Abstract
Human biospecimens are subject to a number of different collection, processing, and storage factors that can significantly alter their molecular composition and consistency. These biospecimen preanalytical factors, in turn, influence experimental outcomes and the ability to reproduce scientific results. Currently, the extent and type of information specific to the biospecimen preanalytical conditions reported in scientific publications and regulatory submissions varies widely. To improve the quality of research utilizing human tissues it is critical that information regarding the handling of biospecimens be reported in a thorough, accurate, and standardized manner. The Biospecimen Reporting for Improved Study Quality (BRISQ) recommendations outlined herein are intended to apply to any study in which human biospecimens are used. The purpose of reporting these details is to supply others, from researchers to regulators, with more consistent and standardized information to better evaluate, interpret, compare, and reproduce the experimental results. The BRISQ guidelines are proposed as an important and timely resource tool to strengthen communication and publications around biospecimen-related research and help reassure patient contributors and the advocacy community that the contributions are valued and respected.
Introduction
Human biospecimens provide the basis for research leading to better understanding of human disease and biology, and discovery of new diagnostics and treatments that are tailored to individual patients with cancer or other diseases. These biological materials are subject to a number of different collection, processing, and storage factors that can significantly alter their molecular composition and consistency. Such preanalytical factors can, in turn, influence experimental outcomes and the ability to reproduce scientific results. A growing number of studies have demonstrated the effects of biospecimen preanalytical factors on molecular measurements.1–7 In biomarker studies, such variations can result in artifacts being misinterpreted as experimental results.6,8 Preanalytical factors can also contribute to false-negative and false-positive results in assays for determining appropriate therapies for cancer patients.9,10 Currently, the extent and type of information specific to the biospecimen preanalytical conditions reported in scientific publications and regulatory submissions varies widely. To improve the quality of research using human specimens it is critical that information regarding the handling of biospecimens be reported in a thorough, accurate, and standardized manner.
The purpose of this paper is to make recommendations for the reporting of data elements for human biospecimens, defined as solid tissues and bodily fluids, used in biomedical studies. Cell lines and biospecimen derivatives such as nucleic acids or proteins, while crucial for biomedical research, are not intended to fall within the scope of these recommendations. The Biospecimen Reporting for Improved Study Quality (BRISQ) recommendations are intended to apply to any study in which human biospecimens are used. This includes biomedical applications such as translational science, biomarker discovery, clinical trials, technology development, and diagnostic-assay and therapeutics development. The recommended data elements would be reported by an author in a journal publication, by a company in a regulatory submission, or by a biorepository distributing biospecimens. It is intended that the list and the elements within it will be interpreted, modified, and applied according to the context of the study being reported. It is also recognized that information corresponding to all data elements may not be available but at least for some categories (described below) the known or unknown status of these elements should be documented.
The list of data elements discussed includes general information for consistent documentation of classes of biospecimens and factors that might influence the integrity, quality, and/or molecular composition of biospecimens. Reporting the details enumerated in the BRISQ list does not guarantee biospecimen quality, and should not be seen as a substitute for empirical quality evaluations. The purpose of reporting these details is to supply others, from researchers to regulatory agencies, with more consistent and standardized information to better evaluate, interpret, compare, and reproduce the experimental results. To maintain consistency with federal regulations on research involving human subjects, information that might enable individual identification of research participants should be withheld.
The BRISQ list has been constructed as an initial step towards defining reporting recommendations. The list will likely evolve as more is learned about the factors that influence biospecimen quality and composition, and in turn their effects on biospecimen analysis. It is envisioned that future iterations of the BRISQ recommendations might include changes to the list of elements and the relative weight thereof in accordance with evidence-based scientific and medical findings and technological developments.
Materials and Methods
A half-day workshop, Development of Biospecimen Reporting Criteria for Publications, was held at the National Cancer Institute (NCI) 2009 Biospecimen Research Network Symposium (http://biospecimens.cancer.gov/meeting/brnsymposium) to initiate a discussion on biospecimen reporting recommendations. Workshop attendees included individuals covering a broad range of expertise: laboratory scientists, clinicians, pathologists, statisticians, patient advocates, biobankers, journal editors, leaders of relevant professional societies, and other stakeholders. The attendees noted that reporting guidelines covering many aspects of biomedical studies already exist, particularly guidelines relevant to experimental design and data reporting.1 It was proposed that the BRISQ recommendations apply to all studies utilizing human biospecimens, and thus complement existing guidelines by filling a niche concerning reporting of biospecimen characteristics and preanalytical variables.
The attendees further proposed that the BRISQ recommendations should broadly encompass solid tissues and bodily fluids, rather than including separate lists for these biospecimen types. It was also agreed that a committee to develop biospecimen reporting recommendations should be formed to take the effort forward. Many of the individuals and disciplines participating in the workshop were included when the BRISQ committee was subsequently formed.
Formulation of the recommendations was based on consideration of what biospecimen information could enable a science reviewer to fully evaluate or replicate a reported study. The preliminary list included the most commonly available data elements. The committee considered the characteristics of the biospecimens themselves as well as numerous preanalytical factors. Types of data elements include the tissue type and the pathology of the sample; patient characteristics that might influence the biospecimens, such as vital and disease states; and the collection and handling of the biospecimens, e.g., the stabilization, shipping, and storage conditions.
