Abstract
Early life presumably required polymerase ribozymes capable of replicating RNA. Known polymerase ribozymes best approximating such replicases use as their catalytic engine an RNA-ligase ribozyme originally selected from random RNA sequences. Here, we report 3.15 Å crystal structures of this ligase trapped in catalytically viable pre-ligation states, with the 3′-hydroxyl nucleophile positioned for in-line attack on the 5′-triphosphate. Guided by metal and solvent-mediated interactions, the 5′-triphosphate hooks into the major groove of the adjoining RNA duplex in an unanticipated conformation. Two phosphates and the nucleophile jointly coordinate an active-site metal ion. Atomic mutagenesis experiments demonstrate that active-site nucleobase and hydroxyl groups also participate directly in catalysis, collectively playing a role that in proteinaceous polymerases is performed by a second metal ion. Thus artificial ribozymes can employ complex catalytic strategies that differ dramatically from those of analogous biological enzymes.
The RNA world hypothesis proposes that primordial organisms lacked both DNA genes and protein enzymes, and instead used RNA as the sole biopolymer of information-storage and catalysis1. Support for this theory comes from the observation that many of life’s most fundamental processes depend on the action of RNA, its precursors or its derivatives1–4. Additional support has come through the study of ribozymes. Although known natural ribozymes catalyze only the cleavage or rearrangement of phosphodiester bonds5 and the formation of peptide bonds4, the diversity of artificial ribozymes produced from in vitro selection experiments has revealed the catalytic potential of RNA to be quite broad6,7. Of particular relevance to the RNA world hypothesis would be ribozymes with the RNA-dependent RNA polymerization activity needed for general RNA replication8,9.
Known catalytic RNAs with activities best approximating that required for general RNA replication derive from the class I ligase, an artificial ribozyme selected from random RNA sequences10,11. This ribozyme rapidly promotes a reaction with chemistry identical to that catalyzed by proteinaceous enzymes that replicate RNA (Fig. 1a): nucleophilic attack by a 3′-hydroxyl on a 5′-triphosphate in the context of Watson–Crick base pairing to form a normal phosphodiester RNA linkage with concomitant release of pyrophosphate10,11. Indeed, ligase variants have been engineered to catalyze primer-extension using exogenous nucleoside triphosphates (NTPs) and an internal template (Fig. 1a, inset)12. Through further engineering and in vitro selection, polymerase ribozymes have been developed in which the ligase provides the catalytic engine for RNA polymerization using NTPs and the information of exogenous RNA templates13–16. Although more efficient with some primer–templates than with others, this primer-extension reaction is general in that all primers and templates tested support detectable extension (provided that the 3′ terminus of the primer pairs to the template)13,15,16. Optimized versions of these polymerase ribozymes can extend a primer by up to 22 nucleotides15, or up to 95 nucleotides when appending to the ribozyme a segment that pairs to a short region of the template16.
Figure 1.
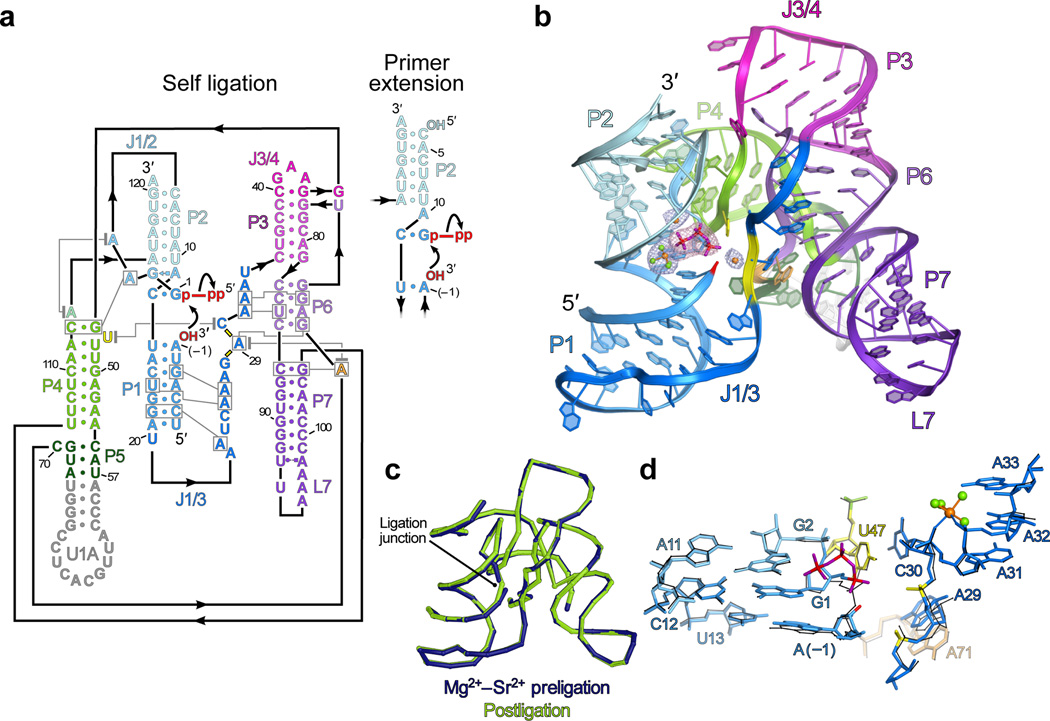
The Class I ligase ribozyme preligation complex. a. Secondary structure of the C47U crystallization construct17, depicted undergoing ligation (curved arrows). The nucleophile, electrophile and leaving group (red), active-site backbone phosphates (yellow bars), and nucleotides added to facilitate crystallization (gray) are indicated. Residue numbering, base pair geometries and tertiary interactions, as in ref. 17. Inset, primer extension catalyzed by a ligase variant resembling the self-ligation construct, except exogenous GTP replaces G1–A4. b. Overview of the Mg2+–Sr2+-preligation structure, peering into the active site. The U1A protein and its cognate loop have been removed from view. The 5′-GTP is shown as sticks. Active site metal ions are shown as spheres (orange, Mg2+; green, water). Meshes are simulated-annealing |Fo| − |Fc| OMIT maps calculated without G1 (magenta, contoured at 5σ) or active-site solvent atoms (dark blue, contoured at 4.5σ). c. Superposition of Mg2+–Sr2+-preligation and product (PDB ID: 3HHN)17 structures. d. Superposition of Mg2+–Sr2+-preligation and product structures (sticks and black lines, respectively) near the active site. A structural metal ion and water molecules are shown as orange and green spheres, respectively; active-site solvent atoms have been removed from view.
We recently reported the crystal structure of the class I ligase self-ligation product17,18. This structure revealed details of the ribozyme architecture and the location of the active site. However, the active site visualized in this post-ligation structure lacked the pyrophosphate leaving group and had no detectable catalytic metal ions. To develop a deeper understanding of the structural basis for RNA-catalyzed RNA polymerization, we determined the crystal structures of the ligase trapped in catalytically viable preligation states and then biochemically interrogated new hypotheses for the functions of active-site residues. These results corrected our model of the ligase catalytic mechanism and showed that the ligase employs a catalytic strategy both more complex than those of other artificial ribozymes19–23 and distinct from that of the proteinaceous polymerases.
