Abstract
The p19ARF tumor suppressor antagonizes Mdm2 to induce p53-dependent cell cycle arrest. Individual TKO (triple knock out) mice nullizygous for ARF, p53, and Mdm2 develop multiple tumors at a frequency greater than those observed in animals lacking both p53 and Mdm2 or p53 alone, demonstrating that p19ARF can act independently of the Mdm2-p53 axis in tumor surveillance. Reintroduction of ARF into TKO mouse embryo fibroblasts (MEFs), but not into those lacking both p53 and ARF, arrested the cell division cycle in the G1 phase. Inhibition of the retinoblastoma protein had no effect on the ability of ARF to arrest TKO MEFs. Thus, in the absence of Mdm2, p19ARF interacts with other targets to inhibit cell proliferation.
Keywords: ARF, p53, Mdm2, DMP1, tumor suppression
Two proteins encoded by the INK4a/ARF locus, p16INK4a and p19ARF, functionally interact with the retinoblastoma protein (RB) and p53 tumor suppressors, respectively. These four proteins are among the most frequently disrupted genes in human cancer. The p53 monomer forms a labile homotetrameric transcription factor that is activated in response to DNA damage and inappropriate mitogenic signaling (Ko and Prives 1996). Among its transcriptional targets are p21Cip1, an inhibitor of cyclin-dependent kinases, and Mdm2, a potent negative regulator of p53. Mdm2 is a multifunctional protein that inhibits p53 activity in several ways: It binds to the p53 transactivation domain to antagonize its activity on DNA (Momand et al. 1992; Oliner et al. 1993), and it fosters p53 degradation (Haupt et al. 1997; Kubbutat et al. 1997), acting as an E3 ligase that directly ubiquitinates p53 (Honda et al. 1997) and enhancing its shuttling to cytoplasmic proteasomes (Roth et al. 1998). In response to cellular stress, stabilization of p53 occurs when its association with Mdm2 is disrupted by posttranslational modifications of the proteins (Giaccia and Kastan 1998) or by interference of Mdm2 function by p19ARF (Kamijo et al. 1998; Pomerantz et al. 1998; Stott et al. 1998; Zhang et al. 1998).
The ARF tumor suppressor acts as a sensor of hyperproliferative signals emanating from oncoproteins and inducers of S-phase entry, such as Myc, E1A, Ras, and E2F-1 (Bates et al. 1998; De Stanchina et al. 1998; Palmero et al. 1998; Zindy et al. 1998). In turn, p19ARF triggers p53-dependent growth arrest in the G1 and G2 phases of the cell cycle or, in the presence of appropriate collateral signals, sensitizes cells to apoptosis (Quelle et al. 1995; Kamijo et al. 1997; De Stanchina et al. 1998; Zindy et al. 1998; Eischen et al. 1999; Jacobs et al. 1999; Schmitt et al. 1999). ARF binds directly to Mdm2, sequestering it in the nucleolus and enabling transcriptionally active p53 to accumulate in the nucleoplasm (Tao and Levine 1999; Weber et al. 1999). ARF also limits the ability of Mdm2 to ubiquitinate p53 in vitro (Honda and Yasuda 1999; Midgley et al. 2000) and in vivo (Xirodimas and Lane, pers. comm.). Binding of ARF to Mdm2 involves two separate domains of both the mouse p19ARF and human p14ARF proteins that interact cooperatively with a central acidic segment of Mdm2 (Lohrum et al. 2000a; Weber et al. 2000). Although ARF localizes to the nucleolus on its own, ARF binding unmasks a cryptic nucleolar localization signal within the carboxy-terminal Mdm2 RING domain, the integrity of which is required for localization of the binary complex to the nucleolus (Lohrum et al. 2000b; Weber et al. 2000). Disruption of the Mdm2 nucleolar localization signal enables Mdm2 to retain ARF in the nucleoplasm. Therefore, the ARF-Mdm2 interaction appears to be bi-directional in that each protein can regulate the subnuclear localization of the other.
Emerging evidence suggests that the ARF-p53-Mdm2 pathway is not strictly linear. Mice engineered to overexpress a c-Myc transgene under the control of the immunoglobulin heavy chain enhancer (Eμ) develop pre-B and B-cell lymphomas, with a majority of the resulting tumors sustaining ARF deletion, p53 mutation, or Mdm2 overexpression (Eischen et al. 1999). However, several tumors that lacked p19ARF or p53 function overexpressed Mdm2, allowing argument against a simple epistatic relationship between the genes. Moreover, premalignant B cells expressing Eμ-Myc and lacking both ARF and p53 proliferated at a faster rate in vitro than cells lacking ARF or p53 alone, implying that p19ARF may functionally interact with proteins other than p53. To address this issue, we generated mice nullizygous for ARF, p53, and Mdm2 and studied the effects of reintroduced ARF in cells lacking its only known target.
