Abstract
Dystrophic cutaneous calcinosis is associated with disorders as common as autoimmune diseases and cancer. To get insight into the pathogenesis of this poorly understood process, we studied the function of SAMD9, a protein of unknown function, recently shown to be deficient in a hereditary form of dystrophic calcification in the skin, known as normophosphatemic familial tumoral calcinosis (NFTC). Consistent with the fact that in NFTC severe inflammatory manifestations always precede cutaneous calcinosis, we found out that SAMD9 is tightly regulated by interferon-γ (IFN-γ). In addition, the SAMD9 promoter was also found to respond strongly to IFN-γ in a luciferase reporter assay. Of interest, we identified a critical 30-bp fragment upstream to the SAMD9 transcription initiation site responsible for driving most of the gene expression. Bioinformatic analysis suggested that SAMD9 function involves interaction with additional protein(s). Using the Ras recruitment system assay and confirmatory immunoprecipitation, we demonstrated that SAMD9 interacts with RGL2. To study the biological importance of this interaction, we assessed the effect of RNA interference-mediated downregulation of this pair of proteins in various cell lines. We found out that downregulation of any of the two protein partners caused increased expression of EGR1, a transcription factor with a known role in the regulation of tissue calcification, inflammation, and cell migration. Supporting the physiological relevance of these data, EGR1 levels were also upregulated in a fibroblast cell line derived from an NFTC patient. In conclusion, our data indicate that SAMD9, an IFN-γ-responsive protein, interacts with RGL2 to diminish the expression of EGR1, a protein of direct relevance to the pathogenesis of ectopic calcification and inflammation.
INTRODUCTION
Ectopic calcification is a pathological process that often accompanies diseases as common as atherosclerosis, cancer, and autoimmune diseases (Touart and Sau, 1998; Budoff et al., 2007), and was found to represent an important predictor of morbidity and mortality in the general population (Chiu et al., 2010; Haas, 2010).
Acquired ectopic calcification in the skin is traditionally classified into metastatic and dystrophic cutaneous calcinosis (Touart and Sau, 1998; Sprecher, 2010). In metastatic calcinosis, as seen in chronic renal failure, sarcoidosis, or hyperparathyroidism, calcium deposits are the direct consequence of elevated blood levels of calcium and/or phosphate; in dystrophic calcinosis, typically associated with autoimmune diseases such as scleroderma and dermatomyositis, as well as with atherosclerosis and cancer (Parwani et al., 2004; Bai et al., 2009), the calcified material always seems to accumulate subsequent to some form of primary tissue damage.
The deciphering of the genetic basis of familial forms of metastatic and dystrophic calcinosis cutis has recently uncovered complex biochemical pathways involved in the pathogenesis of ectopic calcification. The hereditary counterpart of acquired disorders associated with metastatic calcification is termed hyperphosphatemic familial tumoral calcinosis; this was found to result from loss-of-function mutations in one of the following three genes: FGF23, encoding a potent phosphaturic protein, KL, encoding a coreceptor for FGF23, and GALNT3, encoding a glycosyltransferase responsible for mediating a physiologically critical post-translational modification of FGF23 (Topaz et al., 2004; Benet-Pages et al., 2005; Ichikawa et al., 2007).
The inherited equivalent of dystrophic calcification is termed normophosphatemic familial tumoral calcinosis (NFTC). Patients affected with this disease present with a vasculitis-like rash at a young age, followed 3–7 years later by the appearance of calcified masses in the cutaneous and subcutaneous tissues (Topaz et al., 2006). Additionally, these patients display severe inflammatory manifestations in mucosal tissues, suggesting that the pathological basis of their calcification involves abnormal regulation of inflammation. Recently, it has been shown that NFTC is the result of biallelic loss-of-function mutations in the gene SAMD9 (Topaz et al., 2006).
SAMD9 encodes a 1,589 amino-acid protein with no homology to known human proteins except for a paralog protein termed SAMD9-like (SAMD9L) (Topaz et al., 2006; Li et al., 2007). The fact that mutations in SAMD9 are associated with profound inflammatory manifestations led us to investigate the effect of relevant mediators on SAMD9 expression. We showed that tumor necrosis factor-α can induce SAMD9 expression and that both p38 and nuclear factor-κB regulate tumor necrosis factor-α-mediated upregulation of SAMD9 (Chefetz et al., 2008). Independently, another group also suggested that SAMD9 may function physiologically as a tumor suppressor gene (Li et al., 2007), as both SAMD9 and SAMD9L were found to be overexpressed in desmoid tumors transfected with wild-type adenomatous polyposis coli protein. Overexpression of SAMD9 causes apoptosis and reduced proliferation of malignant cells, whereas down-regulation of SAMD9 is associated with increased cellular proliferation and tumor growth both in in vivo and in in vitro models (Li et al., 2007). Interestingly, deletion of a genomic fragment on 7q21.3, spanning the SAMD9 gene, is commonly found in cells from patients with myeloid leukemia and myelodysplastic syndrome, suggesting again that SAMD9 is an inhibitor of tumor progression (Asou et al., 2009). Taken altogether, these data indicate that SAMD9 may have an important role in the pathogenesis of a wide array of pathologies in humans, including ectopic calcification, inflammation, and cancer, suggesting that this protein may be of interest in the treatment of these diverse ailments.
Despite these recent advances, little is currently known about the molecular mechanisms underlying SAMD9’s mode of action. In the present study, we provide evidence indicating that SAMD9 may exert its effects in the context of the interferon-γ (IFN-γ) signaling pathway.