The preliminary list of recommendations was refined by consulting the NCI Biospecimen Research Database (http://brd.nci.nih.gov), an online resource compiling peer-reviewed articles that address biospecimen science. The Biospecimen Research Database’s terminology for scientific literature curation that was deemed relevant was incorporated into the initial BRISQ list. This terminology served as a starting point for discussion at monthly teleconferences by the BRISQ committee.
Results
The committee composed a list of data elements that represent factors believed to often influence biospecimen quality and thus should be considered for reporting, if known or applicable, for the particular study; for example, some list elements will be more applicable to biospecimens collected for a disease specific study than those collected for a population based biospecimen resource. For clarity, these elements are organized according to the lifecycle of the biospecimen (Figure 1), which spans the period immediately prior to removal from the patient through use in a scientific analysis.
Figure 1.

The Lifecycle of the Biospecimen.
The preanalytical phase of the lifecycle of the biospecimen includes each stage from Patient to Distribution. Preanalytical variables are addressed in the BRISQ list.
Many reporting elements were discussed, but only some were approved by consensus for inclusion in the guidelines. The committee was mindful that certain information, while important to report, may not have direct relevance to the biology or condition of the biospecimen, and therefore, would not be under the purview of the BRISQ recommendations. The committee attempted to carefully balance scientific interest in having access to extensive data about biospecimen collection, processing, and storage against practical challenges in obtaining such detailed information. Each reporting element included in the guidelines is backed by evidence that the factor could have an effect on the structural integrity and molecular characteristics of the biospecimen or on the ability to perform certain assays on the biospecimen and obtain reliable results. While the committee recognizes that collection of data about biospecimens can increase the operational costs to collect and use biospecimens, cost was not factored into the exclusion of data elements that were or should be considered necessary.
The elements in the BRISQ list are prioritized into three tiers according to the relative importance of their being reported. The first tier, items recommended to report, includes information such as the organ(s) or the anatomical site from which the biospecimens were derived and the manner in which the biospecimens were collected, stabilized, and preserved; for quick reference, these items are summarized in Table 1. Reporting these items need not be onerous. For example, Beatty et al.11 include most BRISQ Tier 1 items in the following excerpts:
Table 1.
Quick-reference BRISQ Summary/Checklist: Tier 1 items to report if known and applicable.
| Data Elements | Examples | |
|---|---|---|
| ❒ | Biospecimen type | Serum, Urine |
| Solid tissue, whole blood, or another product derived from a human being | ||
| ❒ | Anatomical site | Liver, Antecubital area of the arm |
| Organ of origin or site of blood draw | ||
| ❒ | Disease status of patients | Diabetic, Healthy control |
| Controls or individuals with the disease of interest | ||
| ❒ | Clinical characteristics of patients | Pre-menopausal breast cancer patients |
| Available medical information known or believed to be pertinent to the condition of the biospecimens | ||
| ❒ | Vital State of patients | Postmortem |
| Alive or deceased patient when biospecimens were obtained | ||
| ❒ | Clinical diagnosis of patients | Breast cancer |
| Patient clinical diagnoses (determined by medical history, physical examination, and analyses of the biospecimen) pertinent to the study | ||
| ❒ | Pathology diagnosis | Her2-negative intraductal carcinoma |
| Patient pathology diagnoses (determined by macro and/or microscopic evaluation of the biospecimen at the time of diagnosis and/or prior to research use) pertinent to the study | ||
| ❒ | Collection mechanism | Fine needle aspiration, Pre-operative blood draw |
| How the biospecimens were obtained | ||
| ❒ | Type of stabilization | Heparin, On ice |
| The initial process by which biospecimens were stabilized during collection | ||
| ❒ | Type of long-term preservation | Formalin fixation, freezing |
| The process by which the biospecimens were sustained after collection | ||
| ❒ | Constitution of preservative | 10% neutral-buffered formalin, 10 USP Heparin Units/mL |
| The make-up of any formulation used to maintain the biospecimens in a non-reactive state | ||
| ❒ | Storage temperature | −80 °C, 20 to 25 °C |
| The temperature or range thereof at which the biospecimens were kept until distribution/analysis. | ||
| ❒ | Storage duration | 8 days, 5 to 7 years |
| The time or range thereof between biospecimen acquisition and distribution or analysis. | ||
| ❒ | Shipping temperature | −170 °C to −190 °C |
| The temperature or range thereof at which biospecimens were kept during shipment or relocation. | ||
| ❒ | Composition assessment & selection | Minimum 80% tumour nuclei & maximum 50% necrosis |
| Parameters used to choose biospecimens for the study | ||
“FNA [fine-needle aspiration] specimens were obtained from 55 surgically removed specimens of breast cancer within 1 hour of resection, before tissue fixation. The aspirates were obtained using a 22- to 25-gauge needle and spread directly on slides and fixed in ethanol or formalin or placed in CytoLyt for preparation of ThinPrep slides according to the manufacturer’s protocol. Corresponding FFPE [formalin-fixed, paraffin-embedded] tissue specimens were fixed in 10% neutral buffered formalin for 18 to 24 hours according to routine procedures and embedded in paraffin.”
“All FNA cytologic slides were air dried and stored at room temperature before FISH analysis.”