Results and Discussion
ARF and p53 collaborate in tumor surveillance
Both p53-null and ARF-null mice are highly tumor-prone with mean latencies for survival of 19 and 32 weeks, respectively (Donehower et al. 1992; Kamijo et al. 1999). In mice lacking p53, T-cell lymphomas predominate (∼70%), with the remainder being sarcomas (Jacks et al. 1994). In contrast, ARF-null animals have an appreciably lower incidence of lymphomas (25%) and primarily develop poorly differentiated sarcomas (50%), with carcinomas and gliomas appearing in the others (Kamijo et al. 1997, 1999). These results are compatible with the idea that ARF acts upstream from p53 and is selectively activated in response to only a subset of stress signals that affect p53 function (Sherr 1998). The increased latency for tumor formation observed in ARF-null versus p53-null animals may shift the tumor spectrum away from rapidly lethal lymphomas, allowing the emergence of tumor types not normally seen in p53-null mice.
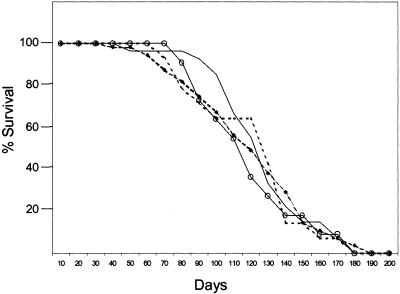
We generated mice deficient for ARF/p53 and ARF/p53/Mdm2 (designated triple knock-out, or TKO, mice) in an attempt to determine whether ARF and p53 might synergize in tumor suppression, either in the presence or absence of Mdm2. We observed no differences in the mean latencies of tumor formation (∼16 weeks) in TKO mice or ARF/p53- or p53/Mdm2-deficient mice, indicating that combinatorial loss of ARF and p53 had little or no effect on survival as compared with animals lacking p53 alone (Fig. 1). Disruption of Mdm2 leads to embryonic lethality, which is rescued on a p53-null background (Jones et al. 1995; Montes de Oca Luna et al. 1995). The spectrum of tumors seen in mice lacking both p53 and Mdm2 is similar to that of mice lacking p53 alone (McDonnell et al. 1999). In contrast, mice lacking both ARF and p53, with or without Mdm2, showed a wider range of tumor types than animals lacking either gene alone, and many developed multiple primary tumors (Table 1). ARF-null animals rarely show hemangiosarcomas and osteogenic sarcomas, but we have never observed teratomas, rhabdomyosarcomas, hepatocellular carcinomas, intestinal adenocarcinomas, or renal cell carcinomas in these animals throughout three years of continuous observation. Four of the 19 TKO mice developed primary tumors arising from three distinct cell types.
Figure 1.
Survival of mice lacking ARF, p53, and/or Mdm2. p53/Mdm2-null (○), ARF/p53-null (dotted line), ARF/p53-null Mdm2 +/- (●), and ARF/p53/Mdm2-null (solid line) mice were observed for 33 weeks. Tumors arising in ARF/p53-null and ARF/p53/Mdm2-null animals are listed in Table 1.
Table 1.
Tumor spectrum in ARF/p53-null and ARF/p53/Mdm2-null mice
| Genotype
|
Tumor typea
|
Number
|
|---|---|---|
| ARF/p53-null | Single tumors | 13 (72%) |
| Lymphoma | 9 | |
| Meningioma | 1 | |
| Osteogenic sarcoma | 1 | |
| Teratoma | 1 | |
| Spindle cell carcinoma (skin) | 1 | |
| Multiple primary tumors | 5 (28%) | |
| Lymphoma | ||
| + Osteogenic sarcoma | 1 | |
| + Hepatocellular carcinoma | 1 | |
| + Hemangiosarcoma | 1 | |
| + Fibrosarcoma | 1 | |
| Intestinal adenocarcinoma | ||
| + Renal cell carcinoma | 1 | |
| ARF/p53/ Mdm2-null | Single tumors | 10 (53%) |
| Lymphoma | 5 | |
| Hemangiosarcoma | 2 | |
| Nerve sheath sarcoma | 1 | |
| Rhabdomyosarcoma | 1 | |
| Osteosarcoma | 1 | |
| Multiple primary tumors | 9 (47%) | |
| Undifferentiated sarcoma | ||
| + Intestinal adenocarcinoma | 1 | |
| Lymphoma | ||
| + Rhabdomyosarcomab | 2 | |
| + Intestinal adenocarcinoma | 1 | |
| + Dermal sarcoma, unknown type | 1 | |
| + Hemangiosarcoma | 1 | |
| + Osteogenic sarcoma | 1 | |
| + Skin basal cell carcinoma | 1 | |
| + Intestinal adenocarcinoma | 1 |
Mice with multiple tumors are grouped under the subheading of one of the primary tumors followed by the diagnosis of the second or third tumor types (indicated by + signs and hierarchical indentation).