RESULTS
IFN-γ regulates SAMD9 expression
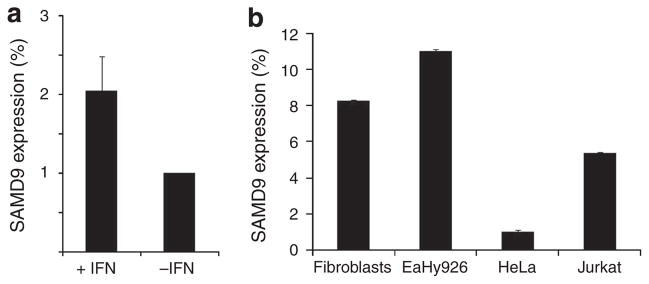
As summarized above, a steadily growing body of evidence suggests that SAMD9 may have an important role in the pathogenesis of inflammation and cancer (Topaz et al., 2006; Li et al., 2007; Chefetz et al., 2008). As IFN-γ is a cytokine involved in both inflammation and cancer (Miller et al., 2009), we studied its effect on SAMD9 expression. Using quantitative real-time RT-PCR (QRT-PCR), we found that SAMD9 transcript levels were upregulated in response to IFN-γ treatment by a factor of 2–2.5 (Figure 1a). Thus, two inflammatory mediators associated with the pathogenesis of inflammation and cancer, IFN-γ and tumor necrosis factor-α (Chefetz et al., 2008), seem to have an important role in the regulation of SAMD9 expression.
Figure 1. Effect of IFN-γ and cell type on SAMD9 expression.
(a) Human fibroblasts were exposed to 10 ng ml−1 of IFN-γ or its vehicle for 24 hours, and SAMD9 expression was measured as described in Materials and Methods. Results are expressed as fold change of expression relative to cells treated with vehicle±SD. (b) SAMD9 expression was measured as described in Materials and Methods in various cell lines. Results are expressed as fold change of expression relative to the levels measured in HeLa cells±SD.
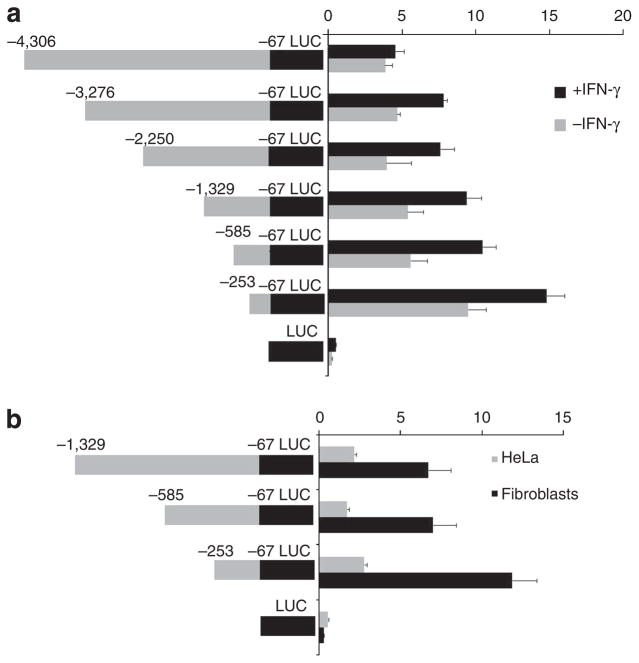
To further investigate the molecular underpinning of the regulation of SAMD9 expression by IFN-γ, transient transfection assays were performed with a luciferase reporter construct carrying various DNA fragments spanning up to 4 kb upstream to the putative SAMD9 transcription start site, in the presence and absence of IFN-γ. All constructs were found to display increased luciferase activity as compared with the empty vector. In addition, all constructs were found to respond to IFN-γ (Figure 2a). As SAMD9 was found by QRT-PCR to be expressed strongly in transformed human fibroblasts, but not in HeLa cells (Figure 1b), we compared its promoter region activity in these two cell types. As shown in Figure 2b, all constructs showed reduced luciferase activity in HeLa cells as compared with human fibroblasts, further supporting the physiological relevance of the luciferase reporter assay system.
Figure 2. Analysis of SAMD9 promoter.
Different DNA fragments of the putative SAMD9 proximal promoter were cloned into pGL3 promoterluciferase (LUC) vector and assayed for luciferase activity as described in Materials and Methods. The locations of the various cloned fragments are given as the position of the nucleotide boundaries (relative to the transcription start site). (a) The constructs were assayed in the presence (+IFNγ) or absence (−IFNγ) of IFN-γ (10 ng ml−1), as well as (b) in human fibroblasts and HeLa cells, two cell lines known to express different levels of SAMD9 transcripts. Results are expressed as mean relative luciferase activity±SD.
Identification of IFN-γ-responsive elements within the SAMD9 promoter
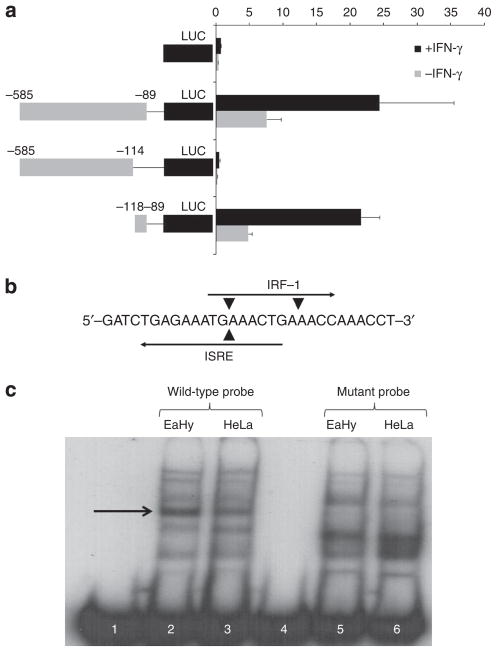
Using a transcription element search software (http://www.cbil.upenn.edu/cgi-bin/tess/tess), we carefully scrutinized the SAMD9 promoter region for candidate transcription element binding sites. Deletion constructs were designed to evaluate the role of these transcription elements in SAMD9 regulation. We found out that most of the IFN-γ-responsive SAMD9 promoter activity resides within a short 30-bp stretch spanning positions −89 to −118 upstream to the SAMD9 transcription start site (Figure 3a). Deletion of this fragment was found to almost completely abolish the promoter activity. In addition, this fragment alone was able to sustain the IFN-γ-responsive promoter activity to the same level as the largest upstream fragment assayed.
