Abstract
Phytochromes are informational photoreceptors through which plants adapt their growth and development to prevailing light conditions. These adaptations are effected primarily through phytochrome regulation of gene expression by mechanisms that remain unclear. We describe a new mutant, hfr1 (long hypocotyl in far-red), that exhibits a reduction in seedling responsiveness specifically to continuous far-red light (FRc), thereby suggesting a locus likely to be involved in phytochrome A (phyA) signal transduction. Using an insertionally tagged allele, we cloned the HFR1 gene and subsequently confirmed its identity with additional alleles derived from a directed genetic screen. HFR1 encodes a nuclear protein with strong similarity to the bHLH family of DNA-binding proteins but with an atypical basic region. In contrast to PIF3, a related bHLH protein previously shown to bind phyB, HFR1 did not bind either phyA or B. However, HFR1 did bind PIF3, suggesting heterodimerization, and both the HFR1/PIF3 complex and PIF3 homodimer bound preferentially to the Pfr form of both phytochromes. Thus, HFR1 may function to modulate phyA signaling via heterodimerization with PIF3. HFR1 mRNA is 30-fold more abundant in FRc than in continuous red light, suggesting a potential mechanistic basis for the specificity of HFR1 to phyA signaling.
Keywords: gravitropism, photomorphogenesis, transcription factor, high irradiance response, T-DNA tagging, light-regulated gene
Plants modify their growth and development in ways that allow them to adapt to their immediate environment. They do so by sensing a variety of environmental parameters and integrating the resulting information into coherent developmental decisions. Light is crucial to a plant's survival, and thus it is not surprising that plants have evolved an intricate set of photoreceptor systems through which they can track this parameter (Kendrick and Kronenberg 1994; Fankhauser and Chory 1997). The regulatory photoreceptors that sense red light (R) and far-red light (FR) are the phytochromes. These molecules undergo photoconversion between two spectroscopically and conformationally distinct forms, Pr (R-absorbing) and Pfr (FR-absorbing), and conversion to Pfr is required for signal transmission. We are interested in the mechanisms by which phytochrome photoconversion effects change in gene expression.
Seedling de-etiolation is not only an important phytochrome-regulated phase of development but also provides a convenient model system for dissecting the molecular basis of phytochrome signal transduction. De-etiolation can be thought of as a switch between two developmental programs: from skotomorphogenesis (or etiolation) in darkness to photomorphogenesis in light (McNellis and Deng 1995). These two programs differ in aspects that range from macroscopic morphology to the expression of a large number of light-regulated genes (Terzaghi and Cashmore 1995).
Though there are five phytochromes in Arabidopsis, designated phyA through phyE (Clack et al. 1994), two of these, phyA and phyB, predominate in the regulation of seedling de-etiolation (Reed et al. 1994). Most aspects of de-etiolation can be induced by either R or FR, with the strongest responses being induced by continuous light (Mancinelli 1994; Smith 1994). phyB predominates in responses to continuous red light (Rc), whereas responses to continuous far-red light (FRc) are exclusively mediated by phyA, providing a useful tool for distinguishing between the two photoreceptor systems (Deng and Quail 1999). The basis for the different photosensory specificities of phyA and phyB is not well understood but may result in part from differences in their abundance in Rc and FRc (Fairchild and Quail 1998; Hennig et al. 1999).
There has recently been a dramatic change in our understanding of phytochrome signal transduction, stemming from two key discoveries. First, it has been shown that, whereas phytochromes are cytoplasmic when synthesized in their Pr forms, they are induced to translocate to the nucleus by photoconversion to Pfr (Sakamoto and Nagatani 1996; Kircher et al. 1999; Yamaguchi et al. 1999). Second, the DNA-binding bHLH protein, PIF3, has been shown to bind phyB in a highly Pfr-preferential manner in vitro (Ni et al. 1999), as well as to interact with both phyA and phyB C-terminal fragments in yeast two-hybrid assays (Ni et al. 1998). The involvement of PIF3 in phyB signaling and, to a lesser extent, phyA signaling in vivo has been corroborated by alterations in light sensitivity observed in seedlings with reduced or increased PIF3 expression (Ni et al. 1998; Halliday et al. 1999). Moreover, PIF3 has been shown to bind specifically to G-box DNA motifs present in various light-regulated promoters, and phyB is induced to bind to DNA-bound PIF3 on conversion to the active Pfr form (Martinez-Garcia et al. 2000). Together, these results suggest that an important form of phytochrome regulation of gene expression is the direct interaction of activated phytochrome with sequence-specific DNA-binding proteins in the nucleus.
Genetic approaches to identification of phytochrome signaling intermediates have also been used, and a considerable number of de-etiolation mutants have been isolated in various genetic screens (Deng and Quail 1999; Nagy and Schäfer 2000; Neff et al. 2000). Some of these mutants are affected specifically in Rc or FRc responsiveness. The photosensory specificity of these mutants suggests loci important to early events in separate phyB and phyA signaling pathways, respectively. Also, two hyper-responsive mutants, spa1 (Hoecker et al. 1998) and eid1 (Büche et al. 2000) have been shown to be specific to phyA signaling by epistasis analysis.
As the genes responsible for the remaining phytochrome signaling mutants are characterized, a clear picture of phytochrome signal transduction may emerge, but only isolated elements are visible now. This picture may include the serine/threonine kinase activity observed in preparations of plant phytochromes (Yeh and Lagarias 1998) and shown to perform Pfr-enhanced phosphorylation of the cytoplasmic phytochrome–interacting protein PKS1 (Fankhauser et al. 1999).
Because previous screens for mutants with a reduced de-etiolation response to FRc were done in nearly saturating light, we reasoned that screening in more limiting FRc fluence rates, with an emphasis on mutants with weak phenotypes, might allow us to detect mutants in loci not previously implicated in phytochrome signaling. These might include mutants in genes of partially redundant function or hemizygous individuals carrying a homozygous lethal mutation. The isolation of mutants in previous FRc-screens has been limited by an inherent difficulty in recovering mutants with a less than complete loss of phyA signaling, as they inevitably bleach and die after transfer to white light (Barnes et al. 1996). We have devised a method for the efficient recovery of all seedlings from FRc, including those with weak phenotypes, thus enabling us to revisit the genetic screen for long-hypocotyl mutants under these light conditions.
We report the isolation of the new mutant, hfr1, that exhibits the desired partially FRc–responsive phenotype. In the process, because of the need to isolate more alleles than the single hfr1-1 allele initially isolated, we also developed a novel, directed genetic screen based on the large-scale fertilization of a mutagenized male-sterile population with hfr1-1 pollen. Using an insertionally tagged allele of hfr1, we have cloned the HFR1 gene and find that it encodes a bHLH protein with strong similarity to PIF3. We explore the light-regulation of the HFR1 gene, the subcellular localization of the HFR1 protein, and the propensity of HFR1 to interact with PIF3, phyA, and phyB. Our results suggest that HFR1 may act in the direct regulation of gene expression hypothesized for phyA.
Results
Isolation of hfr1 mutants
Using a FRc fluence rate below saturation for the de-etiolation response, we screened variously mutagenized populations of Arabidopsis for a long-hypocotyl phenotype and selected seedlings displaying a partial response to the FRc. The progeny of these candidates were tested by germination and growth in darkness and in Rc, as well as in FRc. Of those judged to have a FRc-specific long-hypocotyl phenotype, 13 were assessed for allelism to known FRc long-hypocotyl mutants. Eleven proved to be allelic to phyA, and one to fhy3. One mutant resulting from T-DNA mutagenisis, which we have named hfr1 (long hypocotyl in far-red), shows incomplete linkage to phyA at the top of chromosome I and does not correspond to any other mutant with a FRc-specific long-hypocotyl phenotype (fhy1, fhy3, fin2, far1, pat1).