Lymphomas were of the widely disseminated T-cell type except for one mouse that exhibited lymphoma of distinct B- and T-cell types plus rhabdomyosarcoma.
Although we cannot rigorously exclude the contribution of strain-specific modifiers on the emergence of different tumor types, all parental mice used in our study were originally derived from strain 129/SvJ ES cells inoculated into C57BL/6 blastocysts, generating chimeric animals that were backcrossed onto a C57BL/6 background. Therefore, the emergence of tumors in ARF/p53-null and TKO mice not normally observed in ARF-null, p53-null, or p53/Mdm2 double-null animals, taken together with the similar latencies for tumor development, suggests that ARF and p53 loss contribute independently to some aspects of tumor suppression.
Mdm2- and p53-independent functions of ARF
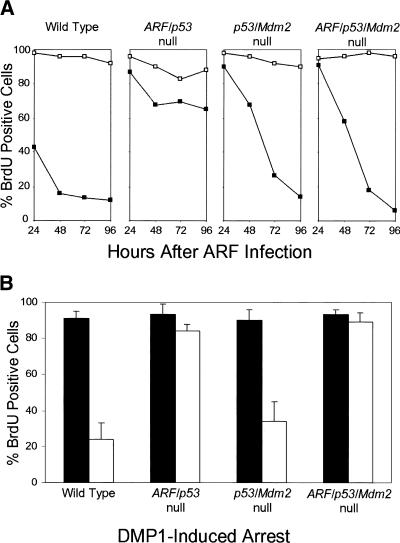
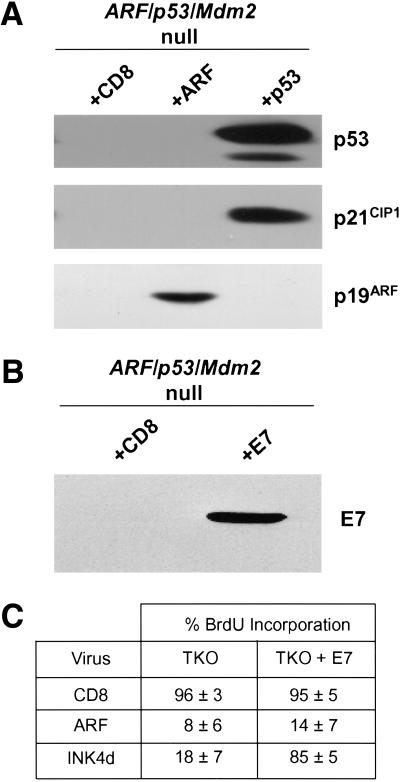
Early-passage mouse embryo fibroblasts (MEFs) explanted from p53-null embryos or cell lines that sustained p53 mutations during establishment are resistant to ARF-induced arrest, underscoring a requirement of functional p53 for ARF activity (Kamijo et al. 1997, 1998; Pomerantz et al. 1998; Stott et al. 1998; Zhang et al. 1998). In agreement with these findings, MEFs lacking both ARF and p53 were resistant to ARF overexpression, whereas 90% of wild-type MEFs arrested within 48 hours of infection with a p19ARF-expressing retrovirus (Fig. 2A). Unexpectedly, >90% of p53/Mdm2-null or ARF/p53/Mdm2-null MEFs failed to incorporate 5-bromodeoxyuridine (BrdU) after ARF was introduced (Fig. 2A); however, the kinetics of ARF-induced arrest were significantly delayed.
Figure 2.
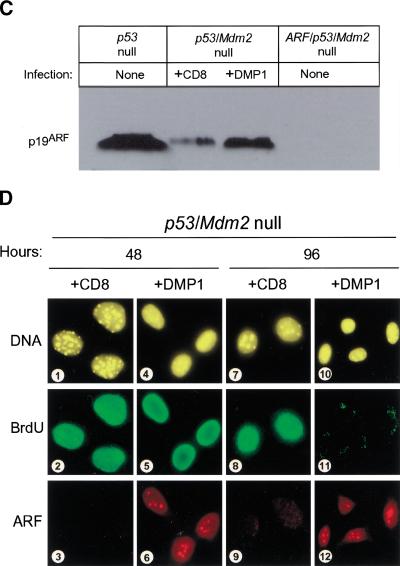
ARF and DMP1 arrest mouse embryo fibroblasts (MEFs) lacking both p53 and Mdm2. (A) Primary MEFs from mice with the indicated genotypes (top) were infected with retroviruses encoding CD8 (□) or p19ARF (▪). Cells labeled with 5-bromodeoxyuridine (BrdU) were analyzed by indirect immunofluorescence (green) and simultaneously scored for p19ARF expression (red) using 3 coverslips and >100 cells per coverslip in 4 independent experiments. (B) Primary MEFs of indicated genotypes (bottom) were infected with retroviruses encoding CD8 (dark bars) or DMP1 (open bars). Cells labeled for 24 hours with BrdU were stained 96 hours postinfection (green) and scored simultaneously for DMP1 expression (red). Error bars indicate standard deviations. (C) Expression of p19ARF protein in MEFs (3T3 strains, each at passage 9) of the indicated genotypes (top). Cells null for both p53 and Mdm2 (middle two lanes) were infected with retroviruses encoding either CD8 or DMP1 as indicated and assayed 96 hours later for ARF protein by direct immunoblotting. Equal quantities of protein (150 μg) were loaded per lane. (D) p53/Mdm2-null MEFs infected with retroviruses encoding CD8 (panels 1–3, 7–9) or DMP1 (panels 4–6, 10–12) were analyzed by immunofluorescence for BrdU incorporation and ARF induction 48 and 96 hours after infection.