Items beneficial to report form the second tier. These are data elements an evaluator might find helpful to know but may be slightly less crucial to the scientific contribution or less likely to be annotated, such as the time from biospecimen excision/acquisition to stabilization. Additional items to report compose the third tier. These include information about conditions that might be useful to know concerning the biospecimens but are not known to be as likely to influence research results or are unlikely to be available to researchers, such as environmental factors to which patients were exposed or the type of storage container in which the biospecimens were kept.
The full BRISQ list featured in Table 2 includes each item and its definition along with additional columns that were designed for an author or reviewer to track where the listed items are reported for a particular study. To the right of the Item Descriptions is a column assigning each item a unique Roman-numeral/letter/number identification code. The far right column provides space to note where each item may be found in a manuscript or application. The far left Apply-to column indicates whether the BRISQ item is applicable to All biospecimen types or is more appropriate for solid Tissue biospecimens or Fluid biospecimens (such as blood, urine, or other fluids). For example, item III.b, “Type of long-term preservation,” is pertinent to all types of biospecimens; item III.b.2, “Time in fixative/preservative solution,” is more relevant to solid tissue than to fluid biospecimens; and item III.c, “Aliquot volume,” applies more often to fluid than to solid tissue biospecimens.
Table 2.
BRISQ table with example references, when available, that exemplify each data element’s influence on experimental results. This is not intended to be an exhaustive list.
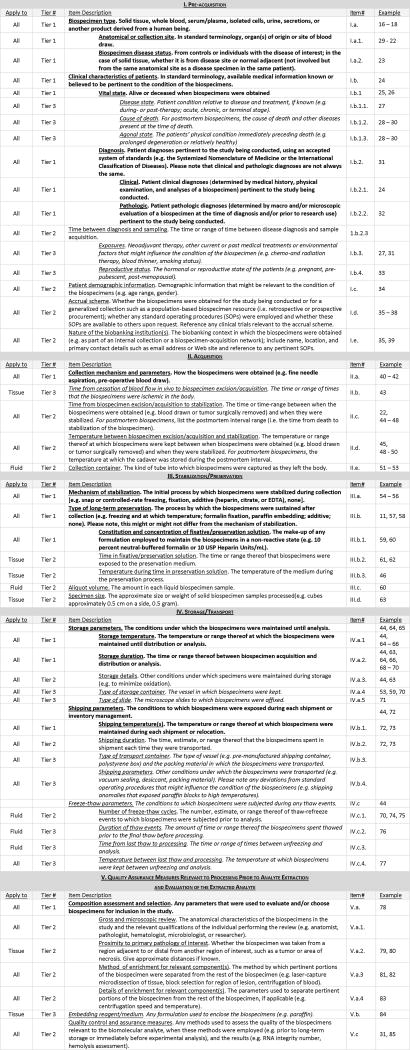
|
When reporting elements of the BRISQ list, standard operating procedures specifying many of the pertinent details, such as blood-collection protocols, may be provided or referenced; any referenced documents should be publicly available. It is preferable that most Tier 1 items relevant to the biospecimen and particular scientific study be reported directly in the intended publication rather than be cited from another document. Detailed descriptions that are too lengthy to be accommodated should be made available as supplemental materials online. Whether the laboratory performing the study was operating under any formal certification or accreditation should be stated if applicable to the study being reported.
The BRISQ committee discussed whether to request information that the biorepository and/or researcher had obtained ethical clearance to collect the biospecimens and perform the study. Clearance from an institutional review board or similar body is important to report in publications, and its reporting is generally required by journals. However, it is not immediately pertinent to the structural integrity and molecular characteristics of the biospecimen and, thus, is not included in the BRISQ recommendations. Similarly, accurate biospecimen-tracking mechanisms are essential to biobanking but not immediately pertinent to the condition of the biospecimen, and thus are also not included in the BRISQ data-elements list.
Surgical parameters, such as type of anesthesia or receipt of blood or other intra-operative infusates, were recognized to be of potential significance to the condition of the biospecimens. However, these data often are not known. When it is available, information about anesthesia and intraoperative treatments that may influence the condition of the biospecimens should be reported. These elements were not included in the BRISQ list because currently such information is rarely available or not required to be recorded as part of biospecimen collection efforts. If or when surgical parameters are determined to be critical through systematic biospecimen research studies these elements will be integrated into future recommendations.
Several preservation parameters known to influence the condition of biospecimens and the results of analyses have been included in the list of recommendations. Researchers should state the rationale for the chosen preservation parameters. For example, if the type and temperature of the biospecimen preservative were selected to optimize stability, extraction, and analysis of a particular analyte, this should be mentioned.
The BRISQ committee recognized the need for greater specificity in the anatomic and histologic details reported concerning solid tissue biospecimens. The committee agreed that the level of detail with which pathology characteristics are reported should be enough to sufficiently address the scientific research question. These characteristics include not only the tissue site of the biospecimen and the relation of the biospecimen to the pertinent clinical diagnosis within the tissue site, but also the composition and pathology within the biospecimen where relevant.
The BRISQ committee included members of the NCI Office of Biorepositories and Biospecimen research (OBBR), participants from the OBBR Biospecimen Research Network Symposium, and members of the International Society for Biological and Environmental Repositories (ISBER) and the committees responsible for the REporting recommendations for tumor MARKer prognostic studies (REMARK)12 and STrengthening the Reporting of OBservational studies in Epidemiology (STROBE)13 guidelines. Essential harmonization with similar efforts underway by these groups is ongoing.