To obtain additional alleles of hfr1 beyond the one initially isolated, hfr1-1, we employed a directed genetic screen (see Materials and Methods). In this approach, the population screened is F1 seed that results from crossing a mutant to a second line, where one parent has been mutagenized. To obtain loss-of-function alleles from a dominant, gain-of-function mutation, the mutant parent is further mutagenized (Timpte et al. 1994); to obtain additional alleles of a recessive, loss-of-function mutation like hfr1-1, the wild-type parent is mutagenized. The method we devised overcomes the primary obstacle to directed screening, namely, the tedious nature of standard techniques for the cross-pollination of Arabidopsis. For our screen, ethylmethanesulfonate-mutagenized, male-sterile plants were fertilized in bulk with pollen from hfr1-1, and 12 F1 progeny with long hypocotyls in FRc were selected for F2 analysis. Two of the twelve mutants lacked wild-type segregants in their progeny, indicating the presence of new hfr1 alleles, designated hfr1-2 and hfr1-3. One, hfr1-2, was isolated in its homozygous state and grown for two generations with selection against deleterious mutations in other loci.
hfr1 mutants are defective in a subset of seedling responses to FRc
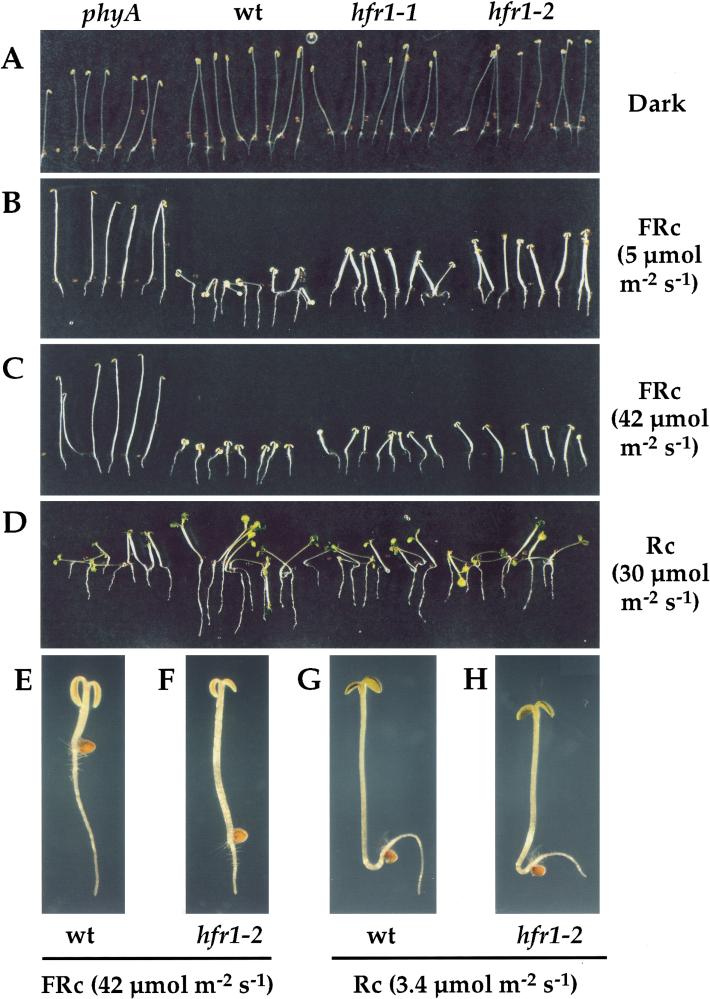
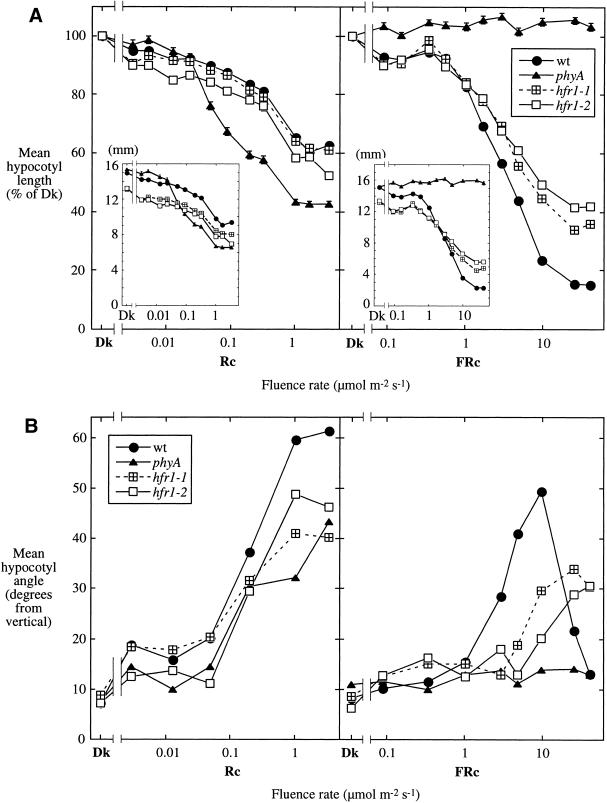
The seedling phenotype of hfr1 mutants is shown in Figure 1. Seedlings of wild-type and mutant phyA and hfr1 seedlings all exhibit a normal etiolated phenotype when grown in complete darkness (Fig. 1A). FRc suppresses hypocotyl elongation in both the wild-type and the hfr1 mutants, but this response is significantly impaired in the hfr1 mutants in moderate and strong FRc (Fig. 1B,C,E,F). This contrasts with the complete blindness to FRc of the phyA mutant (Fig. 1B,C). This effect is FRc specific, as the suppression of hypocotyl elongation in Rc is not altered in the hfr1 mutants (Fig. 1C,G,H). A quantitative examination of hypocotyl elongation responses over a range of FRc and Rc fluence rates corroborates this FRc specificity and shows that hfr1-2 is slightly more impaired in this response than is hfr1-1 (Fig. 2A). The reason for the slightly longer hypocotyls of wild-type and phyA mutants than of the hfr1 mutants in darkness in this experiment has not been determined. However, this difference was not consistently observed in other experiments.
Figure 1.
Visible defects in hfr1 seedling photomorphogenesis. Col-5 wild-type (wt) and mutant (phyA-211 [Reed et al. 1994], hfr1-1 and hfr1-2) seedlings were grown for 4 d in complete darkness or in various light conditions on vertically oriented agar surfaces. They were then photographed without rearrangement. Seedlings grown in (A) darkness, (B) moderate FRc, (C) strong FRc, (D) Rc. (E–H) Representative wt (E,G) and hfr1-2 (F,H) seedlings grown in strong FRc (E,F) or in Rc (G,H). In (E–H), the root/hypocotyl junction is roughly marked by the empty seed coats.
Figure 2.
Hypocotyl responses of hfr1 mutants over a range of Rc and FRc fluence rates. (A) Mean hypocotyl lengths expressed for each line as a percent of the respective value for seedlings grown in darkness (Dk); (insets) the same data expressed as lengths in millimeters. Bars represent the standard error of the mean; where not visible, the bars are eclipsed by a symbol. (B) Mean hypocotyl angle from vertical, a measure of hypocotyl negative gravitropism. An angle of 0° represents maximal negative gravitropism.
A second response to FRc is also strongly affected in the hfr1 mutants: the suppression of hypocotyl negative gravitropism (Poppe et al. 1996; Robson and Smith 1996; Hangarter 1997). Seedling hypocotyls extend vertically, against gravity (negative gravitropism), when grown in darkness (Fig. 1A). Moderate FRc greatly suppresses the hypocotyl negative gravitropism of the wild type but not of the hfr1 or phyA mutants (Fig. 1B). This deficiency, like the reduced suppression of hypocotyl elongation, is FRc specific: the suppression of hypocotyl negative gravitropism by Rc is largely unaffected in hfr1 mutants.
Quantitation of the suppression of hypocotyl negative gravitropism over a range of FRc fluence rates reveals a stronger defect for hfr1-2 than for hfr1-1 (Fig. 2B), as was seen for the suppression of hypocotyl elongation. The hfr1 mutants, unlike phyA, do show some suppression of hypocotyl negative gravitropism in higher fluence rates of FRc. The FRc specificity of the gravitropic phenotype is less absolute, in that there is a somewhat reduced amplitude of Rc response in hfr1 mutants as well as phyA (Fig. 2B). Clearly, the suppression of wild-type hypocotyl negative gravitropism by FRc does not have a monotonic relationship with FRc fluence rate (Fig. 2B), which may explain why the suppression of hypocotyl negative gravitropism by phyA has been considered exclusively a “very low fluence response” (Poppe et al. 1996; Robson and Smith 1996). The data shown here implicate a phyA-mediated “high-irradiance response” in the suppression of hypocotyl negative gravitropism, which is diminished in hfr1 mutants.