Cells lacking p53 express unusually high levels of endogenous p19ARF (Fig 2C, lane 1) (Quelle et al. 1995), which can be suppressed by reintroduction of wild-type p53 (Kamijo et al. 1998; Stott et al. 1998). Surprisingly, endogenous p19ARF was expressed at much lower levels in MEFs lacking both p53 and Mdm2 (Fig. 2C, lane 2; Fig. 2D, panel 3), implying that Mdm2 somehow regulates the p53-ARF feedback loop. Use of cells lacking p53 and Mdm2 therefore provided an opportunity to determine the effects of ARF protein induction on cell cycle progression in response to DMP1, a transcriptional ARF activator (Inoue et al. 1999). Enforced DMP1 expression induced p19ARF to levels that were significantly lower than those observed in p53-null cells (Fig. 2C). Induced ARF was readily detected within the nucleoli of wild-type (not shown) and p53/Mdm2-null MEFs (Fig. 2D, panels 6 and 12). In turn, DMP1 inhibited BrdU incorporation in both cell types by 96 hours postinfection (Fig 2B; Fig 2D, panel 11). In contrast, DMP1 was without effect in ARF/p53-null and TKO MEFs (Fig. 2B), underscoring the dependence of DMP1 on ARF in arresting proliferation (Inoue et al. 1999).
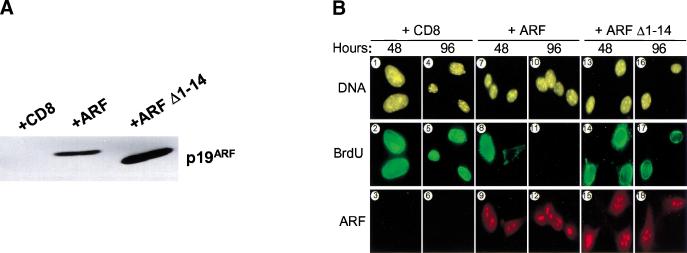
The amino-terminal 14 amino acids of mouse and human ARF are the most conserved residues in the polypeptide and comprise one of two binding sites for Mdm2 (Lohrum et al. 2000a; Weber et al. 2000). In mouse p19ARF, the second Mdm2 interaction domain resides within residues 26–37, the integrity of which is also required for ARF nucleolar localization (Weber et al. 1999). Hence, a p19ARF mutant lacking residues 1–14 (ARF Δ1–14) retains its ability to correctly localize to nucleoli but is unable to sequester Mdm2 and does not activate p53-dependent growth arrest (Weber et al. 2000). To determine whether the same protein interaction domain of ARF was required to arrest proliferation of MEFs lacking Mdm2, we introduced ARF Δ1–14 into TKO cells by retroviral gene transfer. In the experiment shown, the level of ARF Δ1–14 expressed in infected TKO MEFs was higher than that of the full-length ARF protein (Fig. 3A). Both wild-type and mutant proteins localized to nucleoli (Fig. 3B, panels 9, 12, 15, 18), which were independently demarcated by fibrillarin staining (Weber et al. 1999 and data not shown).
Figure 3.
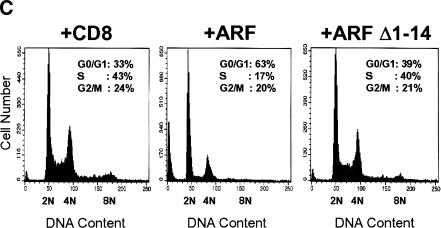
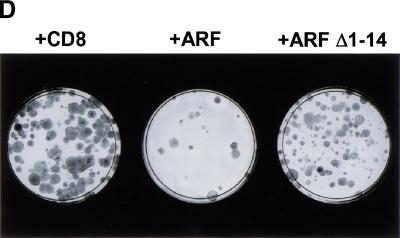
p53-Independent arrest requires the conserved ARF amino-terminal domain. (A) Triple knock out (TKO) mouse embryo fibroblasts (MEFs) infected with retroviruses encoding CD8, ARF, or ARF Δ1–14 were lysed 96 hours after infection, and separated proteins were blotted with affinity-purified polyclonal rabbit antibody to p19ARF. (B) TKO MEFs infected with retroviruses encoding CD8 (panels 1–6), ARF (panels 7–12), or ARF Δ1–14 (panels 13–18) were labeled with 5-bromodeoxyuridine (BrdU) for 24 hours and analyzed 48 and 96 hours postinfection for incorporated BrdU (green) and p19ARF expression (red). DNA was visualized with Hoechst yellow S769121 dye. (C) TKO MEFs (5 × 104) were infected with CD8, ARF, and ARF Δ1–14 retroviruses, stained with propidium iodide 96 hours after infection, and subjected to fluorescence-activated cell sorting to determine DNA content. (D) TKO MEFs were seeded at 300 cells per dish and infected with retroviruses encoding CD8, ARF, or ARF Δ1–14. Colonies were stained 12 days post-infection.