Figure 3. Characterization of a 30-bp IFN-γ-responsive element within the SAMD9 promoter region.
(a) Deletion fragments were used to identify a minimal promoter element between bases −89 and −118 (upstream to the transcription start site) that conferred responsiveness to IFN-γ. The various DNA fragments were cloned into pGL3 promoter-luciferase (LUC) vector and assayed for luciferase activity as described in Materials and Methods. The constructs were assayed in the presence (+IFNγ) or absence (−IFNγ) of IFN-γ (10 ng ml−1). Results are expressed as mean relative luciferase activity±SD. (b) Schematic representation of the designed radio-labeled probe spanning part of the critical IFN-γ-responsive element within the SAMD9 promoter, including the predicted transcription factor recognition sites of IRF-1 and IFN stimulated response element. In addition to the wildtype probe, we generated a probe carrying two A>T transversions at positions −98 and −105, predicted to abolish the activity of the two predicted transcription factor recognition sites (arrowheads). (c) incubation of the wildtype, labeled 30-bp probe with nuclear protein extract from EaHy (lane 2) and HeLa (lane 3) cell lines caused a band shift compared with the probe that was made to run without previous incubation with nuclear extracts (lane 1). Note that the prominent band (arrow) is more apparent in EaHy cells compared with HeLa cells, in accordance with the differential expression of SAMD9 in these two cell lines. The prominent band (arrow) was not visible anymore when the mutant probe was incubated with the nuclear extracts (lanes 4–6).
Bioinformatic analysis revealed the existence of two transcription factor binding sites within this minimal 30-bp fragment, one for IFN regulatory factor 1 (IRF-1) located on the positive strand and the other for IFN stimulated response element located on the negative strand (Figure 3b). This 30-bp fragment caused a band shift when incubated with the nuclear extract derived from transformed human fibroblasts, indicating that it is actually a transcription factor binding sequence (Figure 3c). Incubation of the same probe with the nuclear extract derived from HeLa cells generated a weaker band, as expected given the reduced expression of SAMD9 in this cell line. We then used the TFSEARCH software (http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html) to identify potentially deleterious point mutations capable of abolishing the transcription factor-binding capacity of the critical 30-bp fragment. As predicted, introduction of these mutations within the 30-bp fragment resulted in the disappearance of the band in the band shift assay (Figure 3c), lending further support to the importance of this minimal region for IFN-γ-dependent regulation of SAMD9 expression.
SAMD9 interacts with RGL2
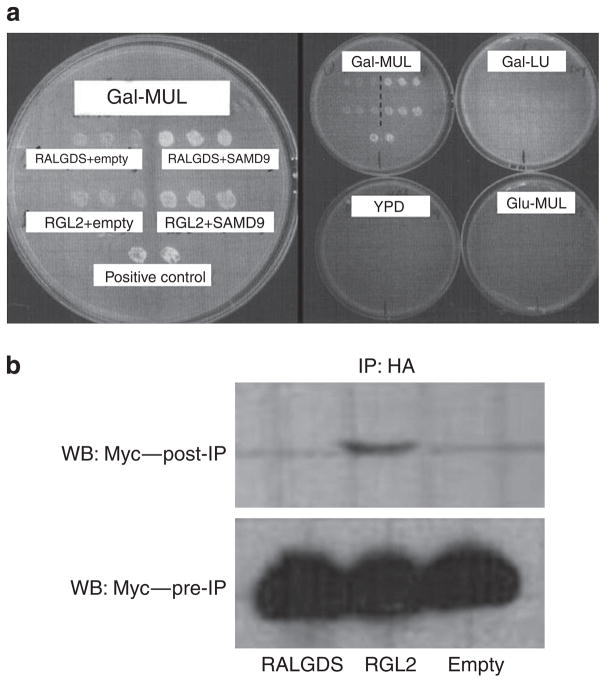
To get further insight into the mechanism of action of SAMD9, we aimed at identifying proteins interacting with SAMD9. For that purpose, we used the Ras recruitment system (Broder et al., 1998; Aronheim, 2001). We chose to screen a cDNA expression library derived from human spleen, given the high level of expression of SAMD9 in this tissue (Li et al., 2007). The SAM domain-containing N-terminal part of SAMD9 was fused in frame with a cDNA encoding the cytoplasmic Ras protein (Holmberg et al., 2002) under the control of the Met425 promoter and served as a bait. The library was composed of cDNAs fused to the v-Src myristoylation signal under the control of the Gal1 promoter (Hubsman et al., 2001). A Cdc25-2 yeast strain was cotransfected with the bait and prey library expression constructs, and selected on glucose plates lacking uracil and leucine. Transformants were replica plated into galactose- containing medium lacking methionine and grown at the restrictive temperature of 36 °C. Yeast colonies growing under these conditions were selected and further analyzed. Plasmid DNA was isolated from the clones and reintroduced into the Cdc25-2 yeast strain with either the specific pMet425-Ras-SAMD9-NT bait or a nonspecific bait. cDNAs conferring to yeast transformants the ability to grow specifically with pMet425-Ras-SAMD9-NT bait at the restrictive temperature, but not with the nonspecific bait, were subsequently characterized by direct DNA sequencing. Out of eight such clones, seven separate clones were found to encode the 131-amino-acid C-terminal part of RGL2 (gene accession number NM_004761.2), whereas one additional clone encoded the 119-amino acid C-terminal part of RALGDS (gene accession number NM_001042368.1). Both RGL2 and RALGDS showed specific interaction with SAMD9 after reintroduction into the yeasts (Figure 4a). RGL2 and RALGDS are two related proteins that function as Ras effectors, and were shown to be involved in in vivo carcinogenesis (Gonzalez-Garcia et al., 2005) and in the immune response to viral infections (Chien et al., 2006).