Discussion
An adage in the business community states, “That which is measured improves. That which is measured and reported improves exponentially.” The BRISQ reporting recommendations represent the product of extensive discussion and input from researchers with varied types of expertise and from many stakeholders, all of whom share the common goal of improving biospecimen reporting and, by extension, fields in which biospecimens are employed. The committee believes that by providing details concerning preanalytical factors that might affect assay results, investigators will further improve the quality of biomedical studies, including research for developing cancer biomarkers for screening, early detection, and treatment.
Adoption of the BRISQ recommendations is expected to help authors, reviewers, editors, and regulatory officials evaluate whether sufficient information about the biospecimens has been provided to enable assessment of the influence of preanalytical biospecimen factors on study results. If reported, this information will allow improved evaluation, interpretation, comparison, and reproduction of the results from studies that employ human biospecimens. Although items in any Tier might not be available or in Tiers 2 or 3 might not be considered significant to report, increased awareness of their potential influence on biospecimen studies might lead to improved tracking and reporting in the future.
The BRISQ recommendations may be implemented by anyone reporting on studies involving biospecimens. Reviewers, editors, and regulatory officials might also employ the list as a tool for evaluating whether sufficient biospecimen information has been included in a manuscript or application. In addition, the recommendations might be employed by investigators requesting biospecimens from a biospecimen resource: essential items on the list might be checked off to indicate the annotation needed for the requested batch of samples. Elements of BRISQ that document preanalytical variables for tissue biospecimens could be economically captured using a reporting system such as the Standard PREanalytical Code, or SPREC, which was recently published by the ISBER Working Group on Biospecimen Science.14
BRISQ reporting items will not necessarily be applicable to every study, and authors and reviewers are urged to use their judgment to decide which factors are essential. It is not always possible for investigators to ascertain every recommended element for every biospecimen, even for Tier 1 items, but unknown elements relevant to the study being reported should be fully acknowledged with a discussion of possible implications that the missing information might have on the study conclusions. Unknown or unreported Tier 1 data elements should not be considered a reason for automatic dismissal of a report or conditional for the award of a grant. The final decision on acceptability of missing Tier 1 information should be specific to the study context.
When consulting the BRISQ list, researchers should evaluate the importance of each item in the context of the study, and adjust their reporting accordingly. An item such as “method of enrichment for relevant components,” listed here as Tier 2 might—for example, in the context of a study comparing the efficacy of various enrichment methods—be essential to report and should thus be considered Tier 1 for that study. The converse may also be true, when, for example, an item listed here as Tier 2—such as “temperature between acquisition and stabilization”—is less pertinent to the study at hand—perhaps because the time at this temperature was negligible—and should be considered Tier 3.
It is hoped that consideration of the BRISQ recommendations will sensitize the biobanking and research communities and their funding agencies to the importance of tracking preanalytical variables, leading to more judicious selection and handling of experimental human specimens and thus improved study quality. Anecdotally, recommendations such as REMARK seem to have had the effect of spurring researchers to consider the recommendations in advance of conducting their investigations, with the result that researchers might take greater care in the design, conduct, and analysis of their studies. The BRISQ committee envisions a similar trajectory for preanalytical biospecimen data elements. Thus, not only might overall quality of publications improve, but the quality of human-biospecimen-dependent investigation in general might improve over time with the formation and adoption of publication recommendations. It is anticipated that biospecimen resources might use these recommendations to improve on their existing standard operating procedures and annotation thereof. Such improvements could include the acquisition of additional relevant biospecimen data based on the BRISQ recommendations and the release of all such data to researchers as a standard procedure. In this way, biospecimen resources might become major players in the universal application of these recommendations.
Patient contribution of biospecimens for research is a voluntary, generous action aimed at helping advance scientific discovery and progress. The research team, pathologist, and biorepository systems, as the stewards of these biospecimens, have a responsibility to be vigilant and persistent in using methods and practices that protect and preserve the highest possible quality biospecimen and associated data. The BRISQ guidelines are proposed as an important and timely resource tool to strengthen communication and publications around biospecimen-related research and help reassure patient contributors and the advocacy community that the contributions are valued and respected. Researchers are further encouraged to strengthen public outreach and education around the use and potential of human biospecimens15 and the biorepository community as these are emerging and potentially misunderstood areas.
Acknowledgments
This project has been funded in whole or in part with Federal Funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
This work was supported in part by NIH grant CA136685 (HUW) carried out at the Lawrence Berkeley National Laboratory under contract DE-AC02-05CH11231.
Footnotes
The EQUATOR project (http://www.equator-network.org/) provides an extensive listing of guidelines for health research.
Contributor Information
Helen M. Moore, Office of Biorepositories and Biospecimen Research, National Cancer Institute
Andrea Kelly, Rose Li and Associates, Inc
Scott D. Jewell, Director, Program for Biospecimen Science Senior Scientific Investigator, Van Andel Research Institute
Lisa M. McShane, Biometric Research Branch, National Cancer Institute
Douglas P. Clark, Johns Hopkins Hospital
Renata Greenspan, U.S. Military Cancer Institute
Daniel F. Hayes, University of Michigan Comprehensive Cancer Center
Pierre Hainaut, International Agency for Research on Cancer, World Health Organization
Paula Kim, Translating Research Across Communities.