TKO cells infected with retroviruses encoding the T-cell coreceptor CD8, wild-type ARF, or ARF Δ1–14 were analyzed for their ability to enter S phase. BrdU was added to the cultures 24 hours before fixation, and cells were stained and examined 48 and 96 hours after infection. Wild-type ARF inhibited almost all BrdU incorporation by 96 hours post-infection (Fig. 3B, panel 11), whereas ARF Δ1–14 was ineffective at preventing S phase entry (Fig. 3B, panels 14 and 17). Analysis of DNA content by flow cytometry revealed that a significant population of TKO MEFs underwent arrest in G1 phase following infection for 96 hours with the retrovirus encoding wild-type ARF (Fig. 3C). In contrast, TKO MEFs infected with viruses encoding control CD8 or ARF Δ1–14 remained distributed throughout the cell cycle. Like MEFs lacking p53 alone, a percentage of the TKO cells were polyploid, yielding a G2/M population with an 8N DNA content. Introduction of wild-type ARF, but not ARF Δ1–14, also reduced the fraction of 8N cells (Fig. 3C), consistent with the ability of ARF to restrict S phase entry. ARF may also sensitize TKO cells to apoptosis, albeit to a significantly lesser extent than p53 (data not shown), as indicated by an increased level of cells with DNA content less than 2N.
To determine whether the effects of ARF were persistent, infected TKO MEFs were cultured at low density and refed every three days. Emerging colonies were stained with Wright-Giemsa solution 12 days postinfection, revealing a significant reduction in colony formation in ARF-infected TKO MEFs but very little effect after introduction of ARF Δ1–14 as compared with CD8 (Fig. 3D). The few colonies that arose from TKO cells infected with wild-type ARF failed to express p19ARF protein as determined by indirect immunofluorescence (data not shown), indicating that these were likely derived from uninfected cells. Although ARF Δ1–14 is severely impaired in its ability to arrest wild-type and TKO MEFs, it does retain some activity. We reproducibly saw a small effect on DNA replication (15% inhibition of BrdU incorporation) and a reduction in the number and size of colonies in TKO MEFs (Fig. 3D), which can likely be attributed to the retained functional ARF domains (Weber et al. 1999, 2000).
Several conclusions emerge from these experiments: (1) ARF can arrest cells lacking both p53 and Mdm2, but not p53 alone. This implies that Mdm2 antagonizes some activity of ARF on targets other than p53; in the absence of Mdm2 this other activity of ARF is revealed. (2) In principle, the ability of overexpressed ARF to accumulate in the nucleolus might nonspecifically interfere with some aspect of nucleolar trafficking required for cell cycle progression. However, nucleolar localization of ARF is not sufficient to induce cell cycle arrest in the absence of both Mdm2 and p53, because ARF Δ1–14 gains access to this compartment but is impaired in halting proliferation. (3) Conversely, the amino-terminal domain of p19ARF, which binds to Mdm2, also appears to be important in ensuring the interaction of ARF with another target required for S phase entry. (4) Finally, although ARF arrests cells containing wild-type Mdm2 and p53 in both G1 and G2, its preferential effects at the G1/S boundary in TKO cells imply that the relevant ARF target protein is required only for G1 exit.
ARF functions are independent of p21Cip1 and RB
Proteins that can interfere with the G1/S transition include the CDK inhibitor p21Cip1. Induction of p21Cip1 was evident after infection of TKO MEFs with a retrovirus-encoding wild-type p53 (Fig. 4A), although the infected cells rapidly underwent apoptosis rather than G1-phase arrest (data not shown). Introduction of ARF into TKO MEFs failed to induce p21Cip1 96 hours after infection, in agreement with previous observations with p53-null cells (Kamijo et al. 1997). Moreover, induction of p21Cip1 is not obligatory for ARF-induced arrest in cells expressing wild-type p53, because MEFs derived from p21Cip1-null or p21Cip1/p27Kip1 double-null mice arrest on ARF overexpression (Groth et al. 2000).
Figure 4.
