Extraction
Nucleic acid extraction from clinical samples should be performed under Biosafety Level 2 (BSL-2) or higher conditions, as these samples are potentially infectious.
The nasal swab sample is transported in viral holding medium. Aliquot 200 μL of the sample into a 1.5 mL Eppendorf microcentrifuge tube.
Optional: pass sample through a 0.22 μm filter (to purify for viral particles)
Optional: pretreat sample with DNAse (to degrade host genomic DNA and purify for protected, encapsidated viral nucleic acid) using the Turbo DNAse kit (Ambion) per the manufacturer's instructions
Extract the sample using the Zymo ZR Viral RNA Extraction Kit or Qiagen Ultrasens Virus Kit per the manufacturer's instructions.
2. Reverse Transcription and 2nd-strand CDNA Synthesis ("Round A")
Add 1 uL 40 pmol / μL Primer A to 4 uL extracted RNA. Heat to 65°C for 5 min in a thermocycler (e.g. GeneAmp PCR System 9700) and then let cool at room temp x 5 min.
Add 5 μL of a master mix consisting of 2 μL 5X RT buffer, 1 μL 12.5 mM dNTP, 1 μL water, 0.5 μL 0.1M DTT, and 0.5 μL SSIII RT (Invitrogen). Incubate at 42°C x 60 minutes.
In a programmable thermocycler, heat to 94°C x 2 min (to abort the reverse transcriptase reaction), cool to 10°C, pulse centrifuge, and then add 5 μL Sequenase mix consisting of 1 μL 5X Sequenase buffer, 3.8 μL water, and 0.15 μL Sequenase (USB Corporation). For 2nd strand synthesis, ramp up from 10°C to 37°C over 8 min, hold 8 min, then heat to 94°C x 2 min (to inactivate Sequenase) and cool to 10°C (hold).
Optional : Rd A sample cDNA can be stored at this point at -20°C.
3. PCR Amplification of Randomly Primed CDNA ("Round B")
Add 5 uL of Rd A sample to 45 μL of a master mix consisting of 5 μL 10X PCR buffer, 1 μL 12.5 mM dNTP, 1 μL 100 pmol / μL primer B, 1 μL KlenTaq LA (Sigma), and 37 μL water.
Run PCR protocol as follows: 94°C, 2 min →(25 cycles of 94°C, 30 s / 50°C, 45 s / 72°C, 1 min) →72°C, 5 min →10°C (hold)
Check the Rd B sample on a 1.5% agarose electrophoresis gel. A smear from approximately 200 - 1000 bp should be visualized (Figure 2A)
4. Cy3 Fluorescent Dye Incorporation
Optional : another sample can be labeled with the Cy5 fluorescent dye and hybridized to the same microarray.
To incorporate aa-dNTP, add 5 uL of Rd B sample to 45 uL of a master mix consisting of 5 uL 10X PCR buffer, 1 uL 12.5 mM aa-dNTP(Invitrogen), 1 uL 100 pmol / uL primer B, 1, 1 uL KlenTaq LA (Sigma), and 37 uL water. Run PCR protocol as follows: 94°C, 2 min →(15cycles of 94°C, 30 s / 50°C, 45 s / 72°C, 1 min) →72°C, 5 min →10°C (hold)
Clean the Rd C sample with the DNA Clean and Concentrator-5 Kit (Zymo Research) per the manufacturer's instructions. Elute in 10 μL.
Add 1 uL of 1M bicarbonate (Sigma) and 1 uL Cy5 (GE Healthcare) to the Rd C sample.
Incubate x 1 hour in the dark.
Clean the Cy3-labeled sample with the DNA Clean and Concentrator-5 Kit (Zymo Research) per the manufacturer's instructions. Elute in 12 μL.
5. Hybridization to Virochip Microarray
Optional: use 1.5 μL to check cDNA concentration and the amount of dye incorporated on a Nanodrop spectrophotometer (for normalization of samples)
In a PCR strip tube add 1 μL 25X fragmentation buffer, 5 μL 5X blocking buffer, 9.5 μL (or amount to normalize) of Cy3-labeled sample, and water to a total volume of 25 μL (Agilent)
Add 25 μL of 2X GEx hybridization buffer (Agilent). Spin down briefly to remove bubbles.
Load 40 μL of the sample onto gasket slide.
Place Agilent-printed Virochip array (University of California, San Francisco / Agilent) (side marked "Agilent" down) on top of gasket slide and fasten screw tightly.
Hybridize overnight in 65°C oven, speed setting at 10 rpm.
To wash arrays, preheat buffer 2 (Agilent) to 37°C. Take apart gasket slide / array sandwich underwater in buffer 1 (Agilent). Wash in buffer 1 x 1 min. Wash in preheated buffer 2 x 1 min. Slowly remove slide from buffer 2, being careful to avoid bubbles (use kimwipe to wick away extra moisture being careful to avoid directly touching active side of array)
Place slide into slide holder and insert into array scanner.
6. Virochip Scanning and Analysis
Scan Cy3-labeled array at 5 μm or 2 μm according to the instrument's standard scanning protocol (Figure 2B).