Figure 4. SAMD9 interacts with RGL2.
(a) To identify SAMD9’s potential partners, we screened a spleen cDNA library using the Ras recruitment system. Cotransfection with pMet425-Ras-SAMD9NT and pMyr expression plasmid encoding either RALGDS or RGL2 myristoylated prey fusion protein resulted in yeast proliferation (left panel). There was no growth in replicate plates in which pMet425-Ras-SAMD9NT promoter was inactivated (Gal-LU), pMyr-RGL2/pMyr-RALGDS promoter was inactivated (Glu-MUL), or both were inactivated (YPD), indicating the specificity of the interaction (right panel). (b) The interaction with RGL2 was further validated using coimmunoprecipitation. HEK-293T cells were cotransfected with pCAN-Myc- SAMD9 and either pcDNA-RGL2-HA or pcDNA-RALGDS-HA. Cell lysate was coimmunoprecipitated with anti-HA and eluted proteins were separated by 10% SDS-PAGE, followed by western blotting and probed with anti-Myc. Total cell lysates of transfected HEK-293T are shown in the lower panel.
To validate the above findings, we used coimmunoprecipitation. The RGL2 C-terminal part fused to an hemagglutinin (HA) tag could immunoprecipitate the SAMD9 N-terminal fragment fused to Myc tag. No such interaction could be demonstrated between the N-terminal part of SAMD9 and RALGDS (Figure 4b).
SAMD9 and RGL2 suppress EGR1 expression
To ascertain the consequences of SAMD9’s interaction with RGL2, we compared global gene expression patterns in cell lines downregulated using specific siRNAs for these two genes. RGL2 downregulation caused upregulation of 11 genes and downregulation of 5 genes by a factor of 2-fold or more. Downregulation of SAMD9 caused an increase in the expression of eight genes (Supplementary Table S1 online). Using QRT-PCR, we found out that one such gene was upregulated following cell transfection with either type of siRNA: EGR1, which encodes the transcription factor early growth response gene-1 (Figure 5a). EGR1 exhibited a more than 4-fold increase in expression following downregulation of SAMD9 or RGL2.
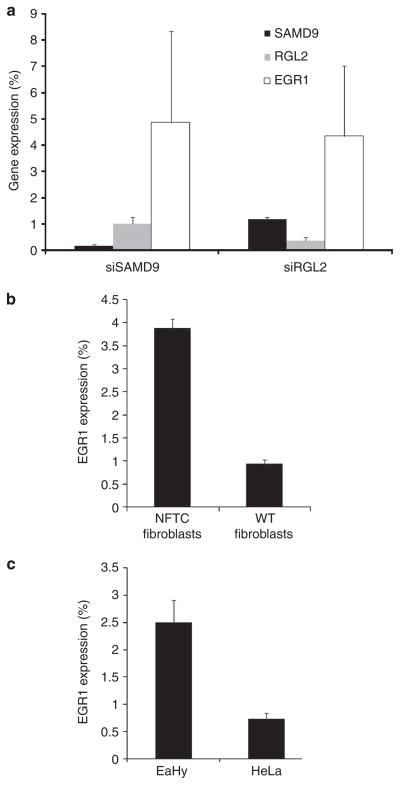
Figure 5. SAMD9 and RGL2 regulate EGR1 expression.
(a) Human fibroblasts were transfected with SAMD9 (siSAMD9)- and RGL2 (siRGL2)-specific siRNA, as well as control siRNA. After 48 hours, RNA was extracted and SAMD9 (black columns), RGL2 (gray columns), and EGR1 (white columns) expression levels were measured as described in Materials and Methods. Results are expressed as fold change of expression relative to cells treated with control siRNA±SD. (b) RNA was extracted from transformed human fibroblasts derived from a healthy individual (WT fibroblasts) and from a patient with normophosphatemic familial tumoral calcinosis (NFTC) (NFTC fibroblasts). EGR1 expression was measured as described in Materials and Methods. Results are expressed as fold change of expression in NFTC relative to normal cells±SD. (c) EaHy and HeLa cells were transfected with an RGL2-specific siRNA, as well as control siRNA. After 48 hours, RNA was extracted and EGR1 expression was measured as described in Materials and Methods. Results are expressed as fold change of expression relative to cells treated with control siRNA±SD. WT, wild type.
Consistent with the above findings, EGR1 levels were 3-fold upregulated in fibroblasts derived from an NFTC patient carrying a loss-of-function mutation in SAMD9 (Topaz et al., 2006) as compared with control fibroblasts (Figure 5b). Additionally, we were able to show that the inhibitory effect of RGL2 on EGR1 expression is SAMD9 dependent. Indeed, downregulation of RGL2 caused an increase in EGR1 RNA levels in endothelial cells, expressing high levels of SAMD9. On the other hand, no effect of RGL2’s downregulation on EGR1 RNA levels was seen in Hela cells expressing SAMD9 at a considerably lower level (Figure 5c). These findings underscore the physiological relevance of SAMD9’s interaction with RGL2 for EGR1 regulation.
DISCUSSION
Mendelian disorders have been recently re-discovered as a unique tool for the delineation of novel biological functions (Antonarakis and Beckmann, 2006). As NFTC is characterized by severe inflammation and ectopic calcification (Topaz et al., 2006), the fact that SAMD9 is defective in this disorder suggests that this protein may have a role in the regulation of both processes (Topaz et al., 2006). Additionally, several laboratory studies suggest that SAMD9 might have a role as a tumor suppressor gene as well (Li et al., 2007; Asou et al., 2009), although no increased incidence of cancerous conditions has been reported among NFTC patients. The purpose of the present work was to further characterize the mode of action of SAMD9.