We have examined other seedling responses to FRc that are absent in phyA null mutants and found them to be unaltered in hfr1-1 over a range of fluence rates similar to those used in Figure 2B (data not shown). These include apical hook opening and cotyledon separation (Liscum and Hangarter 1993), anthocyanin production (Kunkel et al. 1996), and lack of greening in FRc-grown seedlings on transfer to white light (Barnes et al. 1996).
The loss of responsiveness to FRc observed in the hfr1 mutants could, in principle, result from a reduction in phyA protein level or spectral activity (the ability to interconvert between Pr and Pfr forms on absorption of light). No difference in phyA protein levels between wild-type and hfr1-1 was observed in darkness, and a similar pattern of decline in levels for wild-type and mutant was observed in Rc and FRc (data not shown). As the decline in protein level of phyA depends on its spectral activity, we can conclude that HFR1 does not act primarily through the regulation of phyA level or spectral activity.
Molecular cloning of the HFR1 locus
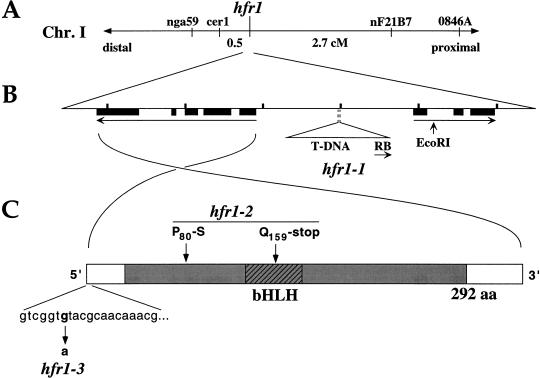
We were able to clone the HFR1 locus by virtue of an inserted T-DNA “tag” in hfr1-1. Though T-DNA-born kanamycin resistance did not cosegregate with the mutation, a set of T-DNA right-border (RB) insertions, detectable by Southern blotting, did cosegregate. The Arabidopsis genomic sequence flanking one of these RB insertions was cloned and found to be physically linked to the nearest genetic marker known to be linked to hfr1 (cer1; Fig. 3A). Complete cosegregation of a codominant marker for the T-DNA junction and hfr1 was observed, indicating a map distance between the T-DNA insertion and hfr1 of <0.07 cM.
Figure 3.
Molecular cloning of the HFR1 locus and identification of lesions in the hfr1 mutants. (A) Genetic linkage of hfr1 to markers at the top end of Chromosome I. (B) Diagram of the 6-kb region surrounding the HFR1 locus that was sequenced in the present study (corresponding to BAC T6A9 coordinates 60330–66530). The length between tick marks is 1 kb. Transcribed regions flanking the hfr1-1 T-DNA insertion are represented by long arrows (exons, solid boxes). The RB and EcoRI site shown are the termini of the flanking region initially isolated from hfr1-1 by PCR walk. In (A,B), centromere distal (and the top of Chromosome I) is to the left side, centromere proximal to the right. (C) Schematic diagram of the mature HFR1 transcript and lesions found in the hfr1-2 and hfr1-3 alleles. The shaded region indicates the predicted coding region for the HFR1 protein.
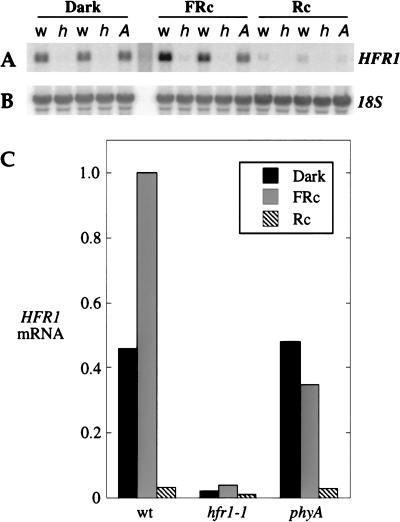
We determined 6 kb of DNA sequence around the T-DNA insertion site (Fig. 3B). This sequence is identical to that recently deposited by the Arabidopsis Genome Project for the BAC T6A9. A candidate HFR1 gene was obtained by analysis of transcribed regions near the T-DNA insertion of hfr1-1. The insertion point was not within a predicted gene, and a probe spanning the insertion point did not allow detection of a transcript in a Northern blot of wild-type RNA. Two transcribed sequences flanking the insertion point, each beginning about 1 kb away, were delineated by a search of the set of Arabidopsis ESTs in GenBank with the 6-kb genomic sequence (Fig. 3B). The centromere-proximal transcript contains regions with strong homology to plant protoporphyrinogen oxidases but lacks an ORF of significant length and thus is unlikely to be the template for a functional enzyme. The centromere-distal transcript has a substantial ORF that includes homology to the bHLH family of DNA-binding proteins. Northern analysis indicated a greatly reduced level of this transcript in hfr1-1 relative to wild type (Fig. 4), making it an excellent candidate to be HFR1. We sequenced the corresponding genomic region in the two hfr1 alleles derived from the directed screen, hfr1-2 and hfr1-3. Both had point mutations in the transcribed region of the putative HFR1 gene, confirming its identity as HFR1. In addition, the 6-kb genomic region depicted in Figure 3B fully complements the hfr1-1 and hfr1-2 mutations when present as a transgene (C.D. Fairchild, M.S. Schumaker, and P.H. Quail, unpubl.).
Figure 4.
HFR1 is a light-regulated gene with strongly reduced expression in the hfr1-1 mutant. Northern blot analysis of total RNA from 3-d-old seedlings grown under the indicated light conditions. (A) HFR1 probe; w, wild-type; h, hfr1-1; A, phyA-211. Duplicates of wild-type and hfr1-1 represent separate RNA preparations. (B) 18S rRNA reprobe of the membrane from (A). (C) HFR1 mRNA levels adjusted for 18S signal and expressed relative to the wild-type FRc value. Wild-type and hfr1-1 values are averages of two samples.
The hfr1-2 allele was found to have two base changes in the transcribed region of HFR1 (Fig. 3C). One, at residue 159, results in a nonsense codon that truncates the predicted HFR1 protein in the loop between helices of the bHLH domain. With this truncation, more than half of the predicted protein would be absent and the bHLH domain inactivated. Thus, based on molecular data, the hfr1-2 mutant would be predicted to have a more complete loss of HFR1 function than has hfr1-1, which produces a lower level of unaltered mRNA, and this prediction is in accord with the physiological data from the two mutants.
The only base change in the transcribed region of hfr1-3 is in the 5′ untranslated region, 6 nucleotides from the longest 5′ end of the HFR1 transcript as determined by RACE (Fig. 3C). Preliminary evidence from Northern blot analysis indicates that the size and abundance of the hfr1-3 mRNA is similar to that of wild type in dark-grown seedlings, suggesting that the hfr1-3 mutation may reduce translation. We have not directly tested this possibility.
HFR1 encodes a bHLH protein with strong similarity to the phy-interactor PIF3
A search of GenBank with the HFR1 sequence revealed that the closest homologs are two Arabidopsis bHLH proteins, PIF3 (Ni et al. 1998) and a protein predicted from genomic sequence, AAD24380. The homology to both of these proteins is highest in the HLH region (>50% identity) but extends in both directions beyond the bHLH domains (Fig. 5A). There is no significant homology between HFR1 and these proteins beyond the region shown. HFR1 lacks the PAS domain of PIF3. A notable difference between them is in the basic region of the bHLH domains, where there appears to be a deletion in HFR1 relative to the others (discussed below). A comparison of HFR1 and representative members of the broader HLH protein family (Fig. 5B) reveals extensive conservation in HFR1 of residues that define the HLH domain (Atchley et al. 1999).
Figure 5.