Results
Structures of Class I ligase preligation complexes
Because the ligase normally undergoes extremely rapid self-ligation18, to visualize an intact, catalytically viable active site, we employed two modifications that each dramatically reduce, but do not completely ablate, catalysis. First, the active-site nucleotide C47 was mutated to uridine, which reduces the catalytic rate by a factor of >104 (ref. 17, Fig. 1a). Second, the preligation complex was reconstituted in a mixture of Ca2+ and Sr2+, which diminishes the self-ligation rate by >105 but allows the enzyme to fold into a conformation indistinguishable (within the resolution limits of Fe•EDTA solvent-accessibility mapping) from that of its active state24. Mg2+-bound crystals were obtained by soaking these crystals (hereafter, “Ca2+–Sr2+-complex” or “Ca2+–Sr2+-preligation” crystals) into isotonic conditions containing Mg2+ (hereafter, the “Mg2+–Sr2+-complex” or “Mg2+–Sr2+-preligation” crystals, Table 1). Crystals soaked without Sr2+, and those soaked with other Mg2+ isosteres (such as Mn2+ or Yb3+) proved intractable. The structure of each of these crystals was solved to 3.15 Å, allowing visualization of the ligase poised prior to catalysis (Fig. 1b, Supplementary Fig. 1a and Supplementary Methods).
Table 1.
Data Collection and refinement statistics
| Ca2+–Sr2+-Preligation Complex |
Mg2+–Sr2+-Preligation Complex |
|
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 58.70, 69.99, 71.86 | 59.19, 70.24, 71.21 |
| α, β, γ(°) | 99.86, 99.73, 103.68 | 99.86, 99.34, 103.81 |
| Resolution (Å) | 30–3.15 (3.26–3.15)a | 40–3.15 (3.26–3.15) |
| RMerge (%) | 9.8 (39.8) | 13.5 (42.9) |
| Mean I/σ (I) | 13.5 (2.1) | 17.5 (3.2) |
| Completeness (%) | 98.4 (94.1) | 99.1 (98.3) |
| Redundancy | 3.3 (2.9) | 7.2 (5.7) |
| Refinement | ||
| Resolution (Å) | 29.9–3.15 | 37.9–3.15 |
| No. Reflections | 18,190 | 18,739 |
| RWork/RFree (%) | 21.2/25.5 | 19.7/24.0 |
| No. atoms | ||
| RNA and Protein | 7231 | 7254 |
| Ligand/ion | 26 | 46 |
| Water | 38 | 51 |
| Mean B-factors (Å2) | ||
| RNA and Protein | 89.9 | 76.4 |
| Ligand/ion | 103.2 | 75.5 |
| Water | 88.0 | 76.0 |
| R.m.s. deviations | ||
| Bond distances (Å) | 0.015 | 0.006 |
| Bond Angles (°) | 1.133 | 1.310 |
Data from one crystal were used for each complex structure.
Values in parenthesis are for highest-resolution shell.
The global conformation of the ligase was remarkably similar in the pre- and post-catalytic states, being more dramatically altered by binding different metals than it was by undergoing ligation (Fig. 1c,d and Supplementary Fig. 1b,c). The Mg2+–Sr2+-complex and product structures superposed with an all-atom root mean square deviation (RMSD) of 0.31 Å, less than the mean precision of either structure (ref. 17), whereas the RMSD between the Ca2+–Sr2+-complex and product structures was 0.76 Å. In the latter case, the majority of the structural perturbations fell near the active site and its environs (Supplementary Fig. 1b–d), or at sites of inner-sphere metal ion coordination and thus attributable to steric effects incurred by Ca2+, which is ~0.3 Å larger than Mg2+ (ref. 25). All of these structural perturbations were alleviated in the active site of the Mg2+–Sr2+ complex (Fig. 1d). The close resemblance between the active site of the Mg2+–Sr2+ complex and that of the product implied that, unlike the analogous variant of the hepatitis delta virus (HDV) ribozyme26, the diminished activity of the C47U mutant was not caused by a gross structural perturbation. Furthermore, as expected from its minor effect on catalysis24, the presence of Sr2+ had no detectable effect on the ribozyme structure, and no peaks corresponding to Sr2+ density were observed in the electron-density maps.
Conformation of the 5′-triphosphate
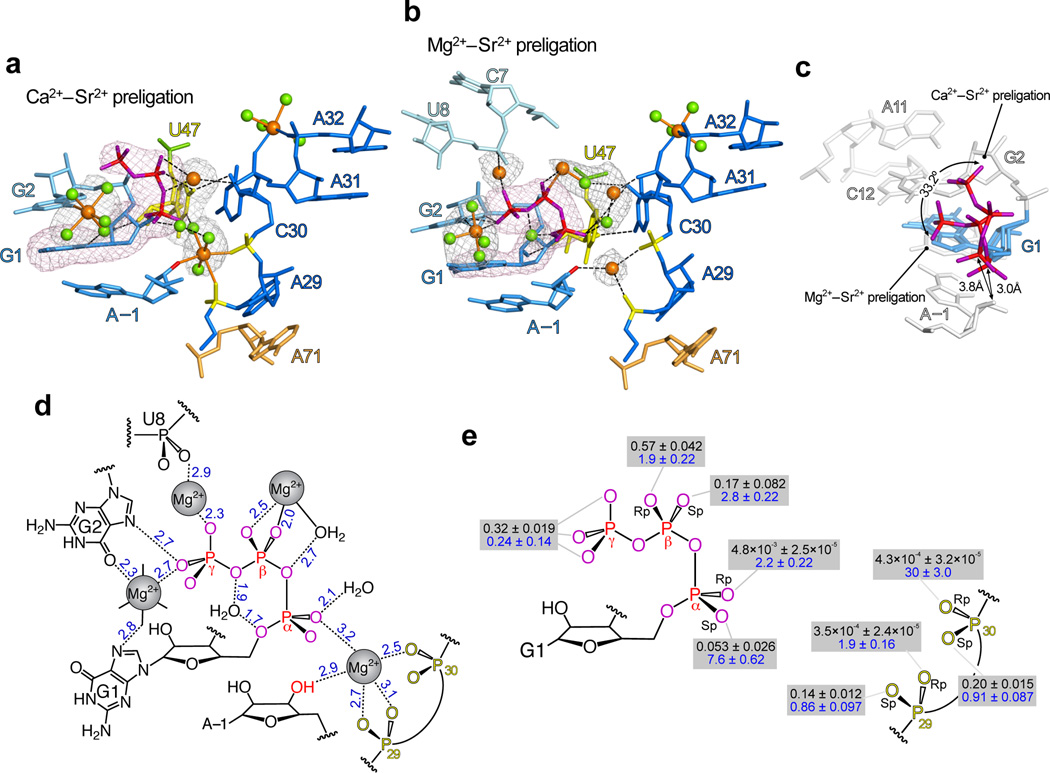
Within the active site of each preligation structure, the 5′-triphosphate forms a hooked conformation, curling at nearly a right angle as it docks the β- and γ-phosphates just above the G1 base (Fig. 2a–c). This conformation is dramatically different from that adopted by deoxynucleoside triphosphates (dNTPs) or NTPs in the active sites of proteinaceous polymerases, wherein the triphosphate is pulled away from the plane of the adjoining base27,28. Moreover, the (d)NTPs in these enzymes are typically Mg2+-bound, with the metal ion coordinated by two or more phosphate groups29. Although the ligase 5′-triphosphate makes no such polydentate contacts, the 5′-γ-phosphate coordinates a Mg2+•4H2O cluster positioned by groups on the Hoogsteen face of G1 and G2 (Fig. 2b,d). If protonated, a γ-phosphate oxygen would also hydrogen bond with the G2 N7, further stabilizing the hooked triphosphate conformation (Fig 2b,d).
