Abstract
Inactivation of cyclin B–Cdc2 kinase at the exit from M phase depends on the specific proteolysis of the cyclin B subunit, whereas the Cdc2 subunit remains present at nearly constant levels throughout the cell cycle. It is unknown how Cdc2 escapes degradation when cyclin B is destroyed. In Xenopus egg extracts that reproduce the exit from M phase in vitro, we have found that dissociation of the cyclin B–Cdc2 complex occurred under conditions where cyclin B was tethered to the 26S proteasome but not yet degraded. The dephosphorylation of Thr 161 on Cdc2 was unlikely to be necessary for the dissociation of the two subunits. However, the dissociation was dependent on the presence of a functional destruction box in cyclin B. Cyclin B ubiquitination was also, by itself, not sufficient for separation of Cdc2 and cyclin B. The 26S proteasome, but not the 20S proteasome, was capable of dissociating the two subunits. These results indicate that the cyclin B and Cdc2 subunits are separated by the proteasome through a mechanism that precedes proteolysis of cyclin B and is independent of proteolysis. As a result, cyclin B levels decrease on exit from M phase but Cdc2 levels remain constant.
Keywords: cyclin B–Cdc2, proteasome, cell cycle, protein unfolding, Xenopus egg extracts
The cyclin B–Cdc2 kinase is a universal regulator of M phase (for review, see Nurse 1990; Nigg 1995). Its activation induces entry into M phase and its inactivation is necessary for exit from M phase. The activity of cyclin B–Cdc2 kinase is regulated primarily by the formation of a complex between the catalytic Cdc2 subunit and the cyclin B regulatory subunit. This complex is stabilized by phosphorylation of Cdc2 on Thr 161, and is kept inactive by inhibitory phosphorylation of Cdc2 on Thr 14 and Tyr 15 (for review, see Nigg 1995). The amount of Cdc2 remains relatively constant throughout the cell cycle, whereas cyclin B accumulates during interphase, reaching a peak at metaphase, and is suddenly destroyed at the exit from M phase (for review, see King et al. 1996; Townsley and Ruderman 1998). The differences in the stability of Cdc2 and cyclin B at the end of M phase is seen even though the two proteins are tightly associated in a complex prior to cyclin B degradation. Although it has not been previously demonstrated, it is probable that the cyclin B–Cdc2 complex must dissociate prior to the degradation of cyclin B subunit so that Cdc2 might escape degradation. However, when the dissociation of cyclin B and Cdc2 takes place and the actual steps involved in the regulation of this dissociation are not known.
The degradation of the mitotic B-type cyclins is performed by ubiquitin–proteasome-mediated proteolysis. The N-terminal region of cyclin B contains a conserved motif called the destruction box, which serves as a signal for ubiquitination and is necessary for cell cycle-regulated proteolysis (Glotzer et al. 1991). The formation of ubiquitin conjugates requires the concerted activity of a series of enzymes that first activate ubiquitin (E1) and then recognize and transfer ubiquitin (E2 and E3) to proteins destined for turnover (for review, see Hershko and Ciechanover 1998). Cyclin B is polyubiquitinated by a specific E3, the multicomponent 20S complex known as the APC/C (anaphase-promoting complex/cyclosome; for review, see Townsley and Ruderman 1998). Neither the activities of E1 nor a specific E2 (E2-C or UBC10) for cyclin B ubiquitination change during the cell cycle (Hershko et al. 1994; King et al. 1995; Sudakin et al. 1995), but the E3-like activity of the APC/C is the target of cell cycle-dependent regulation depending on association with Cdc20 (for review, see Townsley and Ruderman 1998; Morgan 1999). Although the APC/C-dependent polyubiquitination of cyclin B alone may occur in an in vitro reconstitution system (King et al. 1995), it normally occurs in vivo in the cyclin B–Cdc2 complex. However, it has not been determined whether the polyubiquitination by the APC/C is involved in dissociation of the cyclin B–Cdc2 complex or only in targeting cyclin B for degradation.
Polyubiquitinated proteins that are destined for turnover are recognized and degraded by the 26S proteasome (for review, see Baumeister et al. 1998; Rechsteiner 1998). The 26S proteasome can be divided into three subcomplexes. A core subcomplex, the 20S proteasome, is a cylindrical stack consisted of four rings and exhibits proteolytic activity. Each end of the 20S cylinder is capped with another subcomplex, the 19S complex. Proteolytically active sites of the 20S cylinder face an interior chamber that can be entered only through a narrow pore at either end (for review, see Baumeister et al. 1998). Because folded proteins cannot reach this chamber, the 19S complex is thought to be involved in substrate unfolding in addition to recognition of polyubiquitin chain (Glickman et al. 1998). Degradation of polyubiquitinated cyclin B should be executed by the 26S proteasome, and, accordingly, Cdc2 should be dissociated from cyclin B to escape degradation. It remains unclear, however, whether the proteasome is responsible for the dissociation of the cyclin B–Cdc2 complex prior to cyclin B proteolysis.
Similar to the case of the cyclin B–Cdc2 complex, the proteasome can selectively degrade a single domain of various complexes, for example, other types of cyclins in the cyclin–Cdk complex in cell cycle control (for review, see Koepp et al. 1999), securin in the securin–separin complex in sister chromatid separation at the metaphase/anaphase transition (for review, see Nasmyth et al. 2000), IκBα in the IκBα–NFκB complex at the activation of NFκB, and β-catenin in the β-catenin–GSK3β/APC (adenomatous polyposis coli)/Axin complex in the Wnt pathway (for review, see Maniatis 1999). In any case, however, it remains almost unclear how selective degradation by the proteasome is performed for a single domain in the complex.
In the present study, we have studied the relationship between the dissociation of the cyclin B–Cdc2 complex and cyclin B degradation and have determined how Cdc2 escapes degradation even though it is in a complex with cyclin B, which is targeted for proteolysis. As an experimental system, we have used CSF extracts prepared from unfertilized Xenopus eggs arrested at metaphase II, in which a high level of cyclin B–Cdc2 kinase activity is retained and cyclin B destruction can be triggered by the addition of Ca2+ (Murray 1991). We have found that when MG115, an inhibitor of the proteolytic activity of the proteasome, is added to these extracts, although there is a marked inhibition of cyclin B degradation, the cyclin B–Cdc2 kinase activity still decreases. We show that the inactivation of cyclin B–Cdc2 kinase is due to the dissociation of Cdc2 from cyclin B by the 26S proteasome through a mechanism that is not dependent on proteolysis of the cyclin B substrate.
Results
Cdc2 dissociates from cyclin B in the absence of cyclin B destruction
The stability of Cdc2 protein at the time of cyclin B destruction could be explained in two ways. One possible explanation is that a proteolytic cleavage within the cyclin box in cyclin B, which is required for the association of cyclin B with Cdc2 (Kobayashi et al. 1992; Lees and Harlow 1993), separates Cdc2 from cyclin B without degradation of Cdc2. A second possibility is that Cdc2 could be released by some unknown mechanisms prior to, and independent of, cyclin B destruction. To address whether proteolytic activity is indispensable for dissociation of the cyclin B–Cdc2 complex, we utilized extracts prepared from unfertilized Xenopus eggs that are arrested at meiotic metaphase II [cytostatic factor-arrested (CSF) extracts; Murray 1991]. Addition of calcium to the extracts induces cyclin B destruction through the ubiquitin–APC/C-proteasome pathway, leading to the inactivation of cyclin B–Cdc2 kinase (Glotzer et al. 1991).