We initially assessed the effect of IFN-γ on SAMD9 expression, as IFN-γ has been shown to be involved in the pathogenesis of both cancer and inflammation. We could demonstrate that IFN-γ treatment induces the expression of SAMD9 in both endothelial cells and fibroblasts. Very recently, it was found that glioblastoma cell lines show an increase in SAMD9 expression following treatment with type 1 IFNs or incubation with inactivated Sendai virus particle (HVJ-E), suggesting that SAMD9 might have a role in the protection against viral infection (Tanaka et al., 2010). Taken together with the fact that SAMD9 responds to tumor necrosis factor-α treatment (Chefetz et al., 2008), these data position SAMD9 at an important crossroad between major inflammatory pathways. It is worth noting that the SAMD9 paralog, SAMD9L, can also be induced by treatment with type 1 IFNs (Pappas et al., 2009).
Analysis of SAMD9 promoter revealed the existence of a short DNA fragment comprising an IRF-1 transcription factor response element, capable of regulating SAMD9 expression. IRF-1 is a member of a large family of proteins that function as transcription activators of genes induced by IFN α, β, and γ (Schmitz et al., 2007). IRF-1 is an important mediator of apoptosis (Gao et al., 2010) and may have a role in apoptosis induced by SAMD9, as demonstrated in colon cancer cells (Li et al., 2007). There is also ample evidence that IRF-1 functions as a tumor suppressor gene. For example, IRF-1 gene repression is essential for the malignant transformation of hepatitis C virus-infected cells (Zekri et al., 2010); loss of heterozygosity in gastric cancer often involves the IRF-1 gene (Gao et al., 2010); and IRF-1 expression is markedly decreased in esophageal squamous cell carcinoma (Wang et al., 2007).
Our findings suggest that IRF-1 tumor suppressor activity may in part result from its inductive effect on SAMD9 expression. In addition, in several studies myelodysplastic syndrome and leukemia have been linked to both SAMD9 and IRF-1 (Liebermann and Hoffman, 2006; Maratheftis et al., 2006; Asou et al., 2009). Inactivation of IRF-1 is commonly found in myelodysplastic syndrome and leukemia (Liebermann and Hoffman, 2006; Maratheftis et al., 2006). Interestingly, in these same malignancies, a common micro-deletion in chromosome 7q21.3, harboring the SAMD9 locus, has been described (Asou et al., 2009). Moreover, autoimmune manifestations, which are the hallmark of NFTC, are commonly found in myelodysplastic syndrome patients and their pathogenesis is mediated by IRF-1 (Pinheiro et al., 2009).
The SAMD9 protein contains a conserved sterile α-motif (SAM) domain. These domains usually serve as protein interaction domains. In the present case, we found that this domain interacts with the C-terminal part of RGL2. RGL2 is a guanine nucleotide exchange factor that, following activation by GTP-bound Ras, can activate two Ral family members, RalA and RalB (Ferro and Trabalzini, 2010). RalB, the direct downstream target of RGL2, has been shown to have a role in cancer (Chien and White, 2003) and in response to viral infections (Cho et al., 2006) just as SAMD9 (Tanaka et al., 2010).
Both SAMD9 and RGL2 seem to function by inhibiting EGR1 expression. In addition, the inhibitory activity of RGL2 seems to be dependent upon the expression of SAMD9, as it is not detectable in HeLa cells, which are naturally deficient in SAMD9. EGR1 is overexpressed in atherosclerosis in humans as well as mouse models (McCaffrey et al., 2000). A more recent report showed that EGR1 is an important mediator of interleukin-13-induced inflammation as well (Cho et al., 2006). Interestingly, EGR1 was also found to regulate wound healing in a model of scleroderma, an autoimmune disease, which is commonly associated with ectopic calcification (Wu et al., 2009). In another study, EGR1 was found to be the only single gene that was differentially expressed in transforming growth factor-β-treated fibroblasts derived from scleroderma patients as compared with normal fibroblasts (Sargent et al., 2010). Moreover, tissue deposition of calcium phosphate has been associated with increased EGR1 expression (Molloy and McCarthy, 2006). EGR1 has been linked to non-cutaneous inflammatory disorders as well. In Crohn’s disease, another autoinflammatory disorder, EGR1 levels are elevated and, interestingly, RGL2 is downregulated (Fernandez-Becker and Moss, 2009), exactly as in our system.
EGR1 has also been implicated in the pathogenesis of breast, prostate, and lung cancer (Gitenay and Baron, 2009; Fang et al., 2010; Redmond et al., 2010). Apparently, EGR1 is also pivotal for cancer metastasis, as it was downregulated in a clone from a highly metastatic fibrosarcoma cell line that lost its metastatic capacity (Cermak et al., 2010). Metastatic potential was restored following introduction of exogenous EGR1 to the cells (Cermak et al., 2010). One component of the pro-metastatic phenotype induced by EGR1 is the activation of genes that control actin contractility (Cermak et al., 2010). Of interest, we could demonstrate intracellular redistribution of actin filaments following downregulation of SAMD9 or RGL2 (not shown), which was shown to increase EGR1 expression.
In summary, the present data indicate that SAMD9, a molecule associated with the regulation of cutaneous inflammation, ectopic calcification in the skin, and possibly oncogenesis, seems to function along the IFN-γ signaling pathway and regulates, in concert with RGL2, the expression of EGR1, a key mediator of inflammation in health and disease (Figure 6).