Figure 2.
The Class I ligase active site. a. The active site, as observed in the Ca2+–Sr2+-preligation structure. Calcium ions and waters (orange and green spheres, respectively), inner-sphere metal interactions (thin sticks), and outer-sphere interactions and hydrogen bonds (black dashes) are indicated. Meshes are simulated-annealing |Fo| − |Fc| OMIT maps calculated without GTP (magenta, contoured at 5σ) or active site solvent atoms (navy, contoured at 4.5σ). b. The active site, as observed in the Mg2+–Sr2+-preligation structure. As in a, except orange spheres depict Mg2+ ions. c. Reorientation of the 5′-GTP during metal-ion exchange. d. Solvent interactions in active site of the Mg2+–Sr2+-preligation complex. Blue numbers indicate the distance, in angstroms, between atomic centers; the error of these measurements is approximated by the mean crystallographic coordinate precision, 0.4 Å. Waters hydrating the Mg2+ bound by G1 and G2 are represented by black lines. e. Summary of phosphorothioate interference (black, Mgks/Mgkoxy) and metal rescue [blue, (Xks/Xkoxy)/(Mgks/Mgkoxy)] values for oxygens at the active site. Functional groups on the 5′-GTP were interrogated in primer-extension assays, and those at positions 29 and 30 were interrogated in self-ligation assays. Error values are the standard deviations from three independent experiments.
In Mg2+, docking into the major groove places the 5′-triphosphate in a nearly ideal conformation for in-line nucleophilic attack: its α-phosphate is positioned 3 Å from the substrate 3′-hydroxyl, with a ~176° angle formed between these two atomic centers and the [α,β]-bridging oxygen leaving group. Moreover, pulling the β- and γ-phosphates proximal to the G1 base exposes the [α,β]-bridging oxygen, presenting it towards potentially stabilizing functional groups in the active site. In Ca2+, which greatly slows catalysis, this favorable conformation is perturbed. Because the Ca2+•hydrate cluster near G2 doesn’t support close approach of the γ-phosphate and the G2 base (Fig. 2a), the triphosphate is rotated ~33° into the major groove (Fig. 2c), which contorts the angle between nucleophile, electrophile and leaving group to ~154° (Fig. 2a). This conformation also pulls the α-phosphorus to displace it ~1 Å further away from the 3′-hydroxyl nucleophile.
Metal ions in the active site
Although biochemical data17,18 support a model in which the active site A29 and C30 backbone phosphates directly coordinate a catalytic metal ion, electron density for this metal is not observed in the product structure17. This metal was observed in the preligation complexes: during crystallographic refinement, |Fo| − |Fc| difference Fourier maps prominently featured 7.8σ and 4.8σ positive peaks centered within the active sites of the Ca2+–Sr2+ and Mg2+–Sr2+ complexes, respectively (Fig. 2a,b). In the Ca2+–Sr2+ structure, this peak perfectly accommodated a partially hydrated Ca2+ ion, in which the metal was centered between the A29 pro-Sp oxygen, the C30 pro-Rp oxygen and the nucleophilic A(−1) 3′-hydroxyl (Fig. 2a). The corresponding peak in the Mg2+–Sr2+ maps represented a less ideal Mg2+ binding site30, in which the active-site Mg2+ center was ~2.5–2.7 Å from the A(−1) 3′-hydroxyl, C30 pro-Rp oxygen and each of the A29 nonbridging phosphate oxygens (Fig. 2b,d). Moreover, in neither preligation structure was the active site metal ion within inner-sphere distance to the 5′-α-phosphate nonbridging oxygens, even though the ribozyme is substantially inhibited by sulfur substitution of the 5′-α-phosphate pro-Rp oxygen, implying that ligase employs a direct inner-sphere contact to this group31. The discrepancy between the biochemical and crystallographic data might be explained if the phosphorothioate substitution perturbed an outer-sphere contact32, which could be easily accommodated in either structure. Alternatively, since formation of a more ideal active-site Mg2+-binding pocket would require only minor (0.5–1.0 Å) adjustments to nucleotides A(−1), G1, A29 and C30, such a site might have been perturbed by the C47U mutation, or might be fully occupied only during the transition state. Furthermore, the model’s mean coordinate precision (0.4 Å) and the maximum observed inner-sphere bonding distance for Mg2+ (2.45 Å)30 leave open the possibility of direct coordination of the α-phosphate pro-Rp oxygen in the Mg2+–Sr2+ complex.
Natural RNA and DNA polymerases employ a two-metal mechanism in which Metal A activates the 3′-hydroxyl nucleophile for attack, Metal B (aided by a general acid28) facilitates loss of the pyrophosphate leaving group, and both metals stabilize the geometry and charge of α-phosphate during the transition state27,33. If the ligase transition state adopts a more ideal Mg2+-binding site—and one that supports an inner-sphere contact with the α-phosphate—then the active-site metal ion would be analogous to Metal A. However, we did not observe a metal ion analogous to Metal B (Fig. 2a,b), which suggested that its role might be played by other groups. Nonetheless, we did observe an array of solvent and metal interactions that stabilize the hooked conformation of the triphosphate, some of which might also help stabilize a growing negative charge on the β phosphate during the transition state (Fig 2a,b,d). These include the partially hydrated Mg2+ cluster bound to the Hoogsteen faces of GTP1 and G2, a Mg2+ ion jointly coordinated by the GTP1 γ-phoshate and the U8 Pro-Sp-nonbridging oxygen, and a water molecule bound by the GTP1 5′ and [β,γ] bridging oxygens (Fig. 2b,d). An additional partially hydrated Mg2+ ion near the A31 Pro-Sp-nonbridging oxygen bridges the gap between the P6 A-minor triad17 and the 5′-triphosphate, potentially placing a water molecule near the α-Pro-Rp-nonbridging phospate oxygens. Also of potential catalytic relevance, we observed evidence for a partially-hydrated Mg2+ near the β-phosphate, which would place an ordered water molecule within hydrogen-bonding distance of the triphosphate [α,β]-bridging oxygen, the reaction leaving group (Fig. 2b,d).