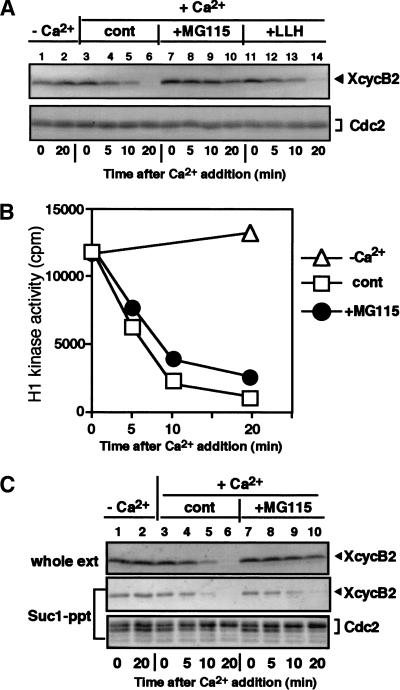
A peptide-aldehyde, MG115 (carbobenzoxyl-leucinyl-leucinyl-norvalinal-H, also called Z-LLnV), has been shown to be a potent inhibitor of the chymotryptic site in the 20S proteasome and calpine (Rock et al. 1994). We examined whether MG115 could block the proteolysis of cyclin B in Xenopus egg extracts. In control extracts, to which calcium was added without MG115, cyclin B2 began to decrease within 10 min and was undetectable by 20 min (Fig. 1A, lanes 3–6). In contrast, the addition of 0.5 mM MG115 10 min prior to calcium addition markedly inhibited the destruction of cyclin B2 (Fig. 1A, lanes 7–10). Because the same concentration of the calpine inhibitor Z-LLH had no effect on cyclin B2 degradation (Fig. 1A, lanes 11–14), the inhibition by MG115 is in all likelihood due to the inhibition of the proteasome. Although ubiquitinated forms of cyclin B2 were not seen when the proteolytic activity of the proteasome was blocked with MG115, this may be due to the isopeptidase activity in Xenopus egg extracts (Mahaffey et al. 1993).
Figure 1.
Dissociation of Cdc2 from cyclin B in Xenopus egg CSF extracts. (A) Inhibition of cyclin B destruction by MG115 in Xenopus egg extracts. Calcium (0.6 mM) was added at time zero to all extracts except controls (lanes 1–2). Ten minutes before calcium addition, 0.5 mM MG115 (proteasome inhibitor) or Z-LLH (calpine inhibitor) was added. At the indicated times, aliquots were taken and immunoblotted with antibodies against Xenopus cyclin B2 (top) or PSTAIR (for Cdc2; bottom). The small loss of cyclin B in the presence of MG115 might reflect the leakage of its effect. (B) Inactivation of cyclin B–Cdc2 kinase in the presence of MG115. (Open triangles) Addition of calcium; (Closed circles) addition of MG115; (Open boxes) control. (C) Dissociation of Cdc2 from cyclin B in the presence of MG115. Xenopus egg CSF extracts were activated by calcium in the presence or absence of 0.5 mM MG115. Samples taken at the indicated times were assayed for H1 kinase activity, recovered by Suc1–Sepharose beads, or immediately mixed with Laemmli's sample buffer. Materials retained on Suc1-beads were eluted with 2× Laemmli's sample buffer. Samples were immunoblotted with antibodies against Xenopus cyclin B2 (top and middle) or PSTAIR (bottom). (Lanes 1,2) Control samples without the addition of calcium.
Even though cyclin B destruction was prevented in the presence of MG115, surprisingly, we found that H1 kinase activity fell within 10 min of calcium addition (Fig. 1B). The rate and extent of cyclin B–Cdc2 kinase inactivation were similar to those in control extracts to which only DMSO had been added. We suspected that a reason for the inactivation of cyclin B–Cdc2 kinase in the presence of MG115 may have been the dissociation of Cdc2 from cyclin B. To examine this possibility further, we first tried to immunoprecipitate cyclin B with its antibodies; however, this immunoprecipitation was unsuccessful. Because these antibodies could readily immunoprecipitate cyclin B–Cdc2 from CSF extracts, these results suggest that cyclin B is in an unusual state in MG115-treated extracts (see Fig. 5, below). Next, we incubated extracts with Suc1–Sepharose, which absorbs Cdc2 and Cdc2-associated polypeptides from extracts (Dunphy et al. 1988), and analyzed the bound proteins with immunoblots. As shown in Figure 1C (lanes 7–10), the addition of calcium in the presence of MG115 led to a significant decrease in the amount of cyclin B2 that was associated with Cdc2, and no cyclin B2 was detectable in Suc1 precipitates at 20 min. Despite the decrease in the amount of cyclin B that was associated with Cdc2 after calcium addition, the total amount of cyclin B2 remained almost the same before and after calcium addition. In parallel, a fraction of Cdc2 shifted to a lower mobility form, possibly due to dephosphorylation of Thr 161 (Fig. 1C, bottom). A similar decrease in the amount of Cdc2-associated cyclin B was also seen when a Myc-tagged version of Cdc2 was immunoprecipitated from extracts (see Fig. 2B, below). Moreover, cyclin B1 behaved identically to cyclin B2 in the presence of MG115 (data not shown). Similar results were obtained with another proteasome inhibitor, MG132 (Rock et al. 1994) (data not shown). Because we have never observed an effect of MG115 in CSF extracts in the absence of calcium addition, these results demonstrate that Cdc2 dissociates from cyclin B in the absence of cyclin B degradation in response to a stimulus that causes the exit from metaphase and that the proteolytic activity of the proteasome is not required for the dissociation.
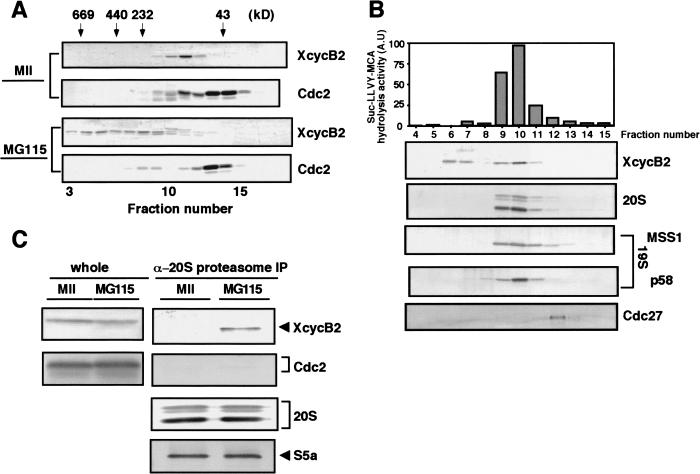
Figure 5.
Cyclin B that has dissociated from Cdc2 remains associated with the 26S proteasome. (A) Cyclin B dissociated from Cdc2 is in high molecular weight complexes. CSF extracts were treated with calcium in the presence of MG115 (MG115-treated extracts), and then fractionated by gel filtration on a Superose 6 column in the presence of 1 mM ATP. Each fraction was tested on immunoblots for the presence of Xenopus cyclin B2 and Cdc2 (PSTAIR). As a control, untreated CSF extracts (MII extracts) were applied to the same column. (B) Cyclin B dissociated from Cdc2 co-elutes with the 26S proteasome. High molecular weight fractions in A containing cyclin B2, but not Cdc2, were pooled and refractionated in the presence of 1 mM ATP by ion-exchange chromatography on Mono Q Sepharose. After elution with increasing concentrations of KCl, each fraction was tested for peptidase activity against Suc-LLVY-AMC, and analyzed by immunoblots with antibodies against Xenopus cyclin B2, the Xenopus 20S proteasome, human MSS1 and p58 (both for the 19S particle), and human Cdc27 (for APC/C). (C) Cyclin B dissociated from Cdc2 is co-precipitated with the 26S proteasome. CSF extracts and MG115-treated extracts were subjected to immunoprecipitation with anti-Xenopus 20S proteasome antibody. The precipitates were probed with antibodies against cyclin B2, PSTAIR (Cdc2), the 20S proteasome, and S5a.
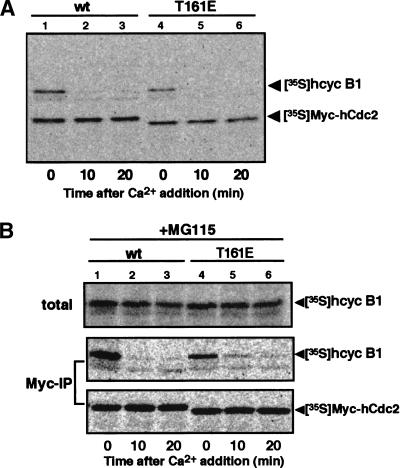
Figure 2.