Elizabeth Mansfield, Food and Drug Administration
Olga Potapova, Cureline, Inc
Peter Riegman, Erasmus MC Tissue Bank
Yaffa Rubinstein, Office of Rare Diseases Research, National Institutes of Health
Edward Seijo, H. Lee Moffitt Cancer Center & Research Institute
Stella Somiari, Windber Research Institute
Peter Watson, Vancouver Island Center, British Columbia Cancer Agency
Heinz-Ulrich Weier, Lawrence Berkeley National Laboratory
Claire Zhu, Division of Cancer Prevention, National Cancer Institute
Jim Vaught, Office of Biorepositories and Biospecimen Research, National Cancer Institute
References
- 1.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161(6):1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore HM, Compton CC, Lim MD, et al. Biospecimen Research Network Symposium: Advancing Cancer Research through Biospecimen Science. Cancer Res. 2009;69:6770–6772. doi: 10.1158/0008-5472.CAN-09-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apweiler R, Aslanidis C, Deufel T, et al. Approaching clinical proteomics: current state and future fields of application in cellular proteomics. Cytometry A. 2009a;75(10):816–32. doi: 10.1002/cyto.a.20779. [DOI] [PubMed] [Google Scholar]
- 4.Apweiler R, Aslanidis C, Deufel T, et al. Approaching clinical proteomics: current state and future fields of application in fluid proteomics. Clin Chem Lab Med. 2009b;47(6):724–44. doi: 10.1515/CCLM.2009.167. [DOI] [PubMed] [Google Scholar]
- 5.Espina V, Muelle C, Edmiston K, et al. Tissue is alive: New technologies are needed to address the problems of protein biomarker pre-analytical variability. Proteomics Clin Appl. 2009;3:874–82. doi: 10.1002/prca.200800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ransohoff DF, Gourlay ML. Sources of Bias in Specimens for Research About Molecular Markers for Cancer. J Clin Oncol. 2010;28(4):698–704. doi: 10.1200/JCO.2009.25.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel KB, Moore HM. Effects of preanalytic variables on the detection of proteins by immunohistochemistry in formalin-fixed, paraffin-embedded tissue Arch. Pathol Lab Med. doi: 10.5858/2010-0702-RAIR.1. In press. [DOI] [PubMed] [Google Scholar]
- 8.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–9. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 9.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF American Society of Clinical Oncology; College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 10.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC American Society of Clinical Oncology; College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch Pathol Lab Med. 2010;134(7):e48–72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 11.Beatty BG, Bryant R, Wang W, et al. HER-2/neu detection in fine-needle aspirates of breast cancer: fluorescence in situ hybridization and immunocytochemical analysis. Am J Clin Pathol. 2004;122(2):246–55. doi: 10.1309/N82C-TQ1J-0XEV-EFQB. [DOI] [PubMed] [Google Scholar]
- 12.McShane LM, Altman DG, Sauerbrei W, et al. Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 13.von Elm E, Altman DG, Egger M, et al. STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008 Apr;61(4):344–9. doi: 10.1016/j.jclinepi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Betsou F, Lehmann S, Ashton G, et al. Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiol Biomarkers Prev. 2010;19(4):1004–11. doi: 10.1158/1055-9965.EPI-09-1268. [DOI] [PubMed] [Google Scholar]
- 15.Secko DM, Preto N, Niemeyer S, et al. Informed consent in biobank research: a deliberative approach to the debate. Soc Sci Med. 2009;68(4):781–9. doi: 10.1016/j.socscimed.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Di Nunno N, Costantinides F, Cina SJ, et al. What is the best sample for determining the early postmortem period by on-the-spot flow cytometry analysis? Am J Forensic Med Pathol. 2002;23(2):173–80. doi: 10.1097/00000433-200206000-00013. [DOI] [PubMed] [Google Scholar]
- 17.Humphreys-Beher MG, King FK, Bunnel B, et al. Isolation of biologically active RNA from human autopsy for the study of cystic fibrosis. Biotechnol Appl Biochem. 1986;8(5):392–403. [PubMed] [Google Scholar]
- 18.Barton RH, Nicholson JK, Elliott P, et al. High-throughput 1H NMR-based metabolic analysis of human serum and urine for large-scale epidemiological studies: validation study. Int J Epidemiol. 2008;37 (Suppl 1):i31–40. doi: 10.1093/ije/dym284. [DOI] [PubMed] [Google Scholar]
- 19.Centeno BA, Enkemann SA, Coppola D, et al. Classification of human tumors using gene expression profiles obtained after microarray analysis of fine-needle aspiration biopsy samples. Cancer. 2005;25; 105(2):101–9. doi: 10.1002/cncr.20737. [DOI] [PubMed] [Google Scholar]
- 20.Hoff-Olsen P, Jacobsen S, Mevåg B, et al. Microsatellite stability in human postmortem tissues. Forensic Sci Int. 2001;119(3):273–8. doi: 10.1016/s0379-0738(00)00443-6. [DOI] [PubMed] [Google Scholar]