To probe for functional metal-ion interactions at the active site, we performed site-directed sulfur substitutions and thiophilic metal-rescue experiments34. We surveyed all nonbridging phosphate oxygens at positions 1, 29 and 30, using GTP analogues in primer-extension reactions (Fig. 1a, inset)12 to survey the oxygens at position 1 and using splint-ligation products to survey oxygens within the ribozyme active site (Fig. 2e and Supplementary Fig. 2). Sulfur substitutions at the G1-α-Pro-Rp, or the A29 and C30-pro-Rp oxygens inhibited catalysis by factors of 103–104, consistent with previous results17,31. Of these groups, the C30-pro-Rp substitution was substantially rescued by adding a thiophilic metal (30-fold rescue in Cd2+), which supported the proposed functional inner-sphere metal contact to this oxygen. Because lack of rescue is difficult to interpret, similar inner-sphere contacts might be important at the other strongly interfering positions. With the exception of the G1-α-pro-Sp site, where phosphorothioate inhibition and metal rescue were modest (inhibition to 0.053 with 7.8-fold rescue), inhibition and metal rescue were weak at all other sites probed (inhibition to 0.17–0.6 with 1.9–2.2-fold rescue). These results generally corroborated the contacts observed crystallographically (Fig. 2d,e and Supplementary Fig. 2) and implied that, if direct metal coordination at the β or γ oxygens plays more than modest roles in substrate binding or catalysis, then these roles are redundant and could only be uncovered by simultaneously substituting more than one oxygen.
The C47 nucleobase participates in catalysis
Based on their proximity to the 5′-triphosphate, two additional functional groups seemed capable of direct catalytic roles (Fig. 3a). In the Mg2+–Sr2+ complex, the U47 O4 group (mutated from the C47 N4 exocyclic amine to slow ligation) is 3.7 Å from the 5′-α-phosphate pro-Rp oxygen, a distance that decreases to 3.1 Å between the C47 N4 and G1 pro-Sp oxygen in the product structure (Supplementary Movie 1). In each structure, the C30 2′-hydroxyl (owing to the unusual 3′-exo ribose conformation35 of C30, a consequence of the P6 A-minor triad17) is within hydrogen-bonding distance of U47 O4 (C47 N4 in the parent). In the Mg2+–Sr2+ complex, the C30 2′-hydroxyl is also within hydrogen-bonding distance of a well-ordered water molecule near the leaving-group oxygen. Hypothesizing that the C47 exocyclic amine and the C30 2′-hydroxyl might perform roles played by Metal B of proteinaceous polymerases, we biochemically interrogated their contributions to catalysis.
Figure 3.
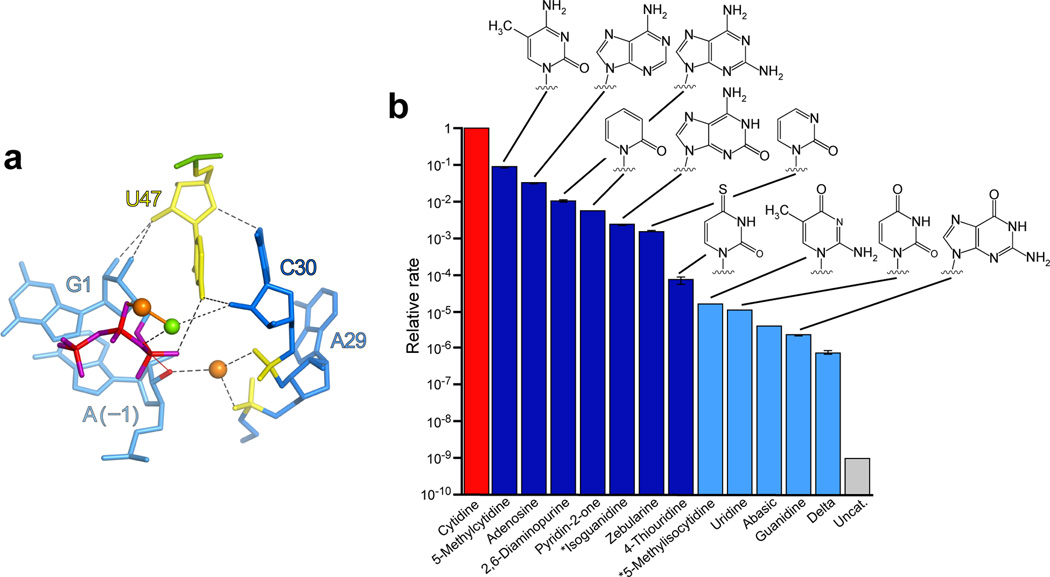
Biochemical interrogation of C47. a. View of the active site observed in the Mg2+–Sr2+ complex, roughly orthogonal to that in Figure 2b, highlighting interactions between C47, C30 and the 5′-triphosphate. Depiction and coloring are as in Figure 2b, with the thin red line indicating the assumed line of nucleophilic attack. b. Self-ligation rates of C47 variants, relative to that of the parental ligase (red bar). The rates of the faster constructs (red and dark blue bars) were measured at pH 6.0. To allow time courses to approach completion within 24 hours, the rates of slower constructs (light blue bars) were measured at pH 8.0 and renormalized, assuming log-linear pH dependencies24. Constructs marked with an asterisk were synthesized with a 2′-deoxy ribose at position 47, with rates normalized to account for this modification. The 4-thiouridine variant exhibited biphasic kinetics in which approximately 10% reacted in a burst phase with a relative rate of 0.23 ± 0.04 (s.d.); shown is the relative rate relevant to the majority of the molecules. Error bars, standard deviation from three independent experiments.
We first assessed the catalytic contribution of functional groups on the C47 base. Replacing C47 with a series of natural or modified bases generated ligase variants with activities spanning six orders of magnitude (Fig. 3b and Supplementary Fig. 4a). Variants that replaced this functional group with a carbonyl were among the slowest, with self-ligation rates 10−6–10−4 that of the unmodified ligase, irrespective of the protonation state of the imine (Supplementary Fig. 4b). Variants that preserved a major-groove exocyclic amine—irrespective of the protonation state of the proximal imine group—were not as dramatically affected, with rates 0.0024–0.09 that of the unmodified ligase. Substitution of C47 with zebularine (which lacks the N4 amino group altogether but is otherwise identical to cytosine) and pyridin-2-one (“P2o,” which lacks both the cytosine N4 and N3 groups) diminished the relative rate to 0.0016 and 0.006, respectively (Fig. 3b and Supplementary Fig. 4a). From these data, we conclude that it is the C47 N4 amine, and not the N3 imine, that participates in catalysis, and that replacing N4 with a carbonyl has an inhibitory effect beyond that of simply loosing the amine. Furthermore, based on the loss of activity observed with the zebularine and pyridin-2-one substitutions, the energetic contribution of the C47 N4 was less than that expected from a general acid-base catalyst, but more than that anticipated from a single hydrogen-bond donor36. Because the C47pyridin-2-one variant was comparably more active yet lacked any pertinent nucleobase functional groups, we focused on this variant in subsequent studies.