Dephosphorylation of Thr 161 on Cdc2 is not necessary for dissociation from cyclin B. (A) Destruction of cyclin B associated with the T161E mutant of Cdc2. (B) Dissociation of the T161E mutant of Cdc2 from cyclin B. To produce the hcyclin B1–Myc–hCdc2 complex, reticulocyte lysates containing 35S-labeled hcyclin B1 and Myc-tagged hCdc2 were incubated in the presence of ATP and, as a source of CAK, starfish oocyte extracts that had been depleted of endogenous starfish Cdc2 and cyclin B. CSF extracts were mixed with hcyclin B1 that had been complexed with either Myc–hCdc2-wt (lanes 1–3) or Myc–hCdc2-T161E (lanes 4–6), and then activated by calcium addition in the absence (A) or presence (B) of 0.5 mM MG115. Samples taken at the indicated times were directly, or after immunoprecipitation with anti-Myc antibody, resolved by SDS-PAGE and analyzed by a Fuji BAS 2000 phosphorimager.
Thr 161 dephosphorylation is unlikely to be necessary for the dissociation of cyclin B–Cdc2
Because the phosphorylation at Thr 161 of Cdc2 is required for its catalytic function and is essential for its stable binding to cyclin (for review, see Morgan 1995), dephosphorylation at Thr 161 could, conceivably, be the cause of the dissociation of the cyclin B–Cdc2 complex. If so, the constitutive phosphorylation of Thr 161 should keep Cdc2 and cyclin B in a stable complex under the experimental conditions described in Figure 1. To examine this possibility, we replaced Thr 161 with glutamic acid (E), which mimics the constitutively phosphorylated state. mRNAs encoding Myc-tagged versions of both wild-type and mutant human Cdc2 (hCdc2-wt and hCdc2-T161E) were translated in reticulocyte lysates in the presence of [35S]methionine. In parallel, we also prepared radiolabeled human cyclin B1 protein (hcyclin B1) in reticulocyte lysates. The hcyclin B1–hCdc2 complexes were prepared by mixing reticulocyte lysates with starfish oocyte extracts that had been depleted of endogenous starfish Cdc2 and cyclin B with Suc1–Sepharose, and incubated in the presence of ATP. The depleted oocyte extracts provided a source of CDK-activating kinase (CAK ) for the phosphorylation of hCdc2-wt on Thr 161 (Fesquet et al. 1993). We confirmed the binding of hCdc2 and hcyclin B1 by immunoprecipitation with antibodies against the Myc epitope and hcyclin B1 (data not shown; see also Fig. 2B, lanes 1,4).
When the hcyclin B1–Myc–hCdc2 complexes were added to CSF extracts, hcyclin B1 bound to either wild-type or mutant hCdc2 was stable during a 30-min incubation in the absence of calcium (data not shown). The addition of calcium to the control CSF extracts caused the complete degradation of hcyclin B1 associated with both hCdc2s within 20 min (Fig. 2A). However, in the presence of MG115, hcyclin B1 showed little degradation after calcium addition, but again the amount of hcyclin B1 that was recovered by anti-Myc antibody was significantly decreased for both Myc–hCdc2-wt and Myc–hCdc2-T161E (Fig. 2B). The extent of dissociation of hcyclin B1 was almost similar for both hCdc2-wt and hCdc2-T161E. It is thus likely that the dephosphorylation of Cdc2 on Thr 161 is not required for either cyclin B degradation or dissociation of the cyclin B–Cdc2 complex.
Dissociation of cyclin B–Cdc2 depends on functional destruction box of cyclin B
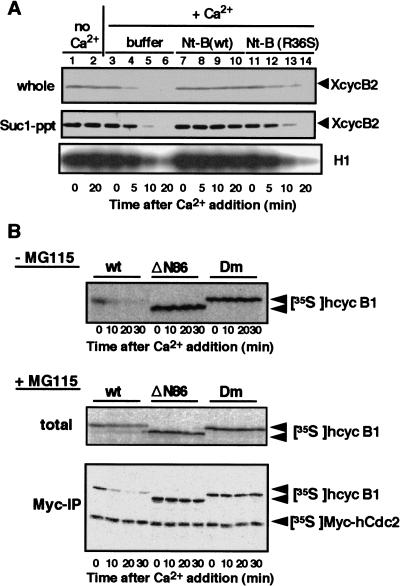
The destruction box located near the N terminus of cyclin B plays a crucial role in targeting cyclin B for destruction (Glotzer et al. 1991). We examined whether the destruction box is required for dissociation of the cyclin B–Cdc2 complex. A peptide coding the first 108 amino acids of Xenopus cyclin B1 [Nt-B(wt)] was produced and added to CSF extracts to compete with the destruction box of endogenous cyclin B. As a control, CSF extracts were mixed with either buffer alone or the R36S version of Nt-B(wt) [Nt-B(R36S)], which is not recognized by the cyclin B destruction system (Glotzer et al. 1991). In contrast to control extracts, when calcium was added to CSF extracts containing Nt-B(wt), cyclin B2 degradation was prevented and the H1 kinase activity, an indicator of the association between Cdc2 with cyclin B2, remained high (Fig. 3A). In fact, Suc1 precipitations confirmed that cyclin B2 remained bound to Cdc2 under these conditions (Fig. 3A, middle).
Figure 3.
Dissociation of Cdc2 from cyclin B requires destruction box of cyclin B. (A) Prevention of dissociation of cyclin B–Cdc2 by the N-terminal fragment of cyclin B1 (Nt-B). CSF extracts were activated with calcium addition in the absence (buffer) or presence of either Nt-B(wt) or Nt-B (R36S), which is not recognized by the cyclin B destruction system. Samples taken at the indicated times were assayed for H1 kinase activity, recovered by Suc1–Sepharose, or mixed directly with Laemmli's sample buffer. After SDS-PAGE, samples were immunoblotted with anti-Xenopus cyclin B2 antibody. (B) A functional destruction box is required for dissociation of cyclin B–Cdc2. 35S-Labeled cyclin B derivatives were produced as hcyclin B1/ΔN86 (ΔN86) in which residues 1–86 were deleted from hcyclin B1, and hcyclin B1-Dm (Dm) in which the invariant Arg and Leu residues in the destruction box of human cyclin B1 were mutated to Ala. These proteins, in a complex with Myc-tagged Cdc2, were added to CSF extracts in the absence or presence of MG115. Samples taken at the indicated times were mixed directly with Laemmli's sample buffer or immunoprecipitated with anti-Myc antibody. These samples were analyzed by autoradiography after SDS-PAGE.
In other experiments, 35S-labeled human cyclin B1 (hcyclin B1) derivatives lacking a functional destruction box were synthesized in reticulocyte lysates: hcyclin B1–ΔN86 (ΔN86) in which residues 1–86 were deleted from hcyclin B1, and hcyclin B1-Dm (Dm) in which the invariant Arg and Leu residues in the destruction box of hcyclin B1 were mutated to Ala (Glotzer et al. 1991). These products were mixed with 35S-labeled Myc-tagged Cdc2 to prepare the cyclin B–Cdc2 complexes. CSF extracts were mixed with these complexes, and then with calcium either in the absence (Fig. 3B, top) or in the presence (Fig. 3B, middle and bottom) of MG115. Both hcyclin B1–ΔN86 and hcyclin B1-Dm were completely stable after the addition of calcium and were still associated with Myc–hCdc2. These observations indicate that the presence of a functional destruction box is essential for dissociation of the cyclin B–Cdc2 complex.
Cdc2 remains associated with ubiquitinated cyclin B
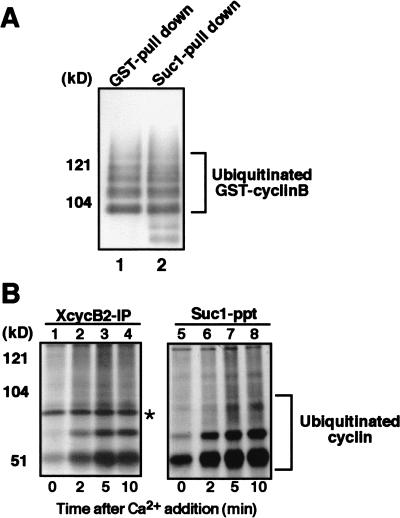
Because both the ubiquitination of cyclin B by the APC/C and the dissociation of the cyclin B–Cdc2 complex are dependent on a functional destruction box, the above results suggest that ubiquitination of cyclin B may be required for dissociation of the cyclin B–Cdc2 complex. To examine whether ubiquitination of cyclin B is sufficient for the dissociation, we performed in vitro ubiquitination of cyclin B that is associated with Cdc2 (see Kotani et al. 1999). To prepare a substrate for ubiquitination, Sf9 cells were infected with baculoviruses encoding mouse GST–cyclin B1 and Cdc2, and the GST–cyclin B1–Cdc2 complex was recovered from cell lysates by glutathione–Sepharose. As an APC/C fraction, anti-Cdc27 immunoprecipitates were recovered from Xenopus egg extracts that were arrested at anaphase by the addition of both calcium and the nondegradable cyclin B fragment to CSF extracts (see Glotzer et al. 1991). Then, the GST–cyclin B1–Cdc2 complex was ubiquitinated in vitro with biotinylated ubiquitin, bacterially expressed recombinant mouse E1 and UBC10 (E2), and the anti-Cdc27 immunoprecipitates (APC/C). Ubiquitin conjugates were detected by the ECL avidin–peroxidase method. As shown in Figure 4A, similar amounts of highly ubiquitinated cyclin B1 were recovered by glutathione–Sepharose (lane 1) and by Suc1–Sepharose (lane 2), indicating that ubiquitinated cyclin B1 remains associated with Cdc2.