- 21.Yang ZW, Yang SH, Chen L, et al. Comparison of blood counts in venous, fingertip and arterial blood and their measurement variation. Clin Lab Haematol. 2001;23(3):155–9. doi: 10.1046/j.1365-2257.2001.00388.x. [DOI] [PubMed] [Google Scholar]
- 22.Heinrich M, Matt K, Lutz-Bonengel S, et al. Successful RNA extraction from various human postmortem tissues. Int J Legal Med. 2007;121(2):136–42. doi: 10.1007/s00414-006-0131-9. [DOI] [PubMed] [Google Scholar]
- 23.Weis S, Llenos IC, Dulay JR, et al. Quality control for microarray analysis of human brain samples: The impact of postmortem factors, RNA characteristics, and histopathology. J Neurosci Methods. 2007;165(2):198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Tantipaiboonwong P, Sinchaikul S, Sriyam S, et al. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics. 2005;5(4):1140–9. doi: 10.1002/pmic.200401143. [DOI] [PubMed] [Google Scholar]
- 25.He S, Wang Q, He J, et al. Proteomic analysis and comparison of the biopsy and autopsy specimen of human brain temporal lobe. Proteomics. 2006;6(18):4987–96. doi: 10.1002/pmic.200600078. [DOI] [PubMed] [Google Scholar]
- 26.Jones RF, Sunheimer R, Friedman H, et al. Comparison of ante- and post-mortem PSA levels for epidemiological studies. Anticancer Res. 2005;25(2B):1263–7. [PubMed] [Google Scholar]
- 27.Pinder SE, Provenzano E, Earl H, Ellis IO. Laboratory handling and histology reporting of breast specimens from patients who have received neoadjuvant chemotherapy. Histopathology. 2007;50(4):409–17. doi: 10.1111/j.1365-2559.2006.02419.x. [DOI] [PubMed] [Google Scholar]
- 28.Tomita H, Vawter MP, Walsh DM, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55(4):346–52. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preece P, Virley DJ, Costandi M, et al. An optimistic view for quantifying mRNA in post-mortem human brain. Brain Res Mol Brain Res. 200319; 116(1–2):7–16. doi: 10.1016/s0169-328x(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 30.Johnston NL, Cervenak J, Shore AD, et al. Multivariate analysis of RNA levels from postmortem human brains as measured by three different methods of RT-PCR. Stanley Neuropathology Consortium. J Neurosci Methods. 1997;7; 77(1):83–92. doi: 10.1016/s0165-0270(97)00115-5. [DOI] [PubMed] [Google Scholar]
- 31.Webster MJ. Tissue preparation and banking. Prog Brain Res. 2006;158:3–14. doi: 10.1016/S0079-6123(06)58001-X. [DOI] [PubMed] [Google Scholar]
- 32.Ellis M, Davis N, Coop A, et al. Development and validation of a method for using breast core needle biopsies for gene expression microarray analyses. Clin Cancer Res. 2002;8(5):1155–66. [PubMed] [Google Scholar]
- 33.Reyna R, Traynor KD, Hines G, et al. Repeated freezing and thawing does not generally alter assay results for several commonly studied reproductive hormones. Fertil Steril. 2001;76:823–5. doi: 10.1016/s0015-0282(01)01986-0. [DOI] [PubMed] [Google Scholar]
- 34.Papale M, Pedicillo MC, Thatcher BJ, et al. Urine profiling by SELDI-TOF/MS: monitoring of the critical steps in sample collection, handling and analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;856(1–2):205–13. doi: 10.1016/j.jchromb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Barnes RO, Parisien M, Murphy LC, et al. Influence of evolution in tumor biobanking on the interpretation of translational research. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3344–50. doi: 10.1158/1055-9965.EPI-08-0622. [DOI] [PubMed] [Google Scholar]
- 36.McIntosh M, Anderson G, Drescher C, et al. Ovarian cancer early detection claims are biased. Clin Cancer Res. 2008 Nov 15;14(22):7574. doi: 10.1158/1078-0432.CCR-08-0623. [DOI] [PubMed] [Google Scholar]
- 37.Sidiropoulos N, Dumont LJ, Golding AC, et al. Quality improvement by standardization of procurement and processing of thyroid fine-needle aspirates in the absence of on-site cytological evaluation. Thyroid. 2009;19(10):1049–52. doi: 10.1089/thy.2009.0161. [DOI] [PubMed] [Google Scholar]
- 38.Thorpe JD, Duan X, Forrest R, et al. Effects of blood collection conditions on ovarian cancer serum markers. PLoS One. 2007 Dec 5;2(12):e1281. doi: 10.1371/journal.pone.0001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karsan A, Eigl BJ, Flibotte S, et al. Analytical and preanalytical biases in serum proteomic pattern analysis for breast cancer diagnosis. Clin Chem. 2005;51(8):1525–8. doi: 10.1373/clinchem.2005.050708. [DOI] [PubMed] [Google Scholar]
- 40.Sung MT, Lin H, Koch MO, et al. Radial distance of extraprostatic extension measured by ocular micrometer is an independent predictor of prostate-specific antigen recurrence: A new proposal for the substaging of pT3a prostate cancer. Am J Surg Pathol. 2007;31(2):311–8. doi: 10.1097/01.pas.0000213359.26003.37. [DOI] [PubMed] [Google Scholar]
- 41.Morrison C, Palatini J, Riggenbach J, et al. Fine-needle aspiration biopsy of non-Hodgkin lymphoma for use in expression microarray analysis. Cancer. 2006;108(5):311–8. doi: 10.1002/cncr.22174. [DOI] [PubMed] [Google Scholar]