Electrostatic Catalysis in the ligase transition state
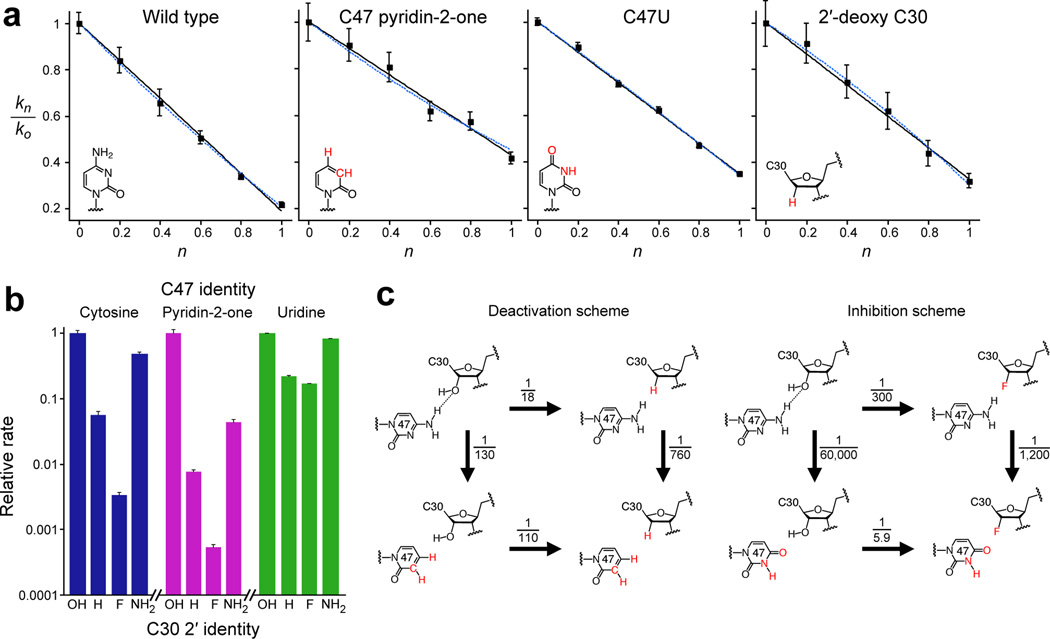
We next determined whether the C47 N4 amine functions electrostatically, through the donation of hydrogen bonds, or as a general acid catalyst, through the donation of protons. Between pH 5.8 and 8.5, self-ligation by both the C47pyridin-2-one and C47U variants exhibited log-linear pH dependence with a slope of 1.0 (Supplementary Fig. 3a), identical to the behavior observed for the parental ligase24. These results suggest that a single net deprotonation event determines the rate of ligase catalysis, regardless of the presence of the C47 N4 group. To more directly address the number of protons in transit during the transition state, we performed proton inventory experiments28,37 on the parental ligase and the two variants (Fig. 4a). For the parental ligase, the dependence of the relative self-ligation rate constant with molar fraction D2O fit well to a line with a slope (Φ) of 0.19 ± 0.01 (Fig. 4a, left), which corresponded to a solvent deuterium kinetic isotope effect (SDKIE)37 of 5.0 ± 0.3, comparable to those observed for some natural polymerases28,37. A higher-order polynomial fit (Fig. 4a, left, dotted line) failed to explain the data with greater significance (F-tests: 0.99 and 0.98 for the single proton and double proton models, respectively; R2 > 0.99 for each)38 and yielded SDKIE values of 4.1 ± 0.4 and 1.15 ± 0.1 for the individual protons transferred, which implied that, were the ligase to shuttle two protons during its transition state, the second proton would contribute insignificantly to the reaction rate. Likewise, proton inventories for the C47P2o and C47U variants could be convincingly fit to only single-proton transfer models, yielding SDKIE values of 2.4 ± 0.1 and 2.9 ± 0.1, respectively (Fig. 4a, middle). That each of these variants had an SDKIE lower than that of the parent might imply that the transition states they stabilize depend less strongly on proton transfer37. Because a linear proton-inventory fit can have alternative interpretations37,38, these data were insufficient to explicitly quantify the number of protons in transit during the transition state (Supplementary Discussion). Nonetheless, because neither the removal of the C47 N4 amino group nor its replacement with an inhibitory carbonyl group changed the apparent number of protons in transit during ligation, these results suggest that C47 functions electrostatically during catalysis. This role differs from that of an active-site cytosine in the HDV ribozyme, for which the N3 group functions as a general-acid catalyst26,39,40.
Figure 4.
Catalytic roles of active-site functional groups. a. Proton inventories for self-ligation by the indicated variants. The solid black line indicates the best fit for a single-proton–transfer model; the dashed blue line indicates the best fit for a two-proton–transfer model28,37,38. Error bars, standard deviation from three independent experiments. b. Relative rates for self-ligation of constructs with the indicated substitutions at the 2′ position of C30, surveyed in combination with the indicated subsitutions at position 47. For each C47 substitution, rates are normalized to that of the C30 2′-OH construct. Error bars, standard deviation from three independent experiments. c. Schemes summarizing functional interaction between the C30 2′-hydroxyl and the C47 N4, showing the effects of removing these groups (left), or replacing them with inhibitory groups (right). Substitutions, relative to the wild type, denoted in red. Numbers indicate the decrease in self-ligation rate, (kright/kleft) or (kbottom/ktop), resulting from the indicated substitution. At pH 6.0, the observed rate of the unmodified ligase (upper left corner of each scheme) was 0.93 ± 0.009 min−1 (s.d.).
Interactions between active-site functional groups
We next investigated the function and energetic contribution of the C30 2′-hydroxyl group. Self-ligation activity by the 2′-deoxy C30 variant was reduced to nearly 5% that of the parent, and followed a log-linear pH dependence between pH 5.8 and 8.5 (Supplementary Fig. 3a, right). This construct also exhibited a linear proton inventory with a measured SDKIE of 3.1±0.2 (Fig. 4a, right), implying that the C30 2′-hydroxyl is not involved in proton-transfer. Substitution of the C30 2′-hydroxyl with a 2′-fluoro group (which can accept but not donate hydrogen bonds) reduced self-ligation activity to 0.3% of the parental activity, whereas substitution with a 2′-amino group (which can donate, but not accept hydrogen bonds) restored activity to within 50% of the parental activity (Fig. 4b, left). These results indicate that the C30 2′-hydroxyl—much like the C47 N4 amine—functions electrostatically, acting as a catalytically important hydrogen-bond donor.