Figure 4.
Cdc2 retains its association with ubiquitinated cyclin B. (A) Cdc2 is in a complex with cyclin B that has been polyubiquitinated in vitro. Purified GST-cyclin B–Cdc2 complex was incubated with biotinylated ubiquitin, E1, E2 (hE2-C), and, as a source of APC/C, anti-Cdc27 immunoprecipitates from Xenopus egg extracts that were arrested at anaphase by the addition of calcium together with nondegradable cyclin B fragment to CSF extracts. Then, GST–cyclin B was pulled down by glutathione–Sepharose 4B, resolved by SDS-PAGE and transferred to PVDF membrane. The membrane was reacted with ExtraAvidin peroxidase at room temperature for 1 h. Ubiquitinated cyclin B was visualized by ECL (lane 1). Alternatively, Cdc2 was recovered by Suc1–Sepharose, followed by ECL detection of ubiquitination (lane 2). (B) Cdc2 is complexed with cyclin B that is ubiquitinated in Xenopus egg extracts. Biotinylated ubiquitin was added to CSF extracts and, then, cyclin B degradation was induced by the addition of calcium. Samples taken at the indicated times were subjected to immunoprecipitation for Xenopus cyclin B2 or recovery of Cdc2 by Suc1–Sepharose beads. Ubiquitinated cyclins were visualized as described in A. (Asterisk) Non-specific band.
Similar results were obtained using Xenopus egg CSF extracts. When biotinylated ubiquitin was added to CSF extracts to detect ubiquitination of cyclin B, we found low levels of ubiquitination on cyclin B even in extracts arrested at metaphase II (Fig. 4B, lanes 1,5). After the release of the arrest by addition of calcium and until the completion of cyclin B degradation, a large increase in the level of ubiquitinated cyclin B was detectable by the biotin–avidin system (Fig. 4B, lanes 2–4 and 6–8) and at lesser extent by anti-cyclin B antibody (data not shown). These ubiquitinated forms of cyclin B were recovered in Suc1 precipitates as well as in anti-cyclin B2 immunoprecipitates (Fig. 4B), indicating that Cdc2 remains associated with ubiquitinated cyclin B. Although it has been reported that Suc1 can bind to the proteasome (Kaiser et al. 1999), this is not the case in Xenopus egg extracts (data not shown), ruling out a possibility that the Suc1 association of cyclin B is via the proteasome. These results indicate that while ubiquitination of cyclin B might be necessary for the dissociation of the cyclin B–Cdc2 complex, it is not sufficient.
Cyclin B tethered by the 26S proteasome is not associated with Cdc2
The foregoing results suggest that the dissociation of the cyclin B–Cdc2 complex might occur after polyubiquitination of cyclin B but before its degradation. We examined the state of cyclin B that had been dissociated from Cdc2 but whose degradation was prevented by MG115 (see Fig. 1C). CSF extracts were treated with calcium in the presence of MG115 to dissociate the cyclin B–Cdc2 complex (MG115-treated extracts), and then the MG115-treated extracts were fractionated by gel filtration on Superose 6. Much of the cyclin B2 was recovered in fractions where complexes of several hundred kilodaltons eluted and were distinct from the fractions containing the majority of Cdc2 (Fig. 5A, bottom). In contrast, both cyclin B and a fraction of Cdc2 co-eluted around 100 kD when control, untreated CSF extracts (MII extracts) were applied to the same column (Fig. 5A, top). A comparison of these gel filtration patterns suggests that cyclin B in MG115-treated extracts is associated with some macromolecular components other than Cdc2. Then, high molecular weight fractions containing cyclin B2, but not Cdc2, were pooled and fractionated on an anion-exchange Mono Q column. Each fraction was assayed for 26S proteasome activity and by immunoblots with antibodies against 20S and 19S (MSS1 and p58) components of the proteasome and against Cdc27, a key component of APC/C. As shown in Figure 5B, a significant amount of cyclin B2 co-eluted with the 26S proteasome activity but not with Cdc27. Both the 20S and the 19S components of the proteasome were detected in fractions containing the 26S proteasome activity, indicating that the 26S proteasome is actually present in these fractions and that the 26S proteasome is possibly associated with cyclin B2. At present we do not know the significance of the cyclin B2 that did not co-elute with the 26S proteasome.
Further evidence for the interaction between the 26S proteasome and cyclin B was obtained by immunoprecipitation of the 20S proteasome from MG115-treated extracts or MII extracts. Immunoprecipitates recovered with an anti-20S proteasome antibody were examined by immunoblotting with anti-cyclin B2 antibody. S5a, a component of the 19S regulatory complex, co-precipitated with the 20S proteasome with an approximately equal stoichiometry from both MII and MG115-treated extracts (Fig. 5C, bottom). This result implies that the 19S regulatory component is almost equally associated with the 20S proteasome to form the 26S complex in both MII and MG115-treated extracts and that immunoprecipitation with the anti-20S proteasome antibody is able to recover the 26S proteasome (see also Fig. 6A). As shown in Figure 5C, anti-20S proteasome immunoprecipitates from MG115-treated extracts contained a significant amount of cyclin B2 but not Cdc2, while those from MII extracts contained neither cyclin B2 nor Cdc2. Taken together, the results in Figure 5B and C support the concept that cyclin B is not associated with Cdc2 in MG115-treated extracts, but is associated with the 26S proteasome. The association of cyclin B with the 26S proteasome may explain why cyclin B could not be immunoprecipitated from MG115-treated extracts by anti-cyclin B antibodies.
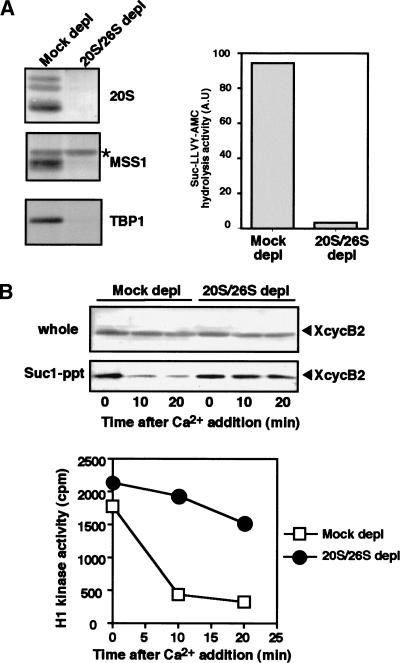
Figure 6.
Dissociation of cyclin B–Cdc2 requires the proteasome. (A) Immunodepletion of the 26S and 20S proteasomes from Xenopus egg extracts. CSF extracts (100 μL) were immunodepleted with anti-Xenopus 20S proteasome or with control rabbit IgG, and then tested for the peptidase activity (right) against Suc-LLVY-AMC in the presence of SDS, which fully activates the 20S peptidase activity. Remaining proteins after the immunodepletion were assayed by immunoblots (left) for components of the 20S proteasome, MSS1 and TBP1 (both for the 19S particle). Note that immunodepletion of the 20S proteasome also removed the 26S proteasome from CSF extracts. (Asterisk) Non-specific band. (B) Dissociation of cyclin B–Cdc2 is prevented in the proteasome-depleted CSF extracts. The 20S/26S proteasome-depleted or mock-depleted CSF extracts were treated with calcium in the presence of 0.5 mM MG115. Samples taken at the indicated times were mixed directly with Laemmli's sample buffer or recovered by Suc1–Sepharose beads for immunoblots with anti-cyclin B2 antibody (top, middle). Separate samples were processed for assay of histone H1 kinase activity (bottom). (Closed circles) 20S/26S proteasome-depleted extracts; (Open squares) mock depleted.