ARF-induced arrest of triple knock out (TKO) cells does not depend on p21Cip1 and is not overridden by HPV E7. (A) TKO mouse embryo fibroblasts (MEFs) infected with retroviruses encoding CD8 or ARF (96 hours postinfection) or wild-type p53 (48 hours postinfection) were lysed, and separated proteins were blotted with antibodies to p53, p21Cip1, and ARF. (B) TKO MEFs infected with retroviruses encoding E7 or CD8 were lysed 48 hours after infection, and separated proteins were blotted with antibody to E7. (C) TKO MEFs infected with MSCV-GFP-IRES or MSCV-GFP-IRES-E7 retroviruses were re-plated 48 hours later and infected with retroviruses encoding CD8, ARF, or p19INK4d. Infected cells were analyzed 96 hours postinfection using an FITC filter to detect GFP-IRES-E7 infected cells (green), immunofluorescence to score 5-bromodeoxyuridine incorporation (red), and antibody to p19ARF followed by Alexa 350 stain to visualize nucleolar ARF (blue). Alternatively, p19INK4d was detected using a rabbit anti-p19INK4d antibody followed by biotinylated anti-rabbit immunoglobulin and streptavidin Texas red to ensure >90% infection efficiency of TKO and E7-containing TKO cells.
Two p53-related proteins, p63 and p73, share significant homology with p53 within their N-terminal transactivation and central DNA-binding domains (Lohrum and Vousden 2000). Although Mdm2 can block transactivation by all three proteins, only p53 is targeted by Mdm2 for degradation (Zeng et al. 1999). Because p63 and p73 are capable of inducing p53-responsive genes such as p21Cip1 in response to various signals (Kaelin 1999), our findings also imply that ARF does not access p21Cip1 through the agency of these p53 family members.
Carnero et al. (2000) provided evidence that ARF might act independently of p53 in MEFs retaining Mdm2 function. Expression of ARF antisense constructs relieved the ability of endogenous p19ARF to induce replicative senescence, whereas subsequent excision of the integrated antisense vector with Cre-recombinase restored the growth suppressive function of ARF even in the presence of a dominant-negative p53 mutant. Additionally, expression of SV40 large T antigen overcame the arrest induced by restored ARF function, suggesting that RB might be targeted by ARF through a p53-independent, but possibly Mdm2-dependent, pathway. Although these results disagree with several published reports that ARF acts upstream from p53 and does not depend on the RB pathway to induce arrest (Quelle et al. 1995; Pomerantz et al. 1998; Stott et al. 1998; Groth et al. 2000), the protocol for reversible antisense suppression is distinctly different from others' use of cells lacking the genes in question.
To explore a possible role for RB in our experimental system, we generated a population of TKO MEFs infected with a retrovirus expressing the green fluorescent protein (GFP) and encoding the human papilloma virus (HPV) E7 protein from an internal ribosomal entry sequence. Expression of E7 protein was documented by immunoblotting (Fig. 4B), and cells overexpressing E7 and GFP or GFP alone were infected with retroviral vectors encoding CD8 or ARF. Three days later, cells pulsed for 24 more hours with BrdU were fixed and analyzed for BrdU incorporation. To control for the effectiveness of E7 in inhibiting RB function, we also infected these cells with a retrovirus encoding p19INK4d, a potent inhibitor of cyclin D-dependent kinases in which the ability to induce G1 phase arrest is RB-dependent (Hirai et al. 1995). Overexpression of E7 had no effect on BrdU incorporation in TKO MEFs but greatly increased the number of cells replicating DNA in the presence of p19INK4d (Fig. 4C). Contrary to its ability to block p19INK4d-induced arrest, E7 was unable to bypass the growth-inhibitory affects of p19ARF (Fig. 4C). Loss of RB function thus appears to have no effect on the kinetics or properties of ARF arrest in the absence of both Mdm2 and p53. Moreover, the failure of E7 to override the ARF-induced G1 block implies that ARF prevents deregulated E2Fs from inducing S phase entry.
The ARF-Mdm2-p53 network
ARF responds to oncogenic signaling by neutralizing functions of Mdm2, thus allowing p53 to accumulate in a transcriptionally active form in the nucleoplasm. In cells that retain Mdm2 but lack p53 function, ARF loses its ability to rapidly halt cell cycle progression in the G1 and G2 phases of the division cycle, indicating that the response of cells to ARF induction is normally p53-dependent. Although cells lacking p53 alone or both p53 and ARF are resistant to ARF-induced arrest, cells lacking p53 and Mdm2 or all three genes are surprisingly more sensitive. In the latter cell types, ARF-induced arrest occurs more slowly than in wild-type MEFs, and the cells accumulate in G1, but not G2. Therefore, when present, Mdm2 antagonizes the ability of ARF to induce cell cycle arrest through targets other than p53.