If the C47 and C30 functional groups act independently, then the effect of altering one group would be unchanged by the status of the other. To test this hypothesis, we generated C47pyridin-2-one and C47U self-ligase variants bearing the same C30 2′-modifications described above. Although each series of variants exhibited the same qualitative trend in behavior, the magnitude of these effects depended on the identity of the C47 base (Fig. 4b, middle and right). In the C47P2o variants, which lacked the C47 N4 amine, C30 2′ substitutions were ~10–20-times more severe than in the parental C47 background. Conversely, in the C47U variants, which replaced the N4 amine with an inhibitory carbonyl group, the activities of all variants fell within one order of magnitude of one another, with the C30 2′-amino modification restoring activity to ~82% that of C47U. Schematic examination of the effects of single- and double-mutations indicated that in the absence of the C30 2′-hydroxyl, catalysis became six-times more dependent on the presence of the C47 N4 amine, and vice versa (Fig. 4c, left). Conversely, when catalysis was quelled by an inhibitory group at either position, it became less dependent on the status of the other group (Fig. 4c, right). These results strongly support a model in which the C47 N4 exocyclic amine donates a hydrogen bond to the C30 2′-hydroxyl, which in turn donates a hydrogen bond during catalysis.
Discussion
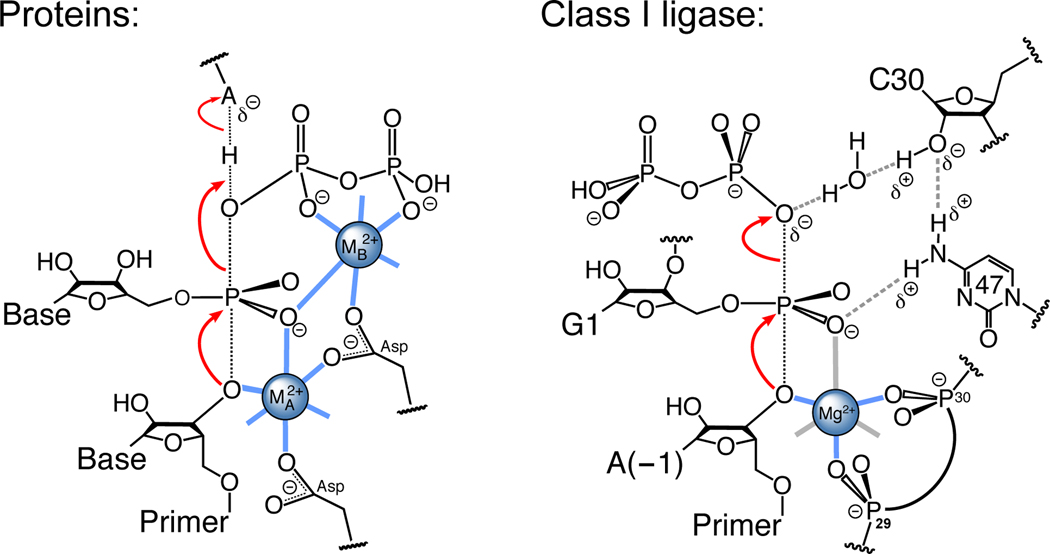
The Class I ligase is the first artificial ribozyme to be crystallographically visualized in a catalytically viable pre-catalytic state and is one of three such ribozymes for which the active site has been biochemically interrogated in detail19,20,23. Our results indicate a model for transition-state stabilization in which the A29 and C30 pro-Rp phosphate oxygens coordinate a catalytic Mg2+ cofactor that activates the 3′-hydroxyl for nucleophilic attack, and the C47 N4 exocylic amine and the C30 2′-hydroxyl participate in a network of hydrogen bonds that stabilizes both the leaving group and the transition-state geometry (Fig. 5). The C47 amine contributes directly to geometric stabilization, forming a hydrogen bond with the 5′-α-phosphate pro-Rp nonbridging oxygen that grows stronger as the reaction proceeds from the ground state to the transition state. The C47 amine also donates a hydrogen bond to the C30 2′-hydroxyl, which contributes a water-mediated partial positive charge to the [α,β]-bridging oxygen thereby helping to stabilize the leaving group.
Figure 5.
Transition-state stabilization by protein and RNA active sites. (Left) Catalysis by proteinaceous polymerases28. Black dotted lines indicate bonds formed or broken during the transition state. Red arrows indicate the reaction direction during polymerization. (Right) Catalysis by the class I ligase. Gray dashed lines denote hydrogen bonds. Thick lines denote inner-sphere contacts with a single catalytic metal ion; those in blue are observed crystallographically, those in gray are either inferred from biochemical results or are presumed to be water (not shown). Red arrows indicate the reaction direction during ligation or polymerization.
The C47 and C30 functional groups are proposed to act through hydrogen bonding rather than proton transfer, because removing either group does not change the proton inventory and because removing the C47 N4 amine does not influence the pH dependence (Fig. 4a and Supplementary Fig. 3a). In contrast, removal of a general-acid catalyst from the active sites of proteinaceous polymerases changes both of these properties28. Our model proposes that a network of hydrogen bonds confers a partial positive charge on a bound water molecule, allowing it to stabilize the developing negative charge on the leaving group during the transition state. Although this same water might eventually contribute a proton to the leaving group, our results suggest that if this transfer happens, it happens well after the transition state and therefore is not rate limiting.
In its use of an acid-bound Mg2+ cofactor to activate the 3′-hydroxyl for nucleophilic attack, the catalytic strategy of the ligase resembles those of proteinaceous polymerases and self-splicing introns (which use two-metal mechanisms; Fig. 5, left)27,28,33,41,42 and that of the ββα-me and HUH nucleases (which use a single active-site metal cofactor)43. However, the manner in which the ligase replaced Metal B—a universal feature of the active sites of natural polymerases27,33—has no known precedent44. Moreover, although catalytic nucleobases function in active sites of some natural catalytic RNAs45, the ligase C47 is the first known example of a catalytic nucleobase acting in an artificial ribozyme.
With regard to mechanisms of natural ribozymes (reviewed in ref. 44), the ligase mechanism most resembles that of the HDV ribozyme, which also uses a metal-ion cofactor to deprotonate its nucleophile and an active-site cytosine to stabilize the leaving group26,39,40,45–47. In HDV, however, this cytosine functions in proton transfer26,39,44,45, utilizing an N3 group with a near-neutral pKa40,47, whereas the ligase uses a cytosine N4 amine and does not perform general acid-base catalysis. Like the ligase mechanism, the GlmS ribozyme mechanism involves hydrogen bonds with both a nucleobase exocyclic amine (G57 N2) and a hydroxyl group (the glucosamine-6-phosphate C1-hydroxyl)48,49. However, both of these groups stabilize the scissile phosphate transition-state geometry rather than the leaving group. Although the ligase mechanism differs from that of the hairpin ribozyme in depending on an active-site metal ion50, its extensive use of electrostatics is reminiscent of the hairpin ribozyme catalytic strategy, which accelerates self-cleavage by forming more hydrogen bonds with the scissile phosphate at the transition state than it does at either ground state51. A water molecule might also play a catalytic role for the hairpin, although it is proposed to function as a specific base, activating the reaction nucleophile52.