The 26S proteasome is necessary for dissociation of cyclin B–Cdc2
All of the foregoing results prompted us to examine a possible involvement of the proteasome in the dissociation of the cyclin B–Cdc2 complex. To address this question, the proteasome was immunodepleted from CSF extracts using anti-20S proteasome antibodies. When compared with the extracts that were mock depleted with a control IgG, those depleted with anti-20S proteasome antibodies had <5% of the 20S proteasome and of the 19S particle, identified by antibodies against MSS1 and TBP1, remaining (Fig. 6A, left). In agreement with the protein levels, the immunodepleted extracts lost >98% of the 26S proteasome activity (Fig. 6A, right). Therefore, because of the shared proteins between the 26S and 20S proteasomes, the anti-20S proteasome immunodepletion from Xenopus egg extracts was able to remove most of the 26S proteasomes.
When calcium was added the 20S/26S proteasome-depleted CSF extracts in the presence of MG115, no significant decrease was observed in cyclin B2 levels of whole extracts and in Suc1 precipitates, or in the histone H1 kinase activity (Fig. 6B), indicating that Cdc2 was still associated with cyclin B2. These results were in marked contrast with those in mock-depleted extracts where an obvious decrease occurred both in cyclin B2 levels of Suc1 precipitates and in histone H1 kinase activity, as already seen in Figure 1B and C. Thus the 26S proteasome seems to be required for the dissociation of Cdc2 from cyclin B.
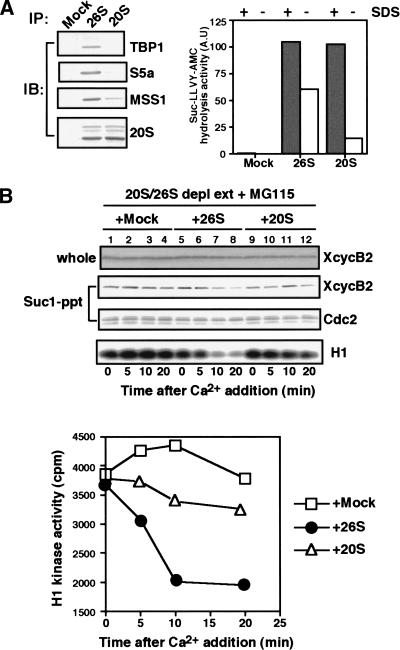
Next, we carried out restoration experiments by adding back the 26S or 20S proteasomes to the 20S/26S proteasome-depleted extracts. The 26S proteasomes were recovered from CSF extracts by anti-20S proteasome antibody-coupled beads, while the 20S proteasomes were recovered by the same beads but from CSF extracts that had been incubated with hexokinase and glucose to deplete ATP and, thereby, convert the 26S proteasomes into 20S proteasomes (see Materials and Methods). In fact, the 26S proteasome precipitates were recognized on immunoblots by the antibodies against the 20S proteasome and several subunits (S5a, TBP1, and MSS1) of the 19S regulatory particle (Fig. 7A). Moreover, they exhibited high levels of peptidase activity both in the presence and in the absence of SDS (Fig. 7B). In contrast, the 20S proteasome precipitates contained approximately the same levels of the proteins common to 26S and 20S proteasomes but significantly reduced levels of proteins from the 19S particle which is only present on 26S proteasomes (Fig. 7A). The 20S proteasomes also lost about 80% of their SDS-insensitive (but not SDS-sensitive) peptidase activity (Fig. 7B) when compared with the 26S proteasomes. The 26S proteasome beads were added back to 20S/26S proteasome-depleted CSF extracts, and then calcium was added in the presence of MG115. In this reconstitution experiment, both the dissociation of Cdc2 from cyclin B and the decrease in histone H1 kinase activity were again observed (Fig. 7B, top, lanes 5–8 and bottom), indicating that the 26S proteasomes restored the dissociation capacity to the depleted extracts. In contrast, the addition of control mock beads (Fig. 7B, top, lanes 1–4) or of the 20S proteasome beads failed to restore the capacity to dissociate Cdc2 from cyclin B, and did not inactivate the histone H1 kinase of the depleted, MG115-treated extracts (Fig. 7B, top, lanes 9–12). Although the effect of the addition of the 19S particle to the 20S/26S proteasome-depleted extracts would be intriguing, this could not be done because of the inability of obtaining the 19S particle at the high concentration found in CSF extracts. Nevertheless, our results demonstrate clearly that 26S proteasomes, and not 20S proteasomes, are likely to be involved in the dissociation of the cyclin B–Cdc2 complex during its inactivation on exit from M phase.
Figure 7.
Dissociation of cyclin B–Cdc2 in proteasome-depleted extracts is restored by the addition of immunopurified 26S, but not 20S, proteasomes. (A) Immunoprecipitates of the 26S or 20S proteasomes. For the 26S proteasome immunoprecipitates, CSF extracts were diluted in buffer A containing 2 mM ATP, incubated for 60 min at 37°C in the same buffer supplemented with an ATP-regenerating system and then subjected to immunoprecipitation with anti-20S proteasome antibody. The 20S proteasomes were immunoprecipitated from CSF extracts that were diluted in buffer A without ATP and then incubated for 60 min at 37°C with an ATP-depleting system (see Materials and Methods). The 26S or 20S proteasome immunoprecipitates were assayed by proteasomal peptidase activity against Suc-LLVY-AMC in the presence or absence of 0.05% SDS (right). Alternatively, the same immunoprecipitates were assayed by immunoblots for the presence of TBP1, S5a, and MSS1 (components of the 19S regulatory particle), and components of the 20S proteasome. (B) Addition of the 26S proteasome to the proteasome-depleted extracts restores dissociation of cyclin B–Cdc2. The 20S/26S proteasome-depleted extracts (see Fig. 6A) were mixed with the 26S proteasome immunoprecipitates (●), the 20S proteasome immunoprecipitates (▵), or control beads (□), and then activated by 0.6 mM calcium in the presence of MG115. Samples were taken at indicated times and analyzed as described in Fig. 6B.
Discussion
By using Xenopus egg extracts that reproduce exit from M phase, the present study demonstrates that Cdc2 dissociates from cyclin B in the absence of cyclin B destruction, and that the dissociation depends on the presence of the 26S proteasome but not on its proteolytic activity. These observations imply that the 26S proteasome is involved in not only the destruction of cyclin B but also in the release of Cdc2 from cyclin B by an activity other than proteolysis. These results may explain why Cdc2 escapes from degradation upon destruction of cyclin B at exit from M phase, even though it is in a tight complex with cyclin B prior to the exit from M phase.
Merit of Xenopus egg extracts system
Although the present results were obtained in Xenopus egg extracts, dissociation of Cdc2 from cyclin B and loss of H1 kinase activity were also observed in starfish oocytes that had been treated with MG115 at exit from metaphase of meiosis I (data not shown). These observations are in marked contrast with previous reports that the proteasome inhibitor causes metaphase arrest accompanied by elevated levels of H1 kinase activity (i.e., no dissociation of cyclin B–Cdc2) in mammalian tissue culture cells and in plant cells (Sherwood et al. 1993; Genschik et al. 1998).
How could the discrepancy between our results and those in somatic cells be explained? It has been well established that there is a dual control for mitotic exit, sister chromatid separation and Cdc2 inactivation, and that both are under the control of the APC/C–proteasome system (for review, see Townsley and Ruderman 1998; Zachariae and Nasmyth 1999). While securin (Pds1/Cut2) binds and prevents separin (Esp1/Cut1) from destroying sister chromatid cohesion, thereby maintaining metaphase (for review, see Nasmyth et al. 2000), the destruction of securin by the APC/C–proteasome in a Cdc20/Fizzy-dependent manner liberates separin, permitting sister chromatid separation. In budding yeast, at least, securin is also involved in preventing the activation of Cdc14, which activates the destruction of cyclin B by the APC/C–proteasome through the dephosphorylation of Cdh1/Hct1, thus coupling sister chromatid separation with Cdc2 inactivation (Cohen-Fix and Koshland 1999; Shirayama et al. 1999; Tinker-Kulberg and Morgan 1999; for review, see Prinz and Amon 1999). However, even if the above coupling system functions normally, one might anticipate that the inhibition of proteolytic activity of the proteasome could still liberate separin from securin, much like Cdc2 from cyclin B in the present study, resulting in the dissociation of sister chromatids, and could also destroy the inhibitory effect of securin on Cdc14, resulting in the inactivation of Cdc2. If so, then the metaphase arrest caused by the proteasome inhibitor in somatic cells might suggest the presence of another proteolysis-dependent system that couples the pathway for dissociation of sister chromatid cohesion with that for destruction of cyclin B.