Sequence comparison of HFR1 to other bHLH proteins. Identical amino acid residues are shaded black; similar residues are shaded grey. (A) Alignment of HFR1 (amino acid residues 105–248) and its closest known homologs, the Arabidopsis protein PIF3 (GenBank accession no. AF100166) and predicted protein AAD24380, over the region of significant sequence similarity. Solid bars indicate prospective monopartite nuclear localization signals in HFR1. (B) Alignment of a more restricted region of HFR1 (amino acid residues 122–194) to a broader set of representative members of the bHLH protein family from various organisms. Arrows indicate key positions where HFR1 is dissimilar to most (perhaps all) known bHLH proteins. GenBank accession nos.: AhR, P30561; Sim, P05709; R-Lc, P13526; Achaete, P10083; Hairy, P14003; MyoD, CAA40000; Arnt, P41739; PHO4, P07270; ID1, P20067. (C) Similarity of the basic region of the HFR1 bHLH domain (amino acid residues 132–153) to the basic regions of the Achaete/Scute bHLH subfamily. In this alignment, only amino acid residues with similarity to HFR1 are shaded. GenBank accession nos.: L-sc, P09774; Scute, P10084; MASH2, P19360; MASH1, P19359; Asense, P09775.
The region amino-terminal of the HLH in HFR1 has a basic character, including residues 23–25 in the alignment (Fig. 5B) that are often basic in DNA-binding bHLH proteins but not in HLH proteins that do not bind DNA, such as ID1 (Atchley et al. 1999). The more amino-terminal part of the basic domain corresponds to the apparent deletion relative to PIF3 and AAD24380. In Figure 5B, these proteins are aligned to ungapped HFR1 to show the lack of colinear similarity. This region of HFR1 is not similar to the corresponding region of most bHLH proteins but does show strong similarity to the basic region of the Achaete/Scute subfamily of animal bHLH proteins (Fig. 5C).
Two amino acid residues of HFR1 that are notably different from most, or perhaps all, bHLH proteins are indicated with arrows in Figure 5B,C. The indicated arginine of HFR1 corresponds to a glutamate in >90% of the known bHLH proteins (Atchley et al. 1999); notable exceptions are AhR, with a serine, and Sim, with an alanine. In no other bHLH is this a basic residue. The other indicated residue, aspartate in HFR1, corresponds to an arginine in about half of the known bHLH proteins (Atchley et al. 1999) or can be hydrophobic (valine in the Achaete/Scute subfamily; Fig. 5C) or hydrophilic (glutamine, threonine), but in no other bHLH is this an acidic residue.
HFR1 is constitutively nuclear localized when transiently expressed in onion epidermal cells
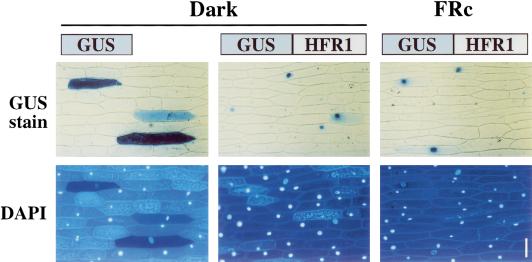
The predicted HFR1 protein contains two potential monopartite nuclear localization signals (Fig. 5A). These signals and the similarity of HFR1 to DNA-binding proteins suggest that HFR1 might function in the nucleus. To test the subcellular localization of HFR1, and the possibility that this localization might be light-regulated, we fused the coding region of HFR1 to the reporter β-glucuronidase (GUS) in a plant expression construct. This construct was transfected into peels of onion epidermis by particle bombardment. Whether the peels were then incubated in darkness or in FRc, the GUS-HFR1 protein was found predominantly in nuclei, in contrast to the cytoplasmic localization of the GUS control (Fig. 6).
Figure 6.
HFR1 protein is constitutively nuclear localized when transiently expressed in onion epidermal cells. Onion epidermal peels were bombarded with constructs for expression of either the GUS reporter only or a GUS-HFR1 chimera. The peels were then incubated in darkness or FRc for 17 h before staining. In the top row, blue color results from GUS activity; below, the fluorescence from DAPI stain shows the position of the nucleus in each cell. Bar, 100 μm.
HFR1 expression is light regulated
HFR1 mRNA levels were assayed by Northern blot analysis of seedlings grown in darkness, FRc, or Rc. The HFR1 mRNA is about 1.3 kb in size, as predicted from 5′-RACE and the poly-A ends of the cDNA clones, and is detectable in all light conditions (Fig. 4A). The mRNA shows more than twofold induction in wild-type seedlings grown in FRc, but a 14-fold decrease in Rc-grown seedlings relative to those grown in darkness (Fig. 4C). Thus, there is 30-fold more HFR1 mRNA in FRc than in Rc. Many genes in seedlings are either induced or repressed by both FRc and Rc compared to darkness (Terzaghi and Cashmore 1995), but to our knowledge, only the HD-Zip-encoding genes ATHB-2 and ATHB-4 have been shown to exhibit a similar induction by FR and suppression by R (Carabelli et al. 1993, 1996). The FRc induction of HFR1 is absent in the phyA mutant, suggesting that this induction is a result of phyA signaling. By contrast, the Rc suppression of HFR1 mRNA levels is like wild type in the phyA mutant, implicating a phytochrome other than phyA in this regulation.
HFR1 lacks affinity for phyA or phyB
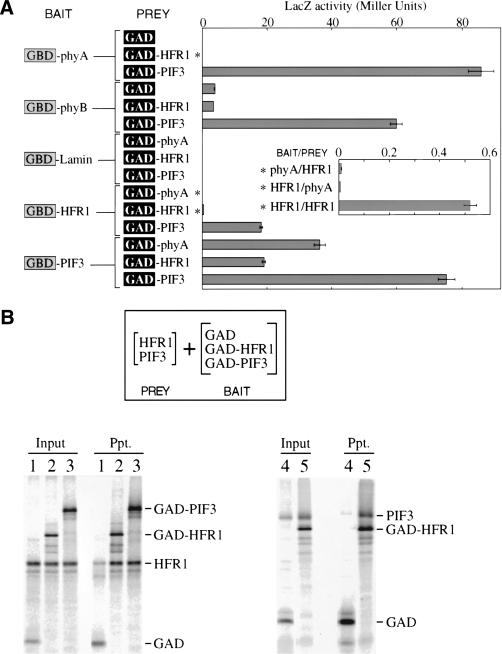
A simple mechanism by which HFR1 might provide FRc specificity in its action is by binding preferentially to phyA itself. We explored the possibility of an HFR1/phyA interaction by two methods used previously to demonstrate interactions between PIF3 and phytochromes: the yeast two-hybrid assay and coimmunoprecipitation. PIF3 has been shown to bind the C-terminal halves of both phyA and phyB in two-hybrid assays (Ni et al. 1998). These interactions are used here as positive controls (Fig. 7A). However, in the analogous experiment with HFR1, there is no indication of an interaction with either phytochrome fragment (Fig. 7A).
Figure 7.
HFR1 protein can bind the phytochrome-interactor PIF3. (A) Two-hybrid assays for interaction in yeast between HFR1, PIF3, phyA C-terminal half, or phyB C-terminal half, along with negative controls nuclear lamin and unfused GAL4 activation domain (GAD). GBD refers to the GAL4 DNA-binding domain. Asterisks mark assay results that are also shown in an expanded view in the inset. Bars represent the standard error of the mean. (B) In vitro binding of HFR1 to PIF3. Shown are autoradiograms of SDS-PAGE separated proteins from immunoprecipitations. The GAL4 activation domain (GAD) was used as an epitope tag in bait constructs in fusion with PIF3 (GAD-PIF3) and HFR1 (GAD-HFR1). Prey HFR1 (HFR1) and PIF3 (PIF3) were expressed without the epitope tag. Bait proteins were immunoprecipitated using antibody to GAD immobilized on beads. Input lanes were loaded with half the fraction of each binding reaction that was loaded for washed precipitate lanes (Ppt.). (Lanes 1–3) precipitation of HFR1 by GAD control (1), GAD-HFR1 (2), and GAD-PIF3 (3). (Lanes 4,5) precipitation of PIF3 by GAD control (4) and GAD-HFR1 (5).