The proposed catalytic strategy of the ligase, which makes extensive use of electrostatic catalysis in addition to a metal-ion cofactor and substrate approximation (Fig. 5, right), is more complex than strategies proposed for less efficient artificial ribozymes that promote similar reactions21,23, suggesting that the relatively complicated catalytic strategy of the ligase might be tied to its catalytic power53. Applying similar structural and biochemical analyses to other ribozymes borne from in vitro selection experiments will increase understanding of the differences between natural and artificial ribozymes and of RNA catalysis more generally. Furthermore, a detailed understanding of the structural contributions to substrate recognition and catalysis by the class I ligase might facilitate efforts to isolate an RNA replicase ribozyme.
Methods
Sample preparation for crystallography
The transcription template for the unreacted C47U mutant ligase, p307HU_C37U, was generated from p307HU (ref. 17) and linearized by EarI digestion before use. RNA was synthesized by T7 in vitro transcription, processed and purified as described previously17, but without removal of the terminal 2′-3′ cyclic phosphate. The ligase substrate oligonucleotide was chemically synthesized (Dharmacon) and gel-purified. The U1A A1-98 Y31H Q36R double mutant54 was expressed and purified as described17.
Crystallization and soaking
Approximately 200 µM unreacted C47U ligase RNA was heated and cooled in water, and then mixed with a 1.1-fold excess of substrate oligonucleotide in calcium annealing buffer (final composition, 5 mM MES, pH 6.0, 10 mM CaCl2, 1 mM DTT). Sample preparation then proceeded as described17. Crystal clusters were grown at 22°C from 1 µL hanging drops containing 16–20% (v/v) 2-methyl-2,4-pentanediol (MPD), 50 mM sodium cacodylate (pH 6.0), 20 mM calcium acetate, 10 mM strontium acetate and 1mM spermine. To generate microseed stocks, these clusters were stabilized by bringing the drop MPD concentration to 30% (v/v), crushed with Seed-Beads (Hampton Research), and serially diluted with stabilization buffer. Diffraction-quality crystals were obtained by mixing these seed stocks with ligase–substrate–U1A samples that had been annealed in the presence of 100 mM KCl, and equilibrating over wells containing 20–24% (v/v) MPD. Crystals were cryo-stabilized by bringing the drop MPD concentration to 30% (v/v) under otherwise isotonic conditions. Stabilized crystals were either mounted in nylon loops and plunged into liquid nitrogen or equilibrated with magnesium prior to freezing. To equilibrate, stabilized drops were serially supplemented with eight two-fold additions of an isotonic solution in which Mg2+ replaced Ca2+, after each addition incubating for 10 minutes and then withdrawing liquid so as to bring the volume back to 4 µL. Following a final two-hour incubation, crystals were frozen as above. Because the ligation rate of the C47U mutant is ~1.1×10−5 min−1 at pH 6.0, between 0.1–0.3% presumably ligated during equilibration with Mg2+.
Data collection, structure solution and refinement
Data were collected at New England Collaborative Access Team (NE-CAT) beamlines 24-ID-C and 24-ID-E at the Advanced Photon Source (APS) and were indexed, scaled and integrated using the HKL2000 software suite55. Each structure was solved independently; each monomer was built separately. Initial phases were obtained by molecular replacement in PHASER56, using as a search model the U1A-bound Class I ligase product structure (PDB ID: 3HHN), in which residues abutting the active site had been removed. The optimal solutions for each search (Z-scores of 26–30) were used as the starting points for further refinement in PHENIX57. Model building was in COOT58. Refinement incorporated rounds of rigid-body and individual ADP strategies, interspersed with rounds of simulated annealing. Active-site atoms were built iteratively, guided by inspection of |Fo| − |Fc| difference maps and the reduction of Rfree values. Residues abutting GTP1 were built first, followed by GTP1, its triphosphate, hydrated metal clusters and finally individual solvent atoms. The final refined Ca2+–Sr2+-complex and Mg2+–Sr2+-complex models have estimated coordinate errors (maximum likelihood57) of 0.47 Å and 0.40 Å, respectively. Structural figures were made in PyMol (http://www.pymol.org, Schrödinger LLC). Additional description of model building and refinement are provided (Supplementary Methods).
Sample preparation for biochemistry
Ligase constructs bearing modified nucleotides were assembled using DNA-splinted RNA ligation60. To enable modular manipulation of all positions near the active site, each ligase (including the unmodified ribozyme) was constructed from five oligonucleotides: in vitro–transcribed 5′- and 3′-fragments spanning residues 1–23 and 52–121 (including the P5 U1A-loop), respectively; a synthetic 10-mer spanning residues 24–33, and synthetic 9-mers spanning residues 34–42 and 43–51. The synthetic oligonucleotides were synthesized with 5′-monophosphates; the 3′-fragment was processed with a hammerhead ribozyme to yield a 5′-hydroxyl and phosphorylated with T4 polynucleotide kinase (New England Biolabs). Each assembly was a single-pot ligation reaction using T4 RNA ligase 2 (New England Biolabs) guided by a DNA that paired to residues 13–61. More information on reactions and oligonucleotide synthesis is provided (Supplementary Methods).
Ribozyme kinetics
Self-ligation and primer-extension assays were performed under single-turnover conditions at 22°C, as described24,31, with final concentrations of 1 µM ribozyme, 0.5 µM oligonucleotide substrate supplemented with trace 5′-32P-radiolabeled sustrate, and 10 mM MgCl2. In primer-extension assays, 1 mM GTP (or analog) was added with an additional 1 mM MgCl2. Reaction timepoints were quenched in EDTA and separated on denaturing urea polyacrylamide gels. Gels were quantified by phosphorimaging (Fujifilm BAS-2500). Data were fit using the program KaleidaGraph (Synergy Software), to a single exponential equation
where Fp(t) is the fraction of product at time t, and FM (the maximum fraction reacted) and kOBS (the observed rate constant) are treated as unknowns. All kinetic experiments were performed in triplicate; reported errors are standard deviations. In experiments calculating relative rates, uncertainties were propagated as follows
where A and B are individual rate constants, R is their ratio, and σA, σB and σR are their associated uncertainties. Additional description of kinetic assays is provided (Supplementary Methods).
Phosphorothioate interference and metal rescue experiments
Interference values were calculated from the apparent first-order rate constants for modified and unmodified constructs in Mg2+ alone:
Rescue values were calculated from interference values in the presence and absence of rescuing metal “X”:
Metal-rescue experiments were attempted at several MgCl2 concentrations, titrating CdCl2, CoCl2, MnCl2 and ZnCl2 as potential rescuing metals. The most substantial rescue was observed when 50 mM MgCl2 was supplemented with 2 mM CdCl2. All interference and rescue values (Fig. 2e) were determined under these conditions using the self-ligation assay, except effects of modifications at the GTP1 α- and β-phosphates, which were determined under these conditions using primer-extension assays (Fig. 1a, inset)31. Values for GTPγS were measured using both assays.