In contrast, securin destruction is not required for progression of the embryonic cell cycle in Xenopus egg extracts but is necessary for sister chromatid separation (Zou et al. 1999), implying that securin degradation and cyclin B destruction are independent. The independence of these two events might constitute the molecular basis for the lack of spindle assembly checkpoint control in Xenopus egg extracts and starfish oocytes (for review, see Murray and Kirschner 1989). Accordingly, even in the absence of the proteasome's proteolytic activity, the pathway toward cyclin B destruction should be able to proceed separately in Xenopus egg extracts, resulting in dissociation of Cdc2 from cyclin B. It is likely that the absence of a spindle assembly checkpoint control in Xenopus egg extracts contributed to our ability to obtain the results we have presented here.
Dissociation of Cdc2 depends on cyclin B ubiquitination rather than Thr 161 dephosphorylation of Cdc2
Because previous studies demonstrated that the phosphorylation of Thr 161 on Cdc2 increases the stability of the cyclin B–Cdc2 complex (for review, see Morgan 1995), one might have anticipated that the dephosphorylation of Thr 161 might contribute to its dissociation. However, contrary to this premise, our results suggest a possibility that dephosphorylation on Thr 161 is not necessary for dissociation of the cyclin B–Cdc2 complex at exit from M phase (Fig. 2). This possibility is consistent with the previous report that, in CSF extracts, okadaic acid prevents both the dephosphorylation of Cdc2 and the drop in H1 kinase activity, but not cyclin B degradation (Lorca et al. 1992). The fact that simply reversing the step that stabilizes the cyclin B–Cdc2 complex does not cause dissociation of the complex once it has formed, provides support to the notion that a positive-acting mechanism releases the two subunits at the end of M phase.
The present results indicate that a functional destruction box is required for dissociation of cyclin B–Cdc2 (Fig. 3). Considering that the destruction box in cyclin B is known to be involved in polyubiquitination of cyclin B (Glotzer et al. 1991), one could postulate that polyubiquitination of cyclin B itself might cause the release of Cdc2 from cyclin B. In accordance with this notion, it has been suggested that the polyubiquitin chain helps to unfold the target proteins (Pickart 1997). However, in the present study polyubiquitination is not by itself sufficient to dissociate Cdc2 from cyclin B (Fig. 4). We do not know to what extent cyclin B is unfolded as a result of ubiquitination. Then, what is the role of the destruction box in the dissociation of cyclin B–Cdc2? Although we cannot exclude the possibility that the destruction box contributes to a function other than polyubiquitination of cyclin B, it is reasonable that polyubiquitination of cyclin B is a way for targeting cyclin B–Cdc2 to the dissociation because the proteasome was required for this dissociation in the present study (Fig. 6) and because a multi-ubiquitin chain is a well-known targeting signal to the proteasome (Thrower et al. 2000). Thus, polyubiquitination of cyclin B is likely to target the complex to the proteasome where Cdc2 is released from cyclin B. Whether or not ubiquitination also plays a role in facilitating the disassembly of the complex by the proteasome remains to be resolved.
Release of Cdc2 from cyclin B by the proteasome
Dissociation of the cyclin B–Cdc2 complex required the 26S proteasome (Figs. 6 and 7) but not its proteolytic activity, indicating that a non-proteolytic function of the proteasome contributes the dissociation. On the other hand, in the presence of an excess amount of recombinant human S5a, which is a multi-ubiquitin chain recognition component of the 26S proteasome (Deveraux et al. 1994), neither cyclin B destruction nor inactivation of Cdc2 kinase occurred in CSF extracts to which calcium had been added (Deveraux et al. 1995; A. Nishiyama, unpubl.). This result implies that the recognition of polyubiquitinated cyclin B by S5a component of the proteasome is not sufficient to dissociate Cdc2 from cyclin B, and exogenously added S5a may actually compete with proteasome-bound S5a for the recognition of polyubiquitinated cyclin B.
Our results show that the ability to dissociate the cyclin B–Cdc2 complex is dependent on the 26S, but not the 20S, proteasome (Fig. 7). This dependence implies that at least the 19S (PA700) regulatory particle may play a key role in the dissociation of the complex. In fact, the 19S particle carries out a nonproteolytic function in nucleotide excision repair (Russell et al. 1999). However, we suspect that the 19S particle alone may not be sufficient for dissociation of cyclin B–Cdc2. Ornithine decarboxylase (ODC), which is degraded by the 26S proteasome without ubiquitination (Murakami et al. 1992), is sequestered by the proteasome, in a process requiring ATP but not the proteolytic activity of the proteasome (Murakami et al. 1999). The 26S complex, but neither the 20S core particle nor the 19S regulatory particle alone, are sufficient for the sequestration of ODC. If the dissociation of Cdc2 from cyclin B occurs in a similar manner, it is likely that the whole 26S complex, but not the 19S regulatory particle alone, might accomplish the dissociation. To support this notion, we have attempted in vitro reconstitution experiments using in vitro polyubiquitinated cyclin B–Cdc2 complex and the purified 26S proteasomes or the 20S or 19S components of the proteasome, but, at present, have been unsuccessful in dissociating the complex, possibly because of the low concentration of multi-ubiquitinated cyclin B.
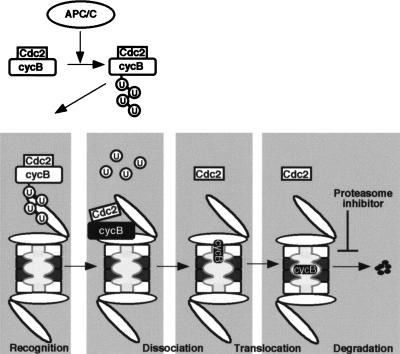
Then, how does the proteasome dissociate Cdc2 from cyclin B? The 19S regulatory particle is composed of two subcomplexes, the “base” and the “lid” (Glickman et al. 1998). The base is located proximal and the lid distal to the 20S core particle. The lid is essential for the recognition, and possibly binding, of polyubiquitinated substrate proteins, whereas the base is likely to promote substrate unfolding through its six distinct AAA (ATPases associated with a variety of cellular activities)-type ATPase components in a chaperone-like manner that is independent of polyubiquitin chains (Braun et al. 1999). In addition, the 20S proteolytic core is composed of two α- and two β-rings (Baumeister et al. 1998). The α-rings discriminate between unfolded and folded proteins, and the β-rings, which constitute the central catalytic core, can cleave substrates proteolytically. On the basis of the above and the results presented in this paper, we propose the following model for degradation of cyclin B (Fig. 8): Initially, polyubiquitin chains of cyclin B that have been added by the APC/C tether the cyclin B–Cdc2 complex to the lid of the 19S particle. Then, because of a chaperone-like function of the base of the 19S particle with the aid of the α-rings in the 20S component, the tethered cyclin B–Cdc2 complex is unfolded. Consequently, Cdc2 dissociates from cyclin B, while cyclin B is translocated into the catalytic core of the 20S component. Lastly, cyclin B is degraded within the lumen of the 20S component by the proteolytic activity of the β-rings. At present, however, we can not discriminate whether Cdc2 is passively separated from cyclin B as a consequence of cyclin B unfolding or whether Cdc2 is actively dissociated from the cyclin B–Cdc2 complex by a chaperone-like activity of the proteasome components.
Figure 8.
A model for release of Cdc2 from cyclin B by the 26S proteasome. First, cyclin B complexed with Cdc2 is polyubiquitinated by an APC/C-dependent pathway at exit from M phase. Second, the lid of the 19S regulatory particle recognizes and tethers polyubiquitinated cyclin B that remains associated with Cdc2. Third, the cyclin B–Cdc2 complex is unfolded and dissociated by the chaperone-like activity of the base of the 19S regulatory particle with the aid of the α ring of the 20S proteasome. The translocation of unfolded cyclin B to the 20S proteasome may be required for further unfolding of cyclin B, and both processes may be coupled to each other. Finally, the unfolded cyclin B is translocated and degraded in the hollow center (β rings of the 20S proteasome) in which all catalytic sites are located. The inhibitory step by MG115 in the present study is indicated by the proteasome inhibitor.