ARF might interfere with the shuttling of other proteins whose nuclear export is required for cell cycle progression, and these interactions are likely to depend on the conserved ARF amino-terminal domain (a.a. 1–14) required for Mdm2 binding. Although ARF binds to Mdm2 with high affinity (Weber et al. 2000), the amounts of Mdm2 expressed in resistant p53-null or ARF/p53 double-null MEFs are negligible when compared with the unusually high levels of endogenous p19ARF protein or those achieved through ectopic ARF expression. It therefore seems unlikely that Mdm2 directly interacts with p19ARF to inhibit its ability to access targets other than p53. Instead, the loss of Mdm2 seems to reset the cell's sensitivity to ARF through some other mechanism.
ARF steadily accumulates as wild-type MEFs are progressively passaged in culture and approach senescence (Zindy et al. 1998), whereas ARF-null MEFs maintain their proliferative capacity and appear to be immortal (Kamijo et al. 1997). Although the ability of ARF to induce arrest in cells lacking both p53 and Mdm2 raises the possibility that replicative limits might be reimposed by Mdm2 loss, these double-null cells, like those lacking p53 alone, do not senesce (McMasters et al. 1996). This may be due to a reduction in the p19ARF level in Mdm2-deficient cells as compared with those in cells lacking only p53 (Fig. 2C).
Many lines of evidence now link ARF-Mdm2-p53 functions through a complex network rather than a simple linear pathway. It has long been appreciated that Mdm2 is a p53-responsive gene (Barak et al. 1993; Wu et al. 1993). Conversely, proliferating cells that lack p53 function alone synthesize relatively low basal levels of Mdm2 but, for still unexplained reasons, gratuitously express supra-physiologic levels of ARF (Quelle et al. 1995) that can again be suppressed by reintroduction of wild-type p53 (Kamijo et al. 1998; Stott et al. 1998). On the other hand, cells lacking both p53 and Mdm2 did not express high levels of p19ARF (Fig. 2C,D). Considering that the one known biochemical function of Mdm2 is to facilitate protein degradation (Honda et al. 1997), we might imagine, for example, that Mdm2 could catalyze the destruction of an ARF negative regulator, such as Bmi-1 or Twist (Jacobs et al. 1999; Maestro et al. 1999). Whatever the mechanism, it seems evident that this regulatory network must depend on other proteins, the actions of which could in part compensate for the missing gene products in ways that we currently do not understand. Nonetheless, at least one ARF target, when regulated in an entirely Mdm2- and p53-independent manner, can be rate limiting for the G1/S transition.
Materials and methods
Interbreeding of mice and tumor analysis
Homozygous Mdm2/p53-null mice (129/Sv × C57BL/6J background) (Montes de Oca Luna et al. 1995) were intercrossed with ARF-null mice (129/Sv × C57BL/6J background) (Kamijo et al. 1997) to generate double-null (p53/mdm2 and p53/ARF) and TKO mice. Genotypes were determined by polymerase chain reaction (PCR)-based assays as above. Moribund mice were killed and necropsied, and tumor tissues fixed in 10% formalin were embedded in paraffin for histopathological analyses (Kamijo et al. 1999).
Cell culture and virus infection
Primary MEFs were maintained in Dulbecco's-modified Eagle's medium with 10% fetal bovine serum, 2 mM glutamine, 0.1 mM non-essential amino acids, 55 μM β-mercaptoethanol, and 10 μg/ml gentamycin (GIBCO/BRL, Gaithersburg, MD). Virus production and infection of cells were performed using retroviral helper and vector plasmids (Roussel et al. 1995) provided by Charles Sawyers (University of California, Los Angeles). Vectors encoding p19ARF and p19ARF Δ1–14 were used as previously described (Weber et al. 2000). p19INK4d cDNA was subcloned into pSRα-MSV-tkCD8 (Hirai et al. 1995), and cDNA for the papilloma virus E7 protein (Slebos et al. 1994) was subcloned into MSCV-IRES-GFP (Hawley et al. 1994).
Immunofluorescence
Primary MEFs from ARF/p53-null, p53/Mdm2-null, and ARF/p53/Mdm2-null backgrounds seeded onto glass coverslips (3 × 104 cells per slip) were infected with retroviruses encoding CD8, ARF, ARF Δ1–14, or p19INK4d. Conversely, ARF/p53/Mdm2-null MEFs (1 × 106) were infected with retroviruses encoding E7, replated 48 hours later (3 × 104 cells per glass coverslip), and infected with retroviruses encoding CD8, ARF, or p19INK4d. BrdU (10 μM) was added to the culture medium 24 hours before fixation. Cells were fixed in methanol/acetone (volume ratio, 1:1) and stained for 1 hr with affinity-purified rabbit anti-p19ARF antibody (Quelle et al. 1995) or rabbit anti-p19INK4d (Hirai et al. 1995), followed by 30-minute exposure to biotinylated anti-rabbit immunoglobulin and strepatavidin-conjugated Texas red (both from Amersham, Arlington Heights, IL). Cells were treated for 10 minutes with 1.5 N HCl and stained for 1 hr with mouse monoclonal antibody to BrdU (Amersham), followed by fluorescein isothiocynate (FITC)-conjugated anti-mouse immunoglobulin (Amersham). DNA was visualized with Hoechst yellow S769121 dye (Molecular Probes, Eugene, OR).