Proton Inventories
All reaction components were reconstituted in both D2O and H2O, and self-ligation reactions were measured under varying molar ratios of the two solvents. Data were fit to the modified Gross-Butler equations37
where kn is the observed rate constant at n molar fraction of D2O, kH2O is the rate constant in pure water, each Φi is the inverse of the of the SKIE for an individual ionizable group, and i is one or two.
Supplementary Material
Acknowledgements
We thank F. Eckstein for the gift of GTP analogs, R.A. Grant, D. Lim, T. Schwartz and K.R. Rajashankar for assistance with data collection and processing, H. Mackie, E. Roesch and J. De Luca for advice on oligonucleotide synthesis, J. Chen, A. Ricardo, K. Frederick, N. Yoder and W. Johnston for help with HPLC purification, E. Spooner for performing mass spectrometry, U. RajBhandary, C. Drennan, J. Piccirilli, J. Szostak and members of the Bartel laboratory for helpful discussions. Supported by National Institutes of Health (NIH) grant GM061835 to D.B. This work is also based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines of the Advanced Photon Source (APS), supported by award RR-15301 from the National Center for Research Resources at the NIH. Use of the APS is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Accession Codes
Atomic coordinates and structure factors are deposited in the Protein Data Bank under accession codes 3R1H and 3R1L for the Ca2+–Sr2+- and Mg2+–Sr2+-preligation complexes, respectively.
Supplementary Information
Supplementary figures 1–5, Supplementary discussion, Supplementary methods, and Supplementary movie 1 are available.
Author Contributions
D.S. and D.B. designed the experiments and wrote the manuscript. D.S. performed the experiments.
Author Information
The authors declare no competing financial interests.
References
- 1.Joyce GF, Orgel LE. Prospects for Understanding the Origin of the RNA World. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World, Second Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 49–77. [Google Scholar]
- 2.Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 3.White HB., 3rd Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 4.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 5.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Li N, Ellington AD. Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers. 2007;4:633–655. doi: 10.1002/cbdv.200790055. [DOI] [PubMed] [Google Scholar]
- 7.Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. Re-creating an RNA Replicase. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World, Second Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 143–162. [Google Scholar]
- 9.McGinness KE, Joyce GF. In search of an RNA replicase ribozyme. Chem Biol. 2003;10:5–14. doi: 10.1016/s1074-5521(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 11.Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 12.Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 13.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence MS, Bartel DP. New ligase-derived RNA polymerase ribozymes. RNA. 2005;11:1173–1180. doi: 10.1261/rna.2110905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-Catalyzed Transcription of an Active Ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 17.Shechner DM, et al. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science. 2009;326:1271–1275. doi: 10.1126/science.1174676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagby SC, Bergman NH, Shechner DM, Yen C, Bartel DP. A class I ligase ribozyme with reduced Mg2+ dependence: Selection, sequence analysis, and identification of functional tertiary interactions. RNA. 2009;15:2129–2146. doi: 10.1261/rna.1912509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuhlmann F, Jaschke A. Characterization of an RNA active site: interactions between a Diels-Alderase ribozyme and its substrates and products. J Am Chem Soc. 2002;124:3238–3244. doi: 10.1021/ja0167405. [DOI] [PubMed] [Google Scholar]
- 20.Serganov A, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat Struct Mol Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson MP, Scott WG. The structural basis of ribozyme-catalyzed RNA assembly. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Murakami H, Suga H, Ferre-D'Amare AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 23.Pitt JN, Ferre-D'Amare AR. Structure-guided engineering of the regioselectivity of RNA ligase ribozymes. J Am Chem Soc. 2009;131:3532–3540. doi: 10.1021/ja8067325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glasner ME, Bergman NH, Bartel DP. Metal ion requirements for structure and catalysis of an RNA ligase ribozyme. Biochemistry. 2002;41:8103–8112. doi: 10.1021/bi012179b. [DOI] [PubMed] [Google Scholar]
- 25.Harding MM. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 26.Chen JH, et al. A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry. 2010;49:6508–6518. doi: 10.1021/bi100670p. [DOI] [PubMed] [Google Scholar]
- 27.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 28.Castro C, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsiao C, et al. Complexes of Nucleic Acids with Group I and II Cations. In: Hud NV, editor. Nucleic Acid-Metal Ion Interactions. Vol. 1. Cambridge, UK: RSC Publishing; 2009. pp. 1–38. [Google Scholar]
- 30.Harding MM. The geometry of metal-ligand interactions relevant to proteins. II. Angles at the metal atom, additional weak metal-donor interactions. Acta Crystallogr D Biol Crystallogr. 2000;56:857–867. doi: 10.1107/s0907444900005849. [DOI] [PubMed] [Google Scholar]
- 31.Glasner ME, Yen CC, Ekland EH, Bartel DP. Recognition of nucleoside triphosphates during RNA-catalyzed primer extension. Biochemistry. 2000;39:15556–15562. doi: 10.1021/bi002174z. [DOI] [PubMed] [Google Scholar]
- 32.Basu S, Strobel SA. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4–P6 domain. RNA. 1999;5:1399–1407. doi: 10.1017/s135583829999115x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sträter N, Lipscomb WN, Klabunde T, Krebs B. Two-metal ion catalysis in enzymatic acyl- and phosphoryl-transfer reactions. Angew. Chem. Int. Ed. Engl. 1996;35:2024–2055. [Google Scholar]
- 34.Frederiksen JK, Piccirilli JA. Identification of catalytic metal ion ligands in ribozymes. Methods. 2009;49:148–166. doi: 10.1016/j.ymeth.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 36.Jencks WP. Catalysis in chemistry and enzymology. xvi. New York: McGraw-Hill; 1969. p. 644. [Google Scholar]
- 37.Venkatasubban KS, Schowen RL. The proton inventory technique. CRC Crit Rev Biochem. 1984;17:1–44. doi: 10.3109/10409238409110268. [DOI] [PubMed] [Google Scholar]
- 38.Schowen KB, Schowen RL. Solvent isotope effects of enzyme systems. Methods Enzymol. 1982;87:551–606. [PubMed] [Google Scholar]
- 39.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 40.Gong B, et al. Direct measurement of a pK(a) near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J Am Chem Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 41.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 42.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat Struct Mol Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fedor MJ. Comparative enzymology and structural biology of RNA self-cleavage. Annu Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 45.Bevilacqua PC, Yajima R. Nucleobase catalysis in ribozyme mechanism. Curr Opin Chem Biol. 2006;10:455–464. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 47.Luptak A, Ferre-D'Amare AR, Zhou K, Zilm KW, Doudna J. Direct pK(a) measurement of the active-site cytosine in a genomic hepatitis delta virus ribozyme. J Am Chem Soc. 2001;123:8447–8452. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- 48.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 49.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nesbitt S, Hegg LA, Fedor MJ. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 51.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 52.Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 54.Ferré-D'Amaré AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collection in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Bullard DR, Bowater RP. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.