In a strict sense, our results showing that the immunodepletion of the 26S proteasome abolished the dissociation of cyclin B–Cdc2 and that the immunoprecipitates restored the dissociation (Figs. 6 and 7) imply that the 26S proteasome itself and/or any component(s) associated with the 26S proteasome is required for the dissociation. A proteasome-associated component that could possibly contribute to the dissociation is Cdc48, also named as valosin-containing protein (VCP), which is an AAA-type ATPase family member (for review, see Confalonieri and Duguet 1995). When the transcription factor NF-κB is activated, IκBα which is complexed with NF-κB and prevents its nuclear translocation, is destroyed in a ubiquitin–proteasome-dependent manner (for review, see Ghosh et al. 1998; Maniatis 1999). In this case, it has been proposed that before the degradation of IκBα, Cdc48 is involved in the dissociation of polyubiquitinated IκBα from NF-κB (Dai et al., 1998). However, it is likely that some additional component, possibly the proteasome itself, may be necessary for the Cdc48 function, because Cdc48 itself may be unable to recognize the polyubiquitin chains because it lacks the ubiquitin-binding motif contained in S5a (Young et al. 1998). On the other hand, a recent report of Thrower et al. (2000) indicates that the addition of Cdc48 does not contribute to the unfolding activity of the proteasome. Further studies will be required to clarify the role, if any, of Cdc48 in the dissociation of cyclin B–Cdc2.
In conclusion, the present study demonstrates that upon exit from M phase, the proteasome, which will degrade polyubiquitinated cyclin B, separates Cdc2 from cyclin B by a mechanism that is not dependent on the proteasome's proteolytic activity. This mechanism ensures that the regulatory component of the complex is degraded while the catalytic subunit is not, and accounts, in part, for the oscillations in cyclin B and the nonoscillations in Cdc2 abundance during the cell cycle. Paradoxically, the proteasome appears to be responsible for both the degradation of cyclin B and the stability of Cdc2.
Materials and methods
Preparation of Xenopus egg extracts
CSF-arrested extracts of Xenopus eggs were prepared according to the method of Murray (1991) with some modifications. Unfertilized eggs were dejellied with 2.5% thioglycolic acid–NaOH (pH 8.2), washed with EGTA-extraction buffer (100 mM KCl, 5 mM MgCl2, 0.5 mM CaCl2, 5 mM EGTA, 20 mM HEPES–KOH at pH 7.5) containing 50 μg/mL cytochalasin B at 15°C and transferred to a microcentrifuge tube. Eggs were packed by brief centrifugation (500g, 10 sec) and after the excess buffer was removed, the eggs were lysed by centrifugation at 15,000g for 10 min at 4°C. The cytoplasmic fraction between the lipid cap and sedimented yolk was recovered and clarified by a second centrifugation at 15,000g for 20 min at 4°C. Egg extracts were supplemented with ATP and creatine phosphate to final concentrations of 1 mM and 10 mM, respectively, preserved on ice and used within 2 h after preparation. CSF extracts were activated by the addition of CaCl2 to a final concentration of 0.6 mM in the presence or absence of 0.5 mM MG115. All incubations of egg extracts were done at 22°C.
Immunodepletion and immunoprecipitation of proteasome from egg extracts
For immunodepletion of 20S/26S proteasomes, 100-μL samples of CSF extracts were treated four times (15 min each, on ice) with 20 μL of protein G–Sepharose beads (Sigma) conjugated with anti-Xenopus 20S proteasome antibodies. For immunoprecipitation of 20S/26S proteasome, 100-μL samples of CSF extracts were diluted with 1.9 mL of either buffer A (20 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 1 mM DTT) supplemented with 2 mM ATP and an ATP-regenerating system (10 mM creatine phosphate and 10 μg/mL creatine kinase) or buffer A supplemented with an ATP-depleting system (10 mM glucose and 1 μg/mL hexokinase) and incubated for 60 min at 37°C. Next, the diluted extracts were treated for 1 h on ice with 6 μg of anti-Xenopus 20S proteasome antibodies conjugated to 10 μl of protein G beads. Protein G beads were then washed three times with buffer A containing 0.15 M NaCl and 0.05% Tween-20, and after equilibration with EB (100 mM KCl, 0.1 mM CaCl2, 5 mM MgCl2, 20 mM HEPES–KOH at pH 7.5) supplemented with 2 mM ATP, added back to the proteasome-depleted extracts.
Fractionation of egg extracts
The cytosolic fraction of egg extracts was obtained by centrifugation at 150,000g for 90 min at 4°C and a 200-μL sample was loaded on a Superose 6 column (HR 10/30) equilibrated with EGTA-extraction buffer supplemented with 1 mM ATP, 1 mM DTT, and 200 mM sucrose. Calibration of the column was performed using thyroglobulin (670 kD), ferritin (440 kD), catalase (230 kD) and ovalbumin (43 kDa) as molecular weight markers. High molecular weight fractions that contained cyclin B, but not Cdc2, were pooled and applied to a Mono Q column (HR 5/5) equilibrated with buffer Q (20 mM Tris-HCl at pH 7.7, 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 200 mM sucrose, 1 mM ATP, 1 mM DTT), washed with buffer Q and eluted with 15 mL of a linear salt gradient (100–500 mM KCl in buffer Q).
Isolation of cyclin B–Cdc2 complexes from egg extracts
Recombinant Suc1 (p13suc1) was expressed in E. coli, purified, and coupled to CNBr-activated Sepharose-4B (Pharmacia), as described (Okumura et al., 1996). To isolate cyclin B–Cdc2 complexes, 5 μL of egg extracts diluted to a total volume of 250 μL with β-GPEB (80 mM β-glycerophosphate, 20 mM EGTA, 5 mM MgCl2 and 20 mM HEPES–KOH at pH 7.5) containing 0.02% NP-40 was added to 10 μL Suc1-beads and incubated for 2 h at 4°C. Suc1–beads were washed sequentially with 500 μL of β-GPEB containing 0.02% NP-40, β-GPEB containing 0.5 M of NaCl and 500 μL of β-GPEB containing 0.02% NP-40.
Preparation of 35S-labeled cyclin B–Cdc2 complexes
Myc-tagged human Cdc2 was constructed by PCR, using a primer with an EcoRI site upstream of the desired start of the Cdc2 protein, and a primer with an XhoI site downstream of a stop codon of Cdc2. The EcoRI–XhoI fragment of Cdc2 was inserted into the pCITE expression vector (Novagen) with a Myc-epitope. A mutant version of human Cdc2 (T161E) was generated using a site-directed mutagenesis system (Mutan K; Takara, Japan), with the oligonucleotide, 5′-GTTCGGGTTTACGAACATGAGGTAGTGACA-3′.
cDNA encoding full-length human cyclin B1 was cloned into the pCITE expression vector. Arginine and leucine in the destruction box of the wild type were changed into alanine (cyclin B1-Dm) with an oligonucleotide, 5′-CGGACTGAGGCCAGCAACAGCTGCTGGGGACATTGG-3′ as described above. Cyclin B1ΔN86 was constructed by PCR with a primer with an NcoI site upstream of the desired start of the protein and a primer with a BamHI site downstream of a stop codon of cyclin B1. The NcoI–BamHI fragment of cyclin B1 was cloned into the pCITE vector.
Messenger RNAs encoding full-length human cyclin B1, cyclin B1ΔN86, cyclin B1 (Dm), Myc–human Cdc2 and Cdc2 (T161E) were transcribed in vitro from the pCITE vectors with T7 RNA polymerase. After removal of the DNA template by digestion with RNase-free DNase, mRNAs were extracted twice each with phenol and chloroform, recovered by ethanol precipitation, and resuspended in H2O at a concentration of 1 mg/mL. To increase the efficiency of translation, 2 μL of RNA was heated for 30 sec at 67°C before in vitro translation in 25 μL of rabbit reticulocyte lysate containing 1 mCi/mL [35S]methionine (Amersham).