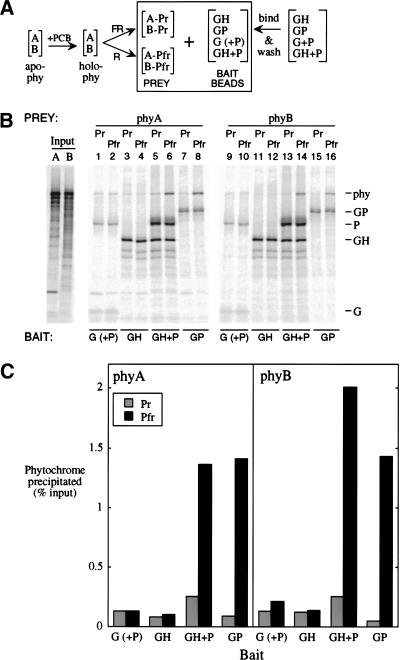
We also looked for evidence of phyA/HFR1 binding in vitro. PIF3 has also been shown to bind full-length phyB in vitro with a dramatic preference for the Pfr form (Ni et al. 1999). Ni et al. used the GAL4 activation domain (GAD) as an epitope tag fused to PIF3 (GAD-PIF3) as “bait,” unfused GAD as a negative control bait, and phyB as prey. They expressed phyB in vitro and combined it with chromophore to make spectrally active holoprotein; bait proteins were produced in Escherichia coli. In the experiment described here, all proteins were expressed in vitro (Fig. 8A). We were able to establish conditions that allow not only the preferential precipitation of phyB Pfr by GAD-PIF3 (Fig. 8B, lanes 15,16; Fig. 8C, right, GP) but also the preferential precipitation of phyA Pfr (Fig. 8B, lanes 7,8; Fig. 8C, left, GP). Under these conditions, even with a greater amount of GAD-HFR1 bait than GAD-PIF3, there is no sign of phyA or phyB binding to GAD-HFR1 (Fig. 8B, lanes 3,4,11,12; Fig. 8C). Thus, both by two-hybrid and immunoprecipitation assays, we have no evidence for a direct interaction between HFR1 and phyA or phyB.
Figure 8.
Both phyA and phyB bind as Pfr in vitro to both the PIF3 homodimer and a HFR1/PIF3 complex. (A) PhyA or phyB apoproteins were expressed separately in vitro (apo-phy), and combined with chromophore (PCB) to make spectrally active holoprotein (holo-phy); portions of each phytochrome solution were then irradiated with either FR or R to form predominantly Pr (A-Pr; B-Pr) or Pfr (A-Pfr; B-Pfr) forms, respectively. Bait beads were prepared by binding GAD-tagged proteins (G; alone or coexpressed with untagged PIF3 [P]), GAD-HFR1 (GH), or GAD-PIF3 (GP) to MAb-Protein A-beads, and then washing to remove loosely bound protein. Bait proteins were labeled at 2% of the specific activity of the phytochromes. (B) Autoradiograms of proteins separated by SDS-PAGE. Input phyA (A) and phyB (B) lanes were loaded with one-fifth the fraction of each binding reaction that was loaded for washed precipitates. (C) Quantitation of the phytochrome precipitated in (B).
HFR1 can bind PIF3
In many cases, HLH proteins can form both homodimers and heterodimers with other HLH proteins. Frequently, the heterodimers involve related HLH domains. PIF3 is the closest known relative to HFR1 and appears to act in phytochrome signaling through a direct interaction with phytochrome. Thus, another possible mechanism by which HFR1 might act in phyA signaling is through interaction with PIF3.
By the yeast two-hybrid assay, positive signals for an HFR1/PIF3 interaction were obtained in both arrangements of the chimeras (Fig. 7A). Positive signals were also obtained from the combinations PIF3/PIF3 and HFR1/HFR1. The HFR1 homodimerization signal is relatively weak but is significantly above the HFR1/phyA background (Fig. 7A, inset). The relative intensity of these signals may not have a simple relationship to the relative affinities of the proteins, as immunoblots with monoclonal antibodies to the GAL4 domains indicate a lower level of expression in yeast for the HFR1 chimeras than for the PIF3 chimeras. The positive results for HFR1 interaction with PIF3 verify the efficacy of the HFR1 two-hybrid chimeras and lend credence to the lack of interaction observed between HFR1 and phyA or phyB.
We were able to confirm the propensity of HFR1 and PIF3 to form a complex by coimmunoprecipitation of the two proteins coexpressed in vitro. GAD was again used as an epitope tag fused to HFR1 (GAD-HFR1) or PIF3 (GAD-PIF3) baits, with unfused GAD as a negative control bait, but here the prey is untagged HFR1 or PIF3. The proteins were expressed in vitro to roughly similar concentrations. HFR1 coprecipitates with either GAD-HFR1 or GAD-PIF3 to a similar extent (Fig. 7B, lanes 1–3). In the reverse arrangement, untagged PIF3 coprecipitates efficiently with GAD-HFR1 (Fig. 7B, lanes 4,5). These results suggest similar affinities for homo- and heterodimerization for HFR1 and PIF3.
Like PIF3, an HFR1/PIF3 complex preferentially binds the Pfr form of phyA and phyB
As HFR1 can bind PIF3 and PIF3 can bind phyA and phyB, it was of interest to test for the ternary complexes HFR1-PIF3-(phyA or phyB). In the same conditions under which we fail to see precipitation of phytochrome by GAD-HFR1 (Fig. 8B, lanes 3,4,11,12; Fig. 8C, GH), the GAD-HFR1/PIF3 complex does preferentially precipitate phyA and phyB Pfr forms (Fig. 8B, lanes 5,6,13,14; Fig. 8C, GH + P), with an efficiency similar to that of GAD-PIF3 alone (Fig. 8B, lanes 7,8,15,16; and Fig. 8C, GP). Almost all of the PIF3 in the GAD-HFR1/PIF3 bait is present by virtue of its association with HFR1 because the GAD-HFR1/PIF3 complex bait was prepared under conditions that remove most of the PIF3 from beads incubated with coexpressed GAD and PIF3 (see Fig. 8B, lanes 1,2,9,10). The negative control bait, GAD with residual PIF3, has scant preferential affinity for the phytochrome Pfr forms (Fig. 8B, lanes 1,2,9,10; Fig. 8C,G [+P]). Thus, like the PIF3 homodimer, the HFR1/PIF3 complex binds preferentially to the Pfr forms of phyA and phyB.
Discussion
Considerable progress has been made in recent years in efforts to define phytochrome signal transduction pathways (Wei and Deng 1999; Nagy and Schäfer 2000; Neff et al. 2000). Genetic and molecular approaches have identified a significant number of components that potentially function as signaling intermediates and have provided evidence of both shared and separate pathway branches for individual phytochrome family members (Soh et al. 1998; Bolle et al. 2000; Büche et al. 2000; Neff et al. 2000; Osterlund et al. 2000). However, lacking until recently was evidence of a contiguous transduction pathway, consisting of identified molecular intermediates, that leads from phytochrome photoconversion to changes in gene expression. The recent discoveries that phytochromes translocate to the nucleus in response to light (Sakamoto and Nagatani 1996; Kircher et al. 1999; Yamaguchi et al. 1999) and that phyB can interact directly with the bHLH protein, PIF3, bound to a DNA target site (Martinez-Garcia et al. 2000) have suggested that one mode of phytochrome signal transduction is the direct transcriptional regulation of target genes. PIF3 was initially isolated as a phytochrome-interacting factor in a yeast two-hybrid screen. Here we have genetically identified a second member of the bHLH family, HFR1, in a screen for components specific to the phyA signaling pathway and have shown that HFR1 can heterodimerize with PIF3. These data support and extend the hypothesis that phytochromes can regulate target genes directly and open up the possibility that they may do so via multiple heterodimerizing members of the bHLH family of transcription factors, which might regulate gene expression in combinatorial fashion.
The observation that loss-of-function mutations in HFR1 result in a FRc-specific phenotype indicates that HFR1 functions positively in the phyA signaling pathway. Furthermore, the normal, or even increased, level of phyA protein and its normal photoconversion activity in hfr1 mutants imply that HFR1 is an authentic signal transducer, rather than a protein involved in phyA synthesis or assembly. In these respects, hfr1 mutants are similar to other FRc-specific, loss-of-response mutants that have been identified previously, far1 (Hudson et al. 1999), fhy1 and fhy3 (Whitelam et al. 1993), fin2 (Soh et al. 1998), and pat1 (Bolle et al. 2000).