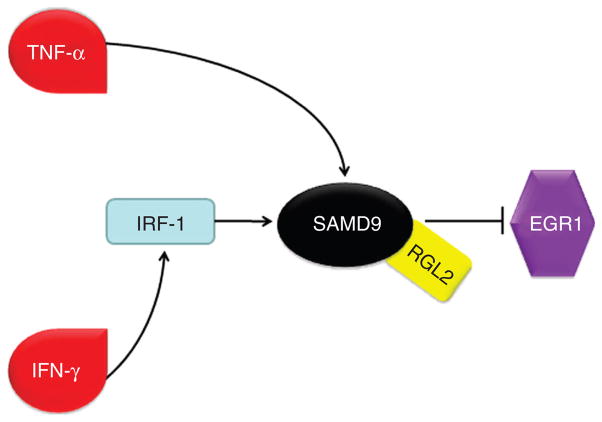
Figure 6. Schematic representation of SAMD9 regulation and downstream effect.
Previous (Chefetz et al., 2008) and current data indicate that SAMD9 expression is tightly regulated by the inflammatory cytokines TNF-α and IFN-γ. Our results suggest that this regulation is mediated in the case of TNF-α by p38 and NF-κB (Chefetz et al., 2008), and in the case of IFN-γ through the IRF-1 transcription factor. Additionally, SAMD9 may regulate inflammation, calcification, and cancer progression through its effect on EGR1 expression, in concert with its partner, RGL2. IFN-γ, interferon-γ; IRF-1, IFN regulatory factor-1; NF-κB, nuclear factor-κB; TNF-α, tumor necrosis factor-α.
MATERIALS AND METHODS
Reagents and antibodies
The following antibodies were used: anti-Myc monoclonal (9E10) and anti-HA monoclonal (12CA5). IFN-γ was obtained from PeproTech (Rocky Hill, NJ). SAMD9-specific (5′-CAAAGCAACCAA UUGCAUA-3′ (dT)) and RGL2-specific siRNA (5′-CUAAUGUAUUC UACGCCAU-3′ (dT)), as well as universal control siRNA, were purchased from Sigma-Aldrich (St Louis, MO).
Plasmids and expression constructs
pGL3 promoter luciferase reporter plasmid (Promega, Madison, WI) was used to insert PCR-amplified MluI–XhoI DNA fragments spanning the different parts of the putative proximal SAMD9 promoter between nucleotide positions −4306 and −67 SAMD9 (relative to the transcription start site). The primers used in these experiments are provided in Supplementary Table S2 online.
A DNA sequence encompassing the SAMD9 SAM domain (amino acids 1–124) was cloned into pMet425-Ras to generate pMet425-Ras-SAMD9-NT, which was then digested using BamHI and XhoI and the insert was subcloned into the mammalian expression plasmid pCAN to generate Myc-tagged SAMD9. The spleen library plasmids expressing RALGDS and RGL2 were digested using EcoRI and XhoI and subcloned into pcDNA-3HA plasmid to generate 3HA-tagged pcDNA-RALGDS and pcDNA-RGL2 plasmids.
Cell cultures and transient transfections
Fibroblasts transformed with a pBABE-H2AGFP construct containing the human telomerase gene, EaHy926 cells, a human umbilical vein endothelial cells-derived transformed endothelial cell line, human embryonic kidney 293T (HEK-293T) and HeLa cells were maintained in DMEM containing 10% FCS and 1% penicillin and streptomycin and grown at 37 °C and 5% CO2. The medium of EaHy926 cells was supplemented with 2% hypoxanthine-aminopterin-thymidine (Biological Industries, Beit Haemek, Israel).
Cells were transfected with the appropriate expression plasmid or siRNA using lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. Fresh medium was added 4–5 hours after transfection and cells were harvested 48 hours after transfection.
QRT-PCR
For QRT-PCR validation of global gene expression results, cDNA was synthesized from 1 μg of total RNA using the Verso cDNA kit (Thermo Scientific, Surrey, UK) and random hexamers. cDNA PCR amplification was carried out using the SYBR Green JumpStart Taq ReadyMix (Sigma, St Louis, MO) on an Mx3000p/5p multifilter system (Stratagene, Cedar Creek, TX) with gene-specific introncrossing oligonucleotide pairs listed in Supplementary Table S3 online. To ensure the specificity of the reaction conditions, at the end of the individual runs, the melting temperature of the amplified products was measured to confirm its homogeneity. Cycling conditions were as follows: 95 °C for 10 minutes, 95 °C for 10 seconds, 62 °C for 25 seconds, and 72 °C for 15 seconds for a total of 40 cycles. Each sample was analyzed in duplicate. For quantification, standard curves were obtained using serially diluted cDNA amplified in the same RT-PCR run. Results were normalized to ACTB mRNA levels.
Promoter activity assay
The luciferase constructs were transfected into fibroblasts using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At 48 hours after transfection, the protein was extracted and assayed for luciferase activity using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Relative luminometer units were read using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA).
Electrophoretic mobility shift assay
Nuclear extraction was performed as previously described (Schreiber et al., 1989), except that the buffers included Protease Inhibitor Cocktail (Sigma-Aldrich). Electrophoretic mobility shift assay was performed with nuclear extracts from EaHy and HeLa cell lines. The lysates were incubated in binding buffer (25mM HEPES (pH 7.9), 150mM KCl, 2% glycerol, 5mM dithiothreitol, and 1mg of poly (dI-dC)) for 5 minutes at room temperature, and 32P-labeled oligonucleotide probes were added (15,000 c.p.m.). After 20 minutes of incubation, the samples were resolved on a 6% nondenaturing polyacrylamide gel in 0.53 TBE buffer and autoradiographed. The 32P-end-labeled DNA probes used spanned nucleotide positions from −118 to −89 (Supplementary Table S2 online) and the same probe carrying A>T transversions at positions −98 and −105.
Ras recruitment system
pMet425-Ras-SAMD9-NT was used as a bait to screen a human spleen expression library. Yeast growth, transfection, and screening protocol were performed as described in Hubsman et al. (2001).