- 42.Schaub S, Wilkins J, Weiler T, et al. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65(1):323–32. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- 43.Smith JL, Pillay SP, de Jersey J, et al. Effect of ischaemia on the activities of human hepatic acyl-CoA:cholesterol acyltransferase and other microsomal enzymes. Clin Chim Acta. 1989;184(3):259–68. doi: 10.1016/0009-8981(89)90059-4. [DOI] [PubMed] [Google Scholar]
- 44.Visvikis S, Schlenck A, Maurice M. DNA extraction and stability for epidemiological studies. Clin Chem Lab Med. 1998;36(8):551–5. doi: 10.1515/CCLM.1998.094. [DOI] [PubMed] [Google Scholar]
- 45.Micke P, Ohshima M, Tahmasebpoor S, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86(2):202–11. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- 46.Burke WJ, O’Malley KL, Chung HD, et al. Effect of pre- and postmortem variables on specific mRNA levels in human brain. Brain Res Mol Brain Res. 1991;11(1):37. doi: 10.1016/0169-328x(91)90018-s. [DOI] [PubMed] [Google Scholar]
- 47.Spruessel A, Steimann G, Jung M, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 2004;36(6):1030–7. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 48.Espina V, Edmiston KH, Heiby M, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics. 2008;7(10):1998–2018. doi: 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Maldegem F, de Wit M, Morsink F, et al. Effects of processing delay, formalin fixation, and immunohistochemistry on RNA Recovery From Formalin-fixed Paraffin-embedded Tissue Sections. Diagn Mol Pathol. 2008;17(1):51–8. doi: 10.1097/PDM.0b013e31814b8866. [DOI] [PubMed] [Google Scholar]
- 50.Langebrake C, Günther K, Lauber J, et al. Preanalytical mRNA stabilization of whole bone marrow samples. Clin Chem. 2007;53(4):5, 87–93. doi: 10.1373/clinchem.2006.078592. [DOI] [PubMed] [Google Scholar]
- 51.Yucel A, Karakus R, Cemalettin A. Effect of blood collection tube types on the measurement of human epidermal growth factor. J Immunoassay Immunochem. 2007;28(1):47–60. doi: 10.1080/15321810601026091. [DOI] [PubMed] [Google Scholar]
- 52.Drake SK, Bowen RA, Remaley AT, et al. Potential interferences from blood collection tubes in mass spectrometric analyses of serum polypeptides. Clin Chem. 2004;50(12):2398–401. doi: 10.1373/clinchem.2004.040303. [DOI] [PubMed] [Google Scholar]
- 53.Preissner CM, Reilly WM, Cyr RC, et al. Plastic versus glass tubes: effects on analytical performance of selected serum and plasma hormone assays. Clin Chem. 2004;50(7):1245–7. doi: 10.1373/clinchem.2004.034108. [DOI] [PubMed] [Google Scholar]
- 54.Frank M, Döring C, Metzler D, et al. Global gene expression profiling of formalin-fixed paraffin-embedded tumor samples: a comparison to snap-frozen material using oligonucleotide microarrays. Virchows Arch. 2007;450(6):699–711. doi: 10.1007/s00428-007-0412-9. [DOI] [PubMed] [Google Scholar]
- 55.Scicchitano MS, Dalmas DA, Bertiaux MA, et al. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J Histochem Cytochem. 2006;54(11):1229–37. doi: 10.1369/jhc.6A6999.2006. [DOI] [PubMed] [Google Scholar]
- 56.Narayanan S. Considerations in the application of selected molecular biology techniques in the clinical laboratory: preanalytical and analytical issues. Rinsho Byori. 1996;(Suppl 103):262–70. [PubMed] [Google Scholar]
- 57.Greer CE, Lund JK, Manos MM. PCR amplification from paraffin-embedded tissues: recommendations on fixatives for long-term storage and prospective studies. PCR Methods Appl. 1991;1(1):46–50. doi: 10.1101/gr.1.1.46. [DOI] [PubMed] [Google Scholar]
- 58.Kouri T, Malminiemi O, Penders J. Limits of preservation of samples for urine strip tests and particle counting. Clin Chem Lab Med. 2008;46(5):703–13. doi: 10.1515/cclm.2008.122. [DOI] [PubMed] [Google Scholar]
- 59.Zsikla V, Baumann M, Cathomas G. Effect of buffered formalin on amplification of DNA from paraffin wax embedded small biopsies using real-time PCR. J Clin Pathol. 2004;57(6):654–6. doi: 10.1136/jcp.2003.013961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferry JD, Collins S, Sykes E. Effect of serum volume and time of exposure to gel barrier tubes on results for progesterone by Roche Diagnostics Elecsys 2010. Clin Chem. 1999;45(9):1574–5. [PubMed] [Google Scholar]
- 61.Macabeo-Ong M, Ginzinger DG, Dekker N, et al. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002;15(9):979–87. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- 62.Miething F, Hering S, Hanschke B, et al. Effect of fixation to the degradation of nuclear and mitochondrial DNA in different tissues. J Histochem Cytochem. 2006;54(3):371–4. doi: 10.1369/jhc.5B6726.2005. [DOI] [PubMed] [Google Scholar]
- 63.Gillio-Tos A, De Marco L, Fiano V, et al. Efficient DNA extraction from 25-year-old paraffin-embedded tissues: study of 365 samples. Pathology. 2007;39(3):345–8. doi: 10.1080/00313020701329757. [DOI] [PubMed] [Google Scholar]
- 64.Sigurdson AJ, Ha M, Cosentino M, et al. Long-term storage and recovery of buccal cell DNA from treated cards. Cancer Epidemiol Biomarkers Prev. 2006;15(2):385–8. doi: 10.1158/1055-9965.EPI-05-0662. [DOI] [PubMed] [Google Scholar]