However, the subset of developmental responses to FRc that are affected in hfr1 mutants differs from the subset affected in other phyA-signaling mutants. The most consistent hfr1 phenotype is limited to a clear, partial loss of FRc-suppression of hypocotyl elongation and negative gravitropism. The hypocotyl elongation and gravitropism defects of hfr1-1 strictly cosegregate, and this correlation is maintained in the hfr1-2 homozygotes that lack the hfr1-1 T-DNA insertion, thereby indicating that both defects result from mutation of HFR1. Except for fin2, which appears to show a loss of FRc suppression of hypocotyl gravitropism similar to hfr1 (Soh et al. 1998), the effects on hypocotyl gravitropism of mutations in other phyA-signaling loci have not been established. In phyA-signaling mutants other than hfr1, a wider range of responses to FRc is affected, including the stimulation of anthocyanin production and the loss of greening on transfer to white light (Barnes et al. 1996; Soh et al. 1998; Hudson et al. 1999; Bolle et al. 2000). These FRc responses are consistently unaffected in hfr1 mutants. The specificity of the hfr1 phenotype for particular FRc responses suggests that HFR1 may direct phyA signals primarily to the subset of phyA-regulated genes that drive these responses.
Although three other genes specific for phyA signaling have been molecularly cloned—SPA1 (Hoecker et al. 1999), FAR1 (Hudson et al. 1999) and PAT1 (Bolle et al. 2000)—HFR1 is the only one of these whose sequence offers an obvious prediction of its biochemical role. The sequence homology of HFR1 to members of the bHLH family of transcription factors, along with potential nuclear localization signals, suggests that it might be a transcription factor. The demonstrated constitutive nuclear localization of transiently expressed HFR1 in onion epidermal cells and the binding affinity of HFR1 for its closest homolog, PIF3, are consistent with this suggestion. PIF3 has similarly been shown to be constitutively nuclear (Ni et al. 1998), and there is more substantial evidence that PIF3 may act as a transcription factor. PIF3 alone can bind DNA, with a strong preference for the G-box core motif and some specificity for one flanking base on either side of it (cCACGTGg), and appears to be involved in the induction of the genes CCA1 and LHY within 1 h of a light signal (Martinez-Garcia et al. 2000). In addition, the PIF3 two-hybrid chimera with the GAL4 DNA-binding domain has transcriptional activation activity in yeast (Ni et al. 1998). However, we have not yet attempted to determine whether HFR1 binds directly to DNA, and in contrast to PIF3, the analogous HFR1 two-hybrid chimera exhibits no detectable intrinsic transcriptional activation activity in yeast.
Does the unusual basic region of HFR1 predict a lack of affinity for DNA or DNA binding with a sequence specificity distinct from that of other bHLH proteins? In all reported cases, other bHLH proteins that have unusual basic regions ultimately have been found to bind DNA with an altered sequence specificity (Littlewood and Evan 1998; Atchley et al. 1999). The two key differences between HFR1 and a consensus basic region (Fig. 4B, arrows) are partly reflected in other bHLH proteins. The highly conserved glutamate residue (arginine at position 22 in Fig. 5B), which is integral to E-box (CAnnTG) sequence recognition (Atchley et al. 1999), is not retained in some members of the bHLH-PAS family. In AhR, it is a serine, in Sim an alanine. The 5′ recognition sequence half-sites preferred by AhR (T(c/t)GC) and Sim (GT(a/g)C) are distinct from the E-box half-sites (CAn) preferred by their heterodimerizing partner Arnt and other conventional bHLH proteins (Sogawa et al. 1995; Swanson et al. 1995). By analogy, HFR1 would be expected to recognize a non-E-box sequence. Similarly, the other unusual HFR1 residue (aspartate at position 26 in Fig. 5B) is an arginine in G-box binding bHLH proteins, including PIF3. Substitutions for this arginine, which is involved in the recognition of the central CG of the G-box (CACGTG), confer specificity for a different central pair within the E-box core (Littlewood and Evan 1998; Atchley et al. 1999). Together, the unusual substitutions in the HFR1 basic region may specify a recognition element that lacks similarity to the G-box preferred by PIF3.
Our demonstration that HFR1 is a PIF3-binding bHLH protein may be the first indication that a network of bHLH proteins is involved in the regulation of plant development by phytochromes. Many animal bHLH proteins act as components of complex regulatory networks that include cross-dimerizing DNA-binding activators and repressors of transcription, as well as HLH inhibitors of bHLH DNA-binding (Littlewood and Evan 1998; Atchley et al. 1999; Massari and Murre 2000). This type of bHLH network has not been demonstrated in plants, though a small number of bHLH proteins have been implicated in various processes, including the regulation of tissue-specific production of anthocyanin by the R/B proteins (Ludwig and Wessler 1990; Lesnick and Chandler 1998), and the induction of a dehydration-response gene by rd22BP1 (Abe et al. 1997). Many more bHLH proteins of unknown function have been revealed by genomic sequencing of Arabidopsis. Perhaps some of these bHLH proteins will be found to be involved in light regulation of gene expression in collaboration with PIF3 and HFR1.
Whatever the nature of HFR1 activity, it appears to be largely specific for responses to FRc, though essentially the same phenotypic responses are induced by Rc in wild-type Arabidopsis. An attractively simple hypothetical mechanism for HFR1 FRc specificity would be for HFR1 to bind phyA specifically. However, we failed to find any evidence for a direct HFR1/phyA interaction, either by yeast two-hybrid or coimmunoprecipitation assays. It remains possible that a direct HFR1/phyA interaction requires a plant-specific posttranslational modification of HFR1 or phyA, but we have no evidence for this.
The HFR1/phytochrome complex that we have demonstrated here requires PIF3, which might act as a bridge between its individual interactors, HFR1 and phytochrome (Fig. 9, middle). The HFR1/PIF3 complex is drawn as a heterodimer rather than a higher-order complex, though we have only very preliminary evidence for this from native gel electrophoresis of proteins expressed in vitro (C.D. Fairchild and P.H. Quail, unpubl.). The evidence for an HFR1/PIF3/phytochrome complex does little to explain the observed FRc specificity of HFR1 function, given that PIF3 binds both phyA and phyB. It is tempting to speculate that in place of, or in addition to PIF3, a phyA-specific binding protein might bridge HFR1 and phyA. Obvious candidates for such a phyA-specific binding protein are the apparently phyA-specific signal transducers that have been identified genetically, but no evidence has yet been presented for a phyA-specific interacting protein.
Figure 9.
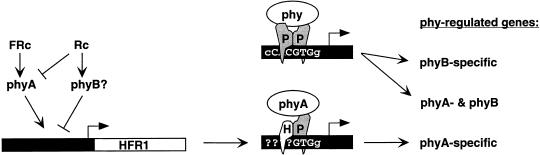
A model for the role of HFR1 in phytochrome signaling. FRc, acting through phyA, enhances HFR1 transcription and Rc, acting through phyB or another phytochrome, suppresses HFR1 transcription. Phytochromes in their Pfr form (phy) translocate to the nucleus, where they are recruited to target gene promoters in genomic DNA (thick black bars, with recognition sequences overlaid and with arrows representing transcription initiation sites) by PIF3 (P). Whereas the PIF3 homodimer recognizes a G-box and potentially regulates gene expression in response to Rc or FRc, the heterodimer of PIF3 and HFR1 (H), formed predominantly in FRc, recognizes a distinct sequence in phyA-specific gene targets.
The 30-fold greater abundance of HFR1 mRNA in FRc relative to Rc that we have observed might be sufficient to explain the FRc specificity of HFR1 action, if this difference in mRNA level translates to a similar difference in HFR1 protein activity in the two light conditions. The role of HFR1 could be to confer a different DNA sequence specificity on the PIF3/phyA complex and, thus, to adjust the gene-regulatory output of phyA (Fig. 9). The apparent FRc-specificity of HFR1 activity might then be determined by a complex, reciprocal regulation of HFR1 abundance by Rc and FRc (Fig. 9).