Long-term colony assay
ARF/p53/Mdm2-null primary mouse embryo fibroblasts were seeded (3 × 102) onto 100-mm diameter dishes and infected with CD8, ARF, or ARF Δ1–14 retroviruses. Spent medium was replenished every three days, and emerging colonies were stained with Wright-Giemsa (Biochemical Sciences, Swedesboro, NJ) 12 days postinfection.
Immunoblotting
Frozen cell pellets were lysed in ice-cold Tween-20 lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 2.5 mM ethyleneglycoltetraacetic acid, 0.1% Tween-20, 1 mM phenylmethylsulfonyl fluoride, 0.4 units/ml aprotinin, 10 μg/ml pepstatin, and 10 μg/ml leupeptin) and sonicated. Samples (200 μg protein per lane) were separated by electrophoresis on denaturing polyacrylamide gels containing sodium dodecyl sulfate and were transferred to Immobilon polyvinylidine difluoride membranes (Millipore, Bedford, MA) preactivated in methanol. ARF, p53, p21, and E7 proteins were visualized by direct immunoblotting using rabbit polyclonal antibodies to the ARF C-terminus (Quelle et al. 1995) or to p53 (Ab421), p21, or E7 (all Santa Cruz Biotech.).
Acknowledgments
We thank Rose Mathew and JinLing Wang for assistance, Sam Lucas and Richard Ashmun for FACS analysis, and Frederique Zindy, Kazushi Inoue, and John Cleveland for helpful suggestions. This work was supported by National Institutes of Health grants CA-63230 (G.P.Z.) and CA-71907 (M.F.R.), Cancer Center Core Grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital. C.J.S. is an investigator and J.D.W. is an associate of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gerard.zambetti@stjude.org; FAX (901) 525-8025.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.827300.
References
- Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild-type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clarke PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumor suppressors pRB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM, Beach DH. p16INK4a and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- De Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L, Harvey AM, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: Emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Groth, A., Weber J,.D., Willumsen, B.M., Sherr, C.J., and Rousse, M.F. 2000. Oncogenic Ras induces p19ARF and growth arrest in mouse embryo fibroblasts lacking p21Cip1 and p27Kip1 without activating cyclin D-dependent kinases. J. Biol. Chem. (In press). [DOI] [PubMed]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of MDM2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci U S A. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Vonchen J-W, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm-2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The p53 gene family. Oncogene. 1999;18:7701–7705. doi: 10.1038/sj.onc.1202955. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Contribution of two independent MDM2-binding domains in p14ARF to p53 stabilization. Curr Biol. 2000a;10:539–542. doi: 10.1016/s0960-9822(00)00472-3. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MHG, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000b;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Lohrum MAE, Vousden KH. Regulation and function of the p53-related proteins: Same family, different rules. Trends Cell Biol. 2000;10:197–202. doi: 10.1016/s0962-8924(00)01736-0. [DOI] [PubMed] [Google Scholar]
- Maestro R, Dei Tos AP, Hamamori Y, Krasnokutky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell TJ, Montes de Oca Luna R, Cho S, Amelse LL, Chavez-Reyes A, Lozano G. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J Pathol. 1999;188:322–328. doi: 10.1002/(SICI)1096-9896(199907)188:3<322::AID-PATH372>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- McMasters KM, Montes de Oca Luna R, Peña JR, Lozano G. Mdm2 deletion does not alter growth characteristics of p53-deficient embryo fibroblasts. Oncogene. 1996;13:1731–1736. [PubMed] [Google Scholar]
- Midgley CA, Desterro JM, Saville MK, Howard S, Sparks A, Hay RT, Lane DP. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liégeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R. The Ink4a tumor suppressor gene product, p19ARF, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel MF, Theodoras AM, Pagano M, Sherr CJ. Rescue of defective mitogenic signaling by D-type cyclins. Proc Natl Acad Sci U S A. 1995;92:6837–6841. doi: 10.1073/pnas.92.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, McCurrach ME, De Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- Slebos RJ, Lee MH, Plunkett BS, Kessis TD, Williams BO, Jacks T, Hedrick L, Kastan MB, Cho KR. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci U S A. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Kuo M-L, Bothner B, DiGiammarino EL, Kriwacki RW, Roussel MF, Sherr CJ. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol. 2000;20:2517–2528. doi: 10.1128/mcb.20.7.2517-2528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, Kaelin WG, Jr, Oren M, Chen J, Lu H. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppressor pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle D, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]