- 65.Atkins D, Reiffen KA, Tegtmeier CL, et al. Immunohistochemical detection of EGFR in paraffin-embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. J Histochem Cytochem. 2004;52(7):893–901. doi: 10.1369/jhc.3A6195.2004. [DOI] [PubMed] [Google Scholar]
- 66.Zhou H, Yuen PS, Pisitkun T, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69(8):1471–6. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad S, Sundaramoorthy E, Arora R, et al. Progressive degradation of serum samples limits proteomic biomarker discovery. Anal Biochem. 2009;394(2):237–42. doi: 10.1016/j.ab.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 68.Isaksson HS, Nilsson TK. Preanalytical aspects of quantitative TaqMan real-time RT-PCR: applications for TF and VEGF mRNA quantification. Clin Biochem. 2006;39(4):373–7. doi: 10.1016/j.clinbiochem.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 69.Kauppinen T, Martikainen P, Alafuzoff I. Human postmortem brain tissue and 2-mm tissue microarrays. Appl Immunohistochem Mol Morphol. 2006;14(3):353–9. doi: 10.1097/00129039-200609000-00016. [DOI] [PubMed] [Google Scholar]
- 70.Rosenling T, Slim CL, Christin C, et al. The effect of preanalytical factors on stability of the proteome and selected metabolites in cerebrospinal fluid (CSF) J Proteome Res. 2009;8(12):5511–22. doi: 10.1021/pr9005876. [DOI] [PubMed] [Google Scholar]
- 71.Paik S, Kim CY, Song YK, Kim WS. Technology insight: Application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat Clin Pract Oncol. 2005;2(5):246–54. doi: 10.1038/ncponc0171. [DOI] [PubMed] [Google Scholar]
- 72.Guder WG. Preanalytical factors and their influence on analytical quality specifications. Scand J Clin Lab Invest. 1999;59(7):545–9. doi: 10.1080/00365519950185328. [DOI] [PubMed] [Google Scholar]
- 73.Timms JF, Arslan-Low E, Gentry-Maharaj A, et al. Preanalytic influence of sample handling on SELDI-TOF serum protein profiles. Clin Chem. 2007;53(4):645–56. doi: 10.1373/clinchem.2006.080101. [DOI] [PubMed] [Google Scholar]
- 74.Chan KC, Yeung SW, Lui WB, et al. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin Chem. 2005;51(4):781–4. doi: 10.1373/clinchem.2004.046219. [DOI] [PubMed] [Google Scholar]
- 75.Fiedler GM, Baumann S, Leichtle A, et al. Standardized peptidome profiling of human urine by magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem. 2007;53(3):421–8. doi: 10.1373/clinchem.2006.077834. [DOI] [PubMed] [Google Scholar]
- 76.Kirk MJ, Hayward RM, Sproull M. Non-patient related variables affecting levels of vascular endothelial growth factor in urine biospecimens. J Cell Mol Med. 2008;12(4):1250–5. doi: 10.1111/j.1582-4934.2008.00182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kueltzo LA, Wang W, Randolph TW, et al. Effects of solution conditions, processing parameters, and container materials on aggregation of a monoclonal antibody during freeze-thawing. J Pharm Sci. 2008;97(5):1801–12. doi: 10.1002/jps.21110. [DOI] [PubMed] [Google Scholar]
- 78.Ginocchio CC, Wang XP, Kaplan MH, et al. Effects of specimen collection, processing, and storage conditions on stability of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1997;35(11):2886–93. doi: 10.1128/jcm.35.11.2886-2893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Braakhuis BJM, Tabor MP, Kummer JA, et al. A genetic explanation of slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63(8):1727–30. [PubMed] [Google Scholar]
- 80.Deng G, Lu Y, Zlotnikov G, et al. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274(5295):2057–9. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 81.Mojica WD, Stein L, Hawthorn L. An exfoliation and enrichment strategy results in improved transcriptional profiles when compared to matched formalin fixed samples. BMC Clin Pathol. 2007;7:7. doi: 10.1186/1472-6890-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Umar A, Dalebout JC, Timmermans AM, et al. Method optimisation for peptide profiling of microdissected breast carcinoma tissue by matrix-assisted laser desorption/ionisation-time of flight and matrix-assisted laser desorption/ionisation-time of flight/time of flight-mass spectrometry. Proteomics. 2005;5(10):2680–8. doi: 10.1002/pmic.200400128. [DOI] [PubMed] [Google Scholar]
- 83.Breit S, Nees M, Schaefer U, et al. Impact of pre-analytical handling on bone marrow mRNA gene expression. Br J Haematol. 2004;126(2):231–43. doi: 10.1111/j.1365-2141.2004.05017.x. [DOI] [PubMed] [Google Scholar]
- 84.Coudry RA, Meireles SI, Stoyanova R, et al. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9(1):70–9. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanchez-Carbayo M, Saint F, Lozano JJ, et al. Comparison of gene expression profiles in laser-microdissected, nonembedded, and OCT-embedded tumor samples by oligonucleotide microarray analysis. Clin Chem. 2003;49(12):2096–100. doi: 10.1373/clinchem.2003.017525. [DOI] [PubMed] [Google Scholar]