Materials and methods
Isolation of mutants
As part of a comprehensive screen of available mutagenized populations for long-hypocotyl mutants in FRc, we used T2 seed from 2000 T-DNA transformed parents, which were a gift from Robert Fischer (University of California, Berkeley). The Fischer lines had been transformed with T-DNA from a vector that was derived from pBI121 (Jefferson et al. 1987) by deletion of the GUS gene. Seedlings were grown in FRc (2–3 μmole m−2 sec−1) for a total of 4–5 d before mutant selection.
In the directed screen for additional alleles of hfr1, F2 seed from a cross of Ler wild type to the male-sterile mutant ms1 (Ler background) was mutagenized in 0.3% or 0.45% (v/v) ethylmethanesulfonate for 13 h and sown onto mesh-covered soil at a rate of 400 per 4-inch pot. All self-fertile plants (three-quarters of the population) were weeded out as they became evident by the presence of elongating siliques, leaving 10–40 male-sterile plants per pot and a mutagenized male-sterile population totaling ∼5000. Concurrently, several flats of hfr1-1 plants were grown as a source of pollen. We found that dragging the flowering, male-sterile plants through a dense stand of young, flowering hfr1-1 was an effective mass cross-pollination method. Cross-pollination was most efficient at midday, when flowers were open to their widest extent. Several rounds of pollination were performed on each pot of male-sterile plants over the course of one week. The result was cross-pollination at a rate of several seeds per inflorescence branch. Seed was collected in pools of two pots each and screened directly for long-hypocotyl mutants in FRc. Leaf tissue samples were taken from the young rosettes of selected putative mutants for small-scale DNA preparation.
New, noncomplementing alleles of hfr1 were confirmed by testing F2 progeny in FRc for the presence of wild-type seedlings (for hfr1-2, >2000 F2 individuals; for hfr1-3, 200).
Genetic mapping and complementation tests
An F2 population resulting from a cross of hfr1-1 to Ler wild-type was used for mapping; DNA for PCR was prepared from leaf samples (Edwards et al. 1991). With the assessment of 16 PCR markers (CAPS and SSLP; Konieczny and Ausubel 1993; Bell and Ecker (1994) in a population of 17 hfr1-1 homozygotes, along with the visible markers er and gl1 in a larger population, hfr1 was mapped to the top of chromosome I. This map position excluded the possibility of allelism to some long-hypocotyl mutants. Others that were linked (phyA) or unmapped (fhy1 and fhy3) failed to complement hfr1-1 in the F1 and were confirmed as nonallelic by the segregation of wild types in the F2 generation.
The expanded mapping population (758 individuals) consisted largely of hfr1 homozygotes and a few homozygous wild type. As hfr1-1 exhibits partial dominance and a subtle phenotype, a larger population of potential hfr1 homozygotes was picked, and then heterozygotes for flanking markers cer1 and nF21B7 were rejected as probable hfr1 heterozygotes. For F2 individuals with an apparent recombination between these markers, the hfr1/hfr1 genotype was confirmed by a lack of wild-type segregants in the F3 population.
Seedling growth and measurements
For seedling growth, seeds were surface sterilized, sown on agar-solidified medium (lacking sucrose) in petri dishes, and germinated in darkness or under defined light conditions as previously described (Hudson et al. 1999). For mutant screening, seed was suspended in sterile 0.15% agar (aq.) and sown densely in horizontal rows. For hypocotyl length and gravitropism measurements, seeds in 0.15% agar were spotted one per 0.5 cm2 in a staggered grid pattern. For RNA and protein extractions, seed was sown on filter paper laid over agar-solidified medium.
For most purposes, seedlings were germinated and grown in vertically oriented petri dishes, such that both hypocotyls and radicals grew along the agar surface, with horizontal R or FR illumination. Seedlings for RNA and protein extractions were grown in petri dishes in the normal, horizontal orientation with illumination from above. For measurement of hypocotyl length and angle-from-vertical, measurements (∼40 per genotype at each fluence rate) were made from digital images of unrearranged seedlings using the program NIH Image.
For recovery of seedlings germinated in FRc for further growth in white light, seedlings were aseptically transferred under dim green light (in some cases after taking digital images in green light) to growth medium containing 1% or 2% sucrose. These seedlings were then kept in darkness for 3 d, exposed to filtered room light for 3 h, and then transferred to full white light. After 1 wk, during which the seedlings partially greened and initiated normal leaves, they were transferred to soil.
Yeast two-hybrid binding assays
HFR1 yeast two-hybrid vectors were constructed from the plasmids pGAD424 and pGBT9. The other two-hybrid constructs, based on the same plasmids, were described previously (Ni et al. 1998). The yeast strain Y187 was transformed with combinations of vectors, and LacZ activity was assayed with ONPG as a substrate according to the Clontech Yeast Protocols Handbook (Clontech). The LacZ activities are the mean of six values from two independent cultures of each vector combination assayed in triplicate.
In vitro binding assays
Immunoprecipitations were performed as previously described (Ni et al. 1999) with the following variations. All proteins for immunoprecipitations were expressed from T7 promoters in the TnT in vitro transcription/translation system (Promega) in the presence of 35S-methionine. The GAD-HFR1 vector was constructed by replacement of PIF3 by HFR1 in the GAD-PIF3 vector. The HFR1 prey construct consisted of the HFR1 coding region in pBluescript (Stratagene). The PIF3 prey was inserted into the NcoI/BamHI sites of the vector pET21 (Novagen) with the addition of a six-His tag at its N terminus. PhyA was expressed from the new plasmid T7-A.BS, which consists of phyA under the control of the T7 promoter and untranslated leader from pET3 in a pBluescript backbone.
For coimmunoprecipitation with PIF3 as prey, the PBS binding buffer (pH 7.2) contained 0.1% (v/v) Tergitol NP-40 (Sigma), 1 mM EDTA, 0.1% BSA, and Complete protease inhibitors. PIF3 and GAD or GAD-HFR1 were coexpressed. Mixtures of expressed proteins were precleared by incubation for 1 h at 4°C with Protein A-agarose in binding buffer, and the supernatants were added to pelleted anti-GAD/Protein A beads. After 4 h at 4°C, beads were pelleted and washed once with 1 mL binding buffer and once with 1 mL binding buffer without BSA or protease inhibitors.
Coimmunoprecipitations with HFR1 as prey were performed similarly, with the exceptions that the binding buffer contained 50 mM Tris-HCl (pH 7.5 at 25°C) in place of PBS (Tris binding buffer), paramagnetic Protein A-beads were used (Dynabeads Protein A; Dynal) in place of Protein A-agarose, and both final washes contained 0.5% Tergitol NP-40.
For coimmunoprecipitations with phyA or phyB as prey, bait beads were prepared and mixed with separately expressed phytochrome. For bait beads, bait proteins expressed individually or in combinations were bound to anti-GAD/Protein A paramagnetic beads in PBS binding buffer. The beads were then washed three times with 1.4 mL PBS binding buffer with 0.5% Tergitol NP-40 and once with Tris binding buffer with 150 mM NaCl. Holo-phytochrome was formed by a 1 : 4 dilution of TnT solution into Tris binding buffer containing 30 μM phycocyanobilin. Precleared phyA or phyB in the Pr or Pfr form was mixed with washed bait beads. After 6 h at 4°C, the beads were collected and washed once with 1.4 mL Tris binding buffer containing 0.5% Tergitol NP-40.
Samples for SDS-PAGE were mixed with SDS sample buffer and either boiled or heated to 65°C for 5 min (paramagnetic beads). Gels were fixed and then dried for autoradiography by phosphor screen (PhosphorImager Storm 860, Molecular Dynamics). For relative quantification of precipitated phytochrome, in-lane backgrounds of mock precipitation controls were subtracted and the amount precipitated expressed as a percent of the precleared input phytochrome.
Subcellular localization
For the determination of subcellular localization of a GUS-HFR1 chimeric protein, the HFR1 was fused to GUS in the vector TEX2. TEX2, onion epidermal cell transient transfection, and staining were as described previously (Ni et al. 1998).