Analyze Virochip microarrays by visual inspection, cluster analysis, or the automated Virochip analysis program, E-Predict11. By each of these three methods, influenza virus is readily detected in the nasal swab sample (from a child with influenza-like illness) (Figure 2C)
7. Representative Results:
When the protocol is done correctly, a smear should be seen on agarose gel electrophoresis after the Rd B step (Figure 2A). The Virochip microarray on the Agilent platform will be difficult to analyze visually (Figure 2B), but if a virus is present it should be readily identified by visual inspection, cluster analysis, and / or E-Predict (Figure 1C). Samples can be spiked with measured quantities of nucleic acid from a known virus (e.g. tobacco mosaic virus; MS2 bacteriophage) to serve as a positive control for the assay.
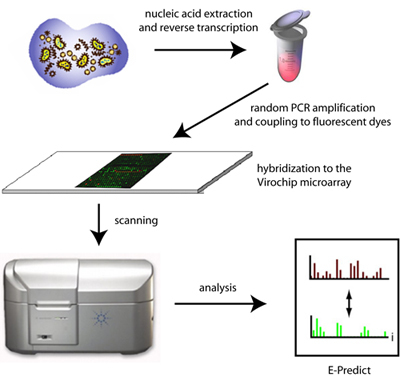 Figure 1. Schematic of Virochip microarray processing and analysis. Extracted nucleic acid from clinical samples are randomly amplified, labeled with fluorescent dyes, and hybridized to the Virochip microarray. Microarrays are scanned at 2 μm resolution and analyzed using a variety of computational tools, including E-Predict.
Figure 1. Schematic of Virochip microarray processing and analysis. Extracted nucleic acid from clinical samples are randomly amplified, labeled with fluorescent dyes, and hybridized to the Virochip microarray. Microarrays are scanned at 2 μm resolution and analyzed using a variety of computational tools, including E-Predict.
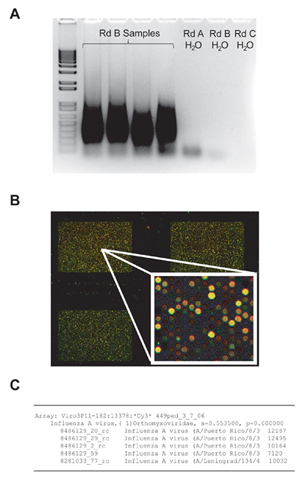 Figure 2. Steps in the Virochip assay. After amplification by random PCR, a smear of 200 - 1000 bp can be visualized by gel electrophoresis (A). (B) Three Virochip microarrays out of the 8 arrays / glass slide are shown, with a small region of one microarray blown-up in the inset on the bottom right corner. (C) Automated microarray viral analysis using E-Predict revealing the presence of influenza A virus in the clinical sample. The high similarity score (s=0.55) corresponds to a p-value that is close to zero, thus making this an extremely significant prediction.
Figure 2. Steps in the Virochip assay. After amplification by random PCR, a smear of 200 - 1000 bp can be visualized by gel electrophoresis (A). (B) Three Virochip microarrays out of the 8 arrays / glass slide are shown, with a small region of one microarray blown-up in the inset on the bottom right corner. (C) Automated microarray viral analysis using E-Predict revealing the presence of influenza A virus in the clinical sample. The high similarity score (s=0.55) corresponds to a p-value that is close to zero, thus making this an extremely significant prediction.
Discussion
The Virochip protocol as described here is complex and requires a meticulous and skilled research technician. Correct reagent concentrations and conditions for PCR, labeling, and hybridization are critical. The Virochip protocol can be easily modified to accommodate analysis of diseased tissues by use of a tissue extraction method such as TRIzol (Invitrogen) for nucleic acid extraction. Probes can be added or deleted as necessary to obtain the desired spectrum and breadth of coverage; for instance, probes for detection of nonviral targets such as bacteria and fungi can be designed and added "on-the-fly". Some applications of the Virochip assay currently under development include broad-spectrum viral diagnostics in the clinical laboratory, outbreak investigation, purity screening of drugs and vaccines, and novel viral pathogen discovery.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank David Wang, Anatoly Urisman, Amy Kistler, Kael Fischer, Patrick Tang, and Alexander Greninger for the initial development and optimization of the Virochip protocol. This work is supported by an NIH K08 grant and Abbott Discovery Award (to CC) and the Howard Hughes Medical Institute (to JLD).
References
- Wang D. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J Clin Microbiol. 2007;45:2340–2343. doi: 10.1128/JCM.00364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43:e71–e76. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY. Utility of DNA microarrays for detection of viruses in acute respiratory tract infections in children. J Pediatr. 2008;153:76–83. doi: 10.1016/j.jpeds.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota PA. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Wang D. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:e2–e2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman A. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:25–25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kistler AL. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol J. 2008;5:e25–e25. doi: 10.1186/1743-422X-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A. 2008;105:14124–14129. doi: 10.1073/pnas.0805968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urisman A. E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol. 2005;6:R78–R78. doi: 10.1186/gb-2005-6-9-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]