Coimmunoprecipitation
At 24 hours after transfection with pCAN-Myc-SAMD9 and pcDNARALGDS- 3HA, pcDNA-RGL2–3HA, or pcDNA-3HA, HEK-293T cells were lysed in whole cell extract buffer (25mM HEPES, pH 7.7, 0.3 M NaCl, 1.5mM MgCl2, 0.2mM EDTA, 0.1% Triton X-100, 0.5mM DTT, 20mM-glycerolphosphate, 0.1mM Na2VO4, 100 gml−1 phenylmethylsulfonyl fluoride, protease inhibitor cocktail 1:100; Sigma-Aldrich). Anti-HA monoclonal antibodies were incubated overnight at 4 °C with 400–600 mg of the HEK-293T protein extract. Protein A Sepharose beads (Sigma Chemical, St Louis, MO) were added to the extracts for 1 hour at 4 °C. After four washes with whole cell extract buffer, the precipitated proteins were eluted using SDS-PAGE sample buffer. Samples were boiled for 3 minutes and the proteins were then separated by 10% SDS-PAGE, followed by transfer to a nitrocellulose membrane. Blots were blocked in 5% dry milk in PBS and washed three times for 5 minutes in PBS. Proteins were detected using antibodies against Myc diluted 1:500.
Global gene expression analysis
An amount of 200 ng of total RNA was amplified into cRNA and biotinylated by in vitro transcription using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX) according to the manufacturer’s protocol. Biotinylated cRNA was purified, fragmented, and subsequently hybridized to an Illumina Human HT-12 v3 BeadChip (Direct Hybridization assay, Illumina, San Diego, CA). The hybridized chip was stained with streptavidin-Cy3 (Amersham, Arlington Heights, IL) and scanned with an Illumina bead array reader. The scanned images were imported into GenomeStudio (Illumina) for data extraction and quality control. Statistical analysis of gene expression in the different treatments was done on the raw bead-level data. The different treatment modalities were compared using one-way analysis of variance test with the SPSS 12.0 software (Chicago, IL). Differences between treatments that were more than 2-fold and with a P-value of less than 0.001 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr Liat Linde (BioRap Technologies, Rappaport Research Institute, Technion, Israel) for expert assistance with the microarray data generation and express our gratitude to Ami Aronheim, Adi Salzberg, Revital Shemer, Nili Avidan, and Sara Selig for their assistance. Dov Hershkovitz is supported by the Etai Sharon Z″l Rambam-Atidim Fellowship Fund for Academic Excellence. This work was supported in part by grants provided by the Israel Science Foundation, Israel Health Ministry, Office of the Chief Scientist, and NIH/NIAMS Grant R01 AR052627.
Abbreviations
- HA
hemagglutinin
- IFN-γ
interferon-γ
- IRF-1
IFN regulatory factor 1
- NFTC
normophosphatemic familial tumoral calcinosis
- QRT-PCR
quantitative real-time RT-PCR
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
References
- Antonarakis SE, Beckmann JS. Mendelian disorders deserve more attention. Nat Rev Genet. 2006;7:277–82. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- Aronheim A. Protein recruitment systems for the analysis of protein +/− protein interactions. Methods. 2001;24:29–34. doi: 10.1006/meth.2001.1153. [DOI] [PubMed] [Google Scholar]
- Asou H, Matsui H, Ozaki Y, et al. Identification of a common microdeletion cluster in 7q21.3 subband among patients with myeloid leukemia and myelodysplastic syndrome. Biochem Biophys Res Commun. 2009;383:245–51. doi: 10.1016/j.bbrc.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Bai Y, Zhou G, Nakamura M, et al. Survival impact of psammoma body, stromal calcification, and bone formation in papillary thyroid carcinoma. Mod Pathol. 2009;22:887–94. doi: 10.1038/modpathol.2009.38. [DOI] [PubMed] [Google Scholar]
- Benet-Pages A, Orlik P, Strom TM, et al. An FGF23 missense mutation causes familial tumoral calcinosis with hyperphosphatemia. Hum Mol Genet. 2005;14:385–90. doi: 10.1093/hmg/ddi034. [DOI] [PubMed] [Google Scholar]
- Broder YC, Katz S, Aronheim A. The ras recruitment system, a novel approach to the study of protein-protein interactions. Curr Biol. 1998;8:1121–4. doi: 10.1016/s0960-9822(98)70467-1. [DOI] [PubMed] [Google Scholar]
- Budoff MJ, Shaw LJ, Liu ST, et al. Long-term prognosis associated with coronary calcification: observations from a registry of 25,253 patients. J Am Coll Cardiol. 2007;49:1860–70. doi: 10.1016/j.jacc.2006.10.079. [DOI] [PubMed] [Google Scholar]
- Cermak V, Kosla J, Plachy J, et al. The transcription factor EGR1 regulates metastatic potential of v-src transformed sarcoma cells. Cell Mol Life Sci. 2010;67:3557–68. doi: 10.1007/s00018-010-0395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefetz I, Ben Amitai D, Browning S, et al. Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-alpha responsive protein. J Invest Dermatol. 2008;128:1423–9. doi: 10.1038/sj.jid.5701203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–6. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YW, Adler SG, Budoff MJ, et al. Coronary artery calcification and mortality in diabetic patients with proteinuria. Kidney Int. 2010;77:1107–14. doi: 10.1038/ki.2010.70. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Kang MJ, Homer RJ, et al. Role of early growth response-1 (Egr-1) in interleukin-13-induced inflammation and remodeling. J Biol Chem. 2006;281:8161–8. doi: 10.1074/jbc.M506770200. [DOI] [PubMed] [Google Scholar]
- Fang L, Min L, Lin Y, et al. Downregulation of stathmin expression is mediated directly by Egr1 and associated with p53 activity in lung cancer cell line A549. Cell Signal. 2010;22:166–73. doi: 10.1016/j.cellsig.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Fernandez-Becker NQ, Moss AC. In silico analysis of T-bet activity in peripheral blood mononuclear cells in patients with inflammatory bowel disease (IBD) In Silico Biol. 2009;9:28. doi: 10.3233/ISB-2009-0410. [DOI] [PubMed] [Google Scholar]
- Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal. 2010;22:1804–10. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Gao J, Senthil M, Ren B, et al. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ. 2010;17:699–709. doi: 10.1038/cdd.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitenay D, Baron VT. Is EGR1 a potential target for prostate cancer therapy? Future Oncol. 2009;5:993–1003. doi: 10.2217/fon.09.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia A, Pritchard CA, Paterson HF, et al. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–26. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Haas MH. The risk of death in patients with a high coronary calcification score: does it include predialysis patients? Kidney Int. 2010;77:1057–9. doi: 10.1038/ki.2010.92. [DOI] [PubMed] [Google Scholar]
- Holmberg C, Katz S, Lerdrup M, et al. A novel specific role for I kappa B kinase complex-associated protein in cytosolic stress signaling. J Biol Chem. 2002;277:31918–28. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- Hubsman M, Yudkovsky G, Aronheim A. A novel approach for the identification of protein-protein interaction with integral membrane proteins. Nucleic Acids Res. 2001;29:E18. doi: 10.1093/nar/29.4.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa S, Imel EA, Kreiter ML, et al. A homozygous missense mutation in human KLOTHO causes severe tumoral calcinosis. J Clin Invest. 2007;117:2684–91. doi: 10.1172/JCI31330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, MacDonald JR, Wei RY, et al. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics. 2007;8:92. doi: 10.1186/1471-2164-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann DA, Hoffman B. Interferon regulatory factor-1 myelodysplasia and leukemia. Leuk Res. 2006;30:1069–71. doi: 10.1016/j.leukres.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Maratheftis CI, Bolaraki PE, Giannouli S, et al. Aberrant alternative splicing of interferon regulatory factor-1 (IRF-1) in myelodysplastic hematopoietic progenitor cells. Leuk Res. 2006;30:1177–86. doi: 10.1016/j.leukres.2005.12.021. [DOI] [PubMed] [Google Scholar]
- McCaffrey TA, Fu C, Du B, et al. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000;105:653–62. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CH, Maher SG, Young HA. Clinical use of interferon-gamma. Ann N Y Acad Sci. 2009;1182:69–79. doi: 10.1111/j.1749-6632.2009.05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy ES, McCarthy GM. Basic calcium phosphate crystals: pathways to joint degeneration. Curr Opin Rheumatol. 2006;18:187–92. doi: 10.1097/01.bor.0000209433.43978.a8. [DOI] [PubMed] [Google Scholar]
- Pappas DJ, Coppola G, Gabatto PA, et al. Longitudinal system-based analysis of transcriptional responses to type I interferons. Physiol Genomics. 2009;38:362–71. doi: 10.1152/physiolgenomics.00058.2009. [DOI] [PubMed] [Google Scholar]
- Parwani AV, Chan TY, Ali SZ. Significance of psammoma bodies in serous cavity fluid: a cytopathologic analysis. Cancer. 2004;102:87–91. doi: 10.1002/cncr.20161. [DOI] [PubMed] [Google Scholar]
- Pinheiro RF, Metze K, Silva MR, et al. The ambiguous role of interferon regulatory factor-1 (IRF-1) immunoexpression in myelodysplastic syndrome. Leuk Res. 2009;33:1308–12. doi: 10.1016/j.leukres.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Redmond KL, Crawford NT, Farmer H, et al. T-box 2 represses NDRG1 through an EGR1-dependent mechanism to drive the proliferation of breast cancer cells. Oncogene. 2010;29:3252–62. doi: 10.1038/onc.2010.84. [DOI] [PubMed] [Google Scholar]
- Sargent JL, Milano A, Bhattacharyya S, et al. A TGFbetaresponsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol. 2010;130:694–705. doi: 10.1038/jid.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz F, Heit A, Guggemoos S, et al. Interferon-regulatory-factor 1 controls Toll-like receptor 9-mediated IFN-beta production in myeloid dendritic cells. Eur J Immunol. 2007;37:315–27. doi: 10.1002/eji.200636767. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, et al. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher E. Familial tumoral calcinosis: from characterization of a rare phenotype to the pathogenesis of ectopic calcification. J Invest Dermatol. 2010;130:652–60. doi: 10.1038/jid.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Shimbo T, Kikuchi Y, et al. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int J Cancer. 2010;126:1982–91. doi: 10.1002/ijc.24965. [DOI] [PubMed] [Google Scholar]
- Topaz O, Indelman M, Chefetz I, et al. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet. 2006;79:759–64. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaz O, Shurman DL, Bergman R, et al. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet. 2004;36:579–81. doi: 10.1038/ng1358. [DOI] [PubMed] [Google Scholar]
- Touart DM, Sau P. Cutaneous deposition diseases. Part II. J Am Acad Dermatol. 1998;39:527–44. doi: 10.1016/s0190-9622(98)70001-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu DP, Chen PP, et al. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 2007;67:2535–43. doi: 10.1158/0008-5472.CAN-06-3530. [DOI] [PubMed] [Google Scholar]
- Wu M, Melichian DS, de la Garza M, et al. Essential roles for early growth response transcription factor Egr-1 in tissue fibrosis and wound healing. Am J Pathol. 2009;175:1041–55. doi: 10.2353/ajpath.2009.090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri AR, Moharram RA, Mohamed WS, et al. Disease progression from chronic hepatitis C to cirrhosis and hepatocellular carcinoma is associated with repression of interferon regulatory factor-1. Eur J Gastroenterol Hepatol. 2010;22:450–6. doi: 10.1097/MEG.0b013e3283329d00. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.