HFR1 mRNA analysis
Northern blot analysis of mRNA levels in seedlings was as described previously (Hoecker et al. 1999). The HFR1 probe was the insert from the cDNA clone EST 209K19T7. By 5′-RACE, the HFR1 transcript was found to extend beyond the 5′-end indicated by this cDNA clone and to, thus, include the hfr1-3 mutation site.
Molecular cloning of HFR1
Genetic linkage of the hfr1-1 mutation to inserted T-DNA was established by Southern hybridization of a RB probe to EcoRI-digested DNA from 10 homozygous hfr1 and nine homozygous wild type from the hfr1-1 × Ler F2. A subset of the eight to 10 RB fragments visible on this blot cosegregated with hfr1-1.
RB-flanking plant DNA was cloned by the splinkerette method of PCR walking (Devon et al. 1995), using nested RB primers and EcoRI-digested hfr1-1 DNA. Two major fragments were characterized. One corresponded to a sequenced portion of the genome distant from hfr1, the other overlapped with the sequenced T7 end of the BAC T6H24, which was anchored at its other end to the sequenced BAC T7I23 at the top end of chromosome I. This flanking region was found to cosegregate with hfr1 in the limited mapping population used for RB segregation analysis by Southern blot.
The wild-type sequence of the genomic region surrounding this flanking sequence was obtained by a combination of inverse PCR and primer walk along two long PCR products to flanking islands of genomic sequence. A three-primer, codominant PCR marker was designed for the junction of this flanking sequence and the T-DNA border: the RB primer CRF2 (CTCCAGAAACCCGCGGCTGAGTG) and flanking region primer c004 (CATCGCACCTGCTCGGTGTAT) amplify a 305-bp fragment from hfr1-1, and c004 and c007-R (AACCAAGAACTGTAATGCACAACGG), a primer from the other side of the T-DNA insertion, amplify a 600-bp fragment from the wild type (2 min, 68°C annealing/extension; Advantage cDNA PCR kit, Clontech). The 48 hfr1-1 × Ler F2 plants exhibiting a recombination between hfr1 and the flanking markers cer1 and nF21B7 were tested with this codominant marker, and complete cosegregation of the T-DNA junction and the hfr1 mutation was observed.
To determine exons and introns, cDNA clones from the transcribed regions flanking the hfr1-1 T-DNA insertion were sequenced: proximal transcript, EST 153A23T7; distal transcript (HFR1), EST 209K19T7 (Arabidopsis Biological Resource Center). The HFR1 coding region for various constructs was derived from EST 209K19T7.
Acknowledgments
We thank Sharon Moran, Jennica Ryu, and Jim Tepperman for technical assistance; Hana Rha, Linda Margossian, and Robert Fischer for the Fischer T-DNA lines and Ute Hoecker for arranging their transfer; Renee Sung for γ-irradiated M2 seed; the Arabidopsis Biological Resource Center in Ohio for seeds and cloned DNA; Enamul Huq for the PIF3 prey construct; Sae Shimizu-Sato for her important contribution to the construction of the phyA expression plasmid; and Matthew Hudson, Laura Williams, and Bassem Al-Sady for critical reading of the manuscript. Supported by NIH grant GM-47475 and the U.S. Department of Agriculture, Current Research Information Service number 5335-21000-006-00D (to P.H.Q.), and Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship DRG-1302 (to C.D.F.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL quail@nature.berkeley.edu; FAX (510) 559-5678.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.82800.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of ArabidopsisMYC and MYB homologs in drought- and abscisic acid–regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH. Far-red light blocks greening of arabidopsis seedlings via a phytochrome A–mediated change in plastid development. Plant Cell. 1996;8:601–615. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua NH. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T. eid1: A new Arabidopsismutant hypersensitive in phytochrome A–dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. The Arabidopsis Athb-2 and Athb-4genes are strongly induced by far-red-rich light. Plant J. 1993;4:469–479. doi: 10.1046/j.1365-313x.1993.04030469.x. [DOI] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2homeobox gene in green plants. Proc Natl Acad Sci. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsisis encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signalling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Devon RS, Porteous DJ, Brookes AJ. Splinkerettes—Improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Quail PQ. The phytochromes: Photosensory perception and signal transduction. In: Greenland AJ, Meyerowitz EM, Steer MW, editors. Control of plant development: Genes and signals. Cambridge: Company of Biologists; 1998. pp. 85–92. [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Hudson M, Ni M, Qin M, Quail PH. poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc Natl Acad Sci. 1999;96:5832–5837. doi: 10.1073/pnas.96.10.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangarter RP. Gravity, light and plant form. Plant Cell Environ. 1997;20:796–800. doi: 10.1046/j.1365-3040.1997.d01-124.x. [DOI] [PubMed] [Google Scholar]
- Hennig L, Bueche C, Eichenberg K, Schäfer E. Dynamic properties of endogenous phytochrome A in Arabidopsisseedlings. Plant Physiol. 1999;121:571–577. doi: 10.1104/pp.121.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A–specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. Gus fusions: β glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3908. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. In: Photomorphogenesis in plants. 2nd ed. Kendrick RE, Kronenberg GHM, editors. Dordrecht: Kluwer; 1994. [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schaefer E, Nagy F. Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsismutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T, Neuhaus G, Batschauer A, Chua NH, Schäfer E. Functional analysis of yeast-derived phytochrome A and B phycocyanobilin adducts. Plant J. 1996;10:625–636. doi: 10.1046/j.1365-313x.1996.10040625.x. [DOI] [PubMed] [Google Scholar]
- Lesnick ML, Chandler VL. Activation of the maize anthocyanin gene a2is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 1998;117:437–445. doi: 10.1104/pp.117.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Hangarter RP. Light-stimulated apical hook opening in wild-type Arabidopsis thalianaseedlings. Plant Physiol. 1993;101:567–572. doi: 10.1104/pp.101.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Evan GI. Helix-loop-helix transcription factors. Oxford: Oxford University Press; 1998. [Google Scholar]
- Ludwig SR, Wessler SR. Maize R gene family tissue-specific helix-loop-helix proteins. Cell. 1990;62:849–852. doi: 10.1016/0092-8674(90)90259-h. [DOI] [PubMed] [Google Scholar]
- Mancinelli AL. The physiology of phytochrome action. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in plants. 2nd ed. Dordrecht: Kluwer; 1994. pp. 211–269. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element–bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, Deng XW. Light control of seedling morphogenetic pattern. Plant Cell. 1995;7:1749–1761. doi: 10.1105/tpc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schäfer E. Nuclear and cytosolic events of light-induced, phytochrome-regulated signaling in higher plants. EMBO J. 2000;19:157–163. doi: 10.1093/emboj/19.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- ————— Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Poppe C, Hangarter RP, Sharrock RA, Nagy F, Schaefer E. The light-induced reduction of the gravitropic growth-orientation of seedlings of Arabidopsis thaliana (L.) Heynh. is a photomorphogenic response mediated synergistically by the far-red-absorbing forms of phytochromes A and B. Planta (Heidelberg) 1996;199:511–514. doi: 10.1007/BF00195180. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsisdevelopment. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, Smith H. Genetic and transgenic evidence that phytochromes A and B act to modulate the gravitropic orientation of Arabidopsis thalianahypocotyls. Plant Physiol. 1996;110:211–216. doi: 10.1104/pp.110.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Smith H. Sensing the light environment: The functions of the phytochrome family. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in plants. 2nd ed. Dordrecht: Kluwer; 1994. pp. 377–416. [Google Scholar]
- Sogawa K, Nakano R, Kobayashi A, Kikuchi Y, Ohe N, Matsushita N, Fujii-Kuriyama Y. Possible function of Ah receptor nuclear translocator (Arnt) homodimer in transcriptional regulation. Proc Natl Acad Sci. 1995;92:1936–1940. doi: 10.1073/pnas.92.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Timpte C, Wilson AK, Estelle M. The axr2-1 mutation of Arabidopsis thalianais a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138:1239–1249. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng X-W. Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsisto human. Trends Genet. 1999;15:98–103. doi: 10.1016/s0168-9525(98)01670-9. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. phytochrome A null mutants of Arabidopsisdisplay a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]