To obtain 35S-labeled cyclin B1–Cdc2 complexes, reticulocyte lysates containing cyclin B1 (4.5 μL) and Myc–Cdc2 (1.5 μL) were added to Suc1-treated starfish oocyte extracts (3 μL) supplemented with 5 mM MgCl2 and 1mM ATP and incubated for 60 min at 25°C. The mixture was aliquotted, frozen in liquid nitrogen, and stored at −80°C. Starfish oocyte extracts were prepared as described (Fesquet et al. 1993) and treated twice with Suc1–Sepharose beads (Okumura et al. 1996) to increase the binding efficiency of exogenously added cyclin B1 and Myc–Cdc2. To isolate the complexes of 35S-labeled Myc–Cdc2 and 35S-labeled cyclin B1 from starfish oocyte extracts or Xenopus egg extracts to which the starfish extracts had been added, extracts containing the 35S-labeled proteins were diluted with 3 volumes of EB and treated with c-Myc (9E10) or human cyclin B1 antibodies (Santa Cruz). The mixture was diluted further with 3 volumes of EB and added to protein G–Sepharose beads. After rotation for 1 h at 4°C, protein G–Sepharose beads were washed once in low salt buffer (100 mM NaCl, 5 mM EDTA, 1% Triton X-100, 20 mM Tris-HCl at pH 7.4), twice in high salt buffer (1 M NaCl, 5 mM EDTA, 1% Triton X-100, 20 mM Tris-HCl at pH 7.4) and again once in low salt buffer. The associated proteins were eluted with SDS sample buffer. Samples were subjected to SDS-PAGE and analyzed by an phosphorimage analyzer (Fujix BAS2000; Fuji Photo Film).
Ubiquitination of cyclin B in complex with Cdc2
Recombinant mouse E1, human E2-C, and biotinylated bovine ubiquitin were prepared essentially as described previously (Funabiki et al. 1997). The APC/C was isolated using rabbit anti-human Cdc27 antibodies from CSF extracts that had been activated by the addition of CaCl2 in the presence of a nondegradable fragment of Xenopus cyclin B2 (ΔN85), which maintained a high level of Cdc2 activity after degradation of the endogenous cyclin B. As a substrate for ubiquitination, mouse GST–cyclin B1 and Cdc2 were expressed in a baculovirus system and their complexes were purified using glutathione–Sepharose 4B (Pharmacia). To perform in vitro ubiquitination of substrates, a 50-μL reaction mix (50 mM Tris-HCl at pH 7.4, 5 mM MgCl2, 2 mM DTT, 2 mM ATP), containing 1 μg of mouse E1, 3 μg of human E2-C, APC/C immunoprecipitated from 50 μL of CSF extracts by the use of 7.5 μL of anti-human Cdc27-coupled protein A beads (Bio-Rad), 15 μg of biotinylated bovine ubiquitin and 2 μg of mouse GST–cyclin B1–Cdc2, were incubated for 30 min at 25°C. Substrates were recovered by glutathione–Sepharose 4B or Suc1–Sepharose and washed three times. The samples were subjected to 7.5% SDS-PAGE, and the ubiquitinated GST–cyclin B were detected by the ECL avidin–peroxidase method (Amersham).
Recombinant N-terminal fragments of cyclin B
A peptide containing the N-terminal amino acids (1–108) of Xenopus cyclin B1 was fused to the polyhistidine tag in the pTrcHis plasmid vector (Invitrogen). Arginine in the destruction box of the wild type was changed into serine (R36S) by using the QuickChange site-directed mutagenesis kit (Stratagene) with synthetic primers, 5′-CCAGGGTTGAGACCTAGTACTGCC TTGGGAGACATTG-3′, 5′-CAATGTCTCCCAAGGCAGTA CTAGGTCTCAACCCTGG-3′. The polyhistidine-fused proteins were expressed in Escherichia coli BL21, and affinity purified with nickel–agarose beads (His-Bind Resin, Novagen) according to the product manual.
Immunoblotting
Antibodies against TBP1, MSS1, p58, and the Xenopus 20S proteasome were prepared as previously described (Tanaka et al. 1988; Kominami et al. 1997; Tanahashi et al. 1998). Anti-Xenopus cyclin B1 and B2 antisera and anti-PSTAIR antibodies were kindly provided from Dr. J.L. Maller (University of Colorado) and Drs. M. Yamashita and Y. Nagahama (National Institute for Basic Biology, Japan), respectively. Anti-Cdc27 monoclonal antibody was purchased from MBL (TL-C40920). Anti-human S5a antibody was raised against full-length human S5a expressed in E. coli by the use of the pET system (Y. Nagai, unpubl.).
All the samples were run on SDS–polyacrylamide gels and blotted onto an Immobilon membrane (Millipore) according to Towbin et al. (1979). After blocking with 5% skimmed milk, the membrane was incubated with primary antibodies for 1 h at room temperature. The membrane was then incubated with alkaline phosphatase-conjugated or horseradish peroxidase-conjugated secondary antibodies. Reacted proteins were detected by a BCIP/NTB phosphatase substrate system (KPL) or ECL (Amersham ).
Assays for histone H1 kinase and proteasome activities
For histone H1 kinase assay, egg extracts were quickly frozen in liquid nitrogen and stored at −80°C. Frozen extracts were thawed by adding 9 volumes of ice-cold β-GPEB. Ten microliters of diluted extract was mixed with 20 μL of reaction buffer containing 80 mM β-glycerophosphate (pH 7.4), 20 mM MgCl2, 0.6 mM ATP, 30 μg/mL leupeptin, 30 μg/mL aprotinin, 0.6 mg/mL histone H1 and 1 mCi [γ-32P]ATP, and incubated for 30 min at 25°C. Reactions were stopped by the addition of SDS-sample buffer and boiling for 2 min. Histone H1 was separated by SDS-PAGE and stained with Coomassie blue. The band was excised and 32P incorporation was quantitated in the gel slice by the Cerenkov method. Alternatively, the gel was autoradiographed with X-ray film (X-OMAT, Kodak) at −80°C. For the proteasome activity assay, protein samples were examined for their ability to degrade Suc-LLVY-AMC in solution in the presence of 1 mM ATP or in the absence of 1mM ATP and 0.05% SDS, as described previously (Murakami et al. 1999).
Acknowledgments
We thank Drs. Mari Iwabuchi for cyclin B mutants, Sayaka Tahara for Myc-tagged constructs, James L. Maller and Yukiko Nagai for antibodies, Hiroyuki Kawahara for invaluable discussion and Manfred J. Lohka for critical reading of the manuscript. This study was supported by the scientific grants from Ministry of Education, Science and Culture, Japan and the CREST Research Project of Japan Science and Technology Corporation to T. K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tkishimo@bio.titech.ac.jp; FAX 81–45–924–5738.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.823200.
References
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: Inhibition of anaphase initiation and regulation of mitotic exit. Genes & Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri F, Duguet M. A 200-amino acid ATPase module in search of a basic function. BioEssays. 1995;17:639–650. doi: 10.1002/bies.950170710. [DOI] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Deveraux Q, van Nocker S, Mahaffey D, Vierstra R, Rechsteiner M. Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26 S protease subunit S5a. J Biol Chem. 1995;270:29660–29663. doi: 10.1074/jbc.270.50.29660. [DOI] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Fesquet D, Labbe JC, Derancourt J, Capony JP, Galas S, Girard F, Lorca T, Shuttleworth J, Doree M, Cavadore JC. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, Yanagida M. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. Cell cycle -dependent proteolysis in plants. Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell. 1998;10:2063–2076. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen LH, Luca FC, Ruderman JV, Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- Kaiser P, Moncollin V, Clarke DJ, Watson MH, Bertolaet BL, Reed SI, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes & Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Stewart E, Poon R, Adamczewski JP, Gannon J, Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Harper JW, Elledge SJ. How the cyclin became a cyclin: Regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- Kominami K, Okura N, Kawamura M, DeMartino GN, Slaughter CA, Shimbara N, Chung CH, Fujimuro M, Yokosawa H, Shimizu Y, et al. Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1–1. Mol Biol Cell. 1997;8:171–187. doi: 10.1091/mbc.8.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lees EM, Harlow E. Sequences within the conserved cyclin box of human cyclin A are sufficient for binding to and activation of cdc2 kinase. Mol Cell Biol. 1993;13:1194–1201. doi: 10.1128/mcb.13.2.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T, Labbe JC, Devault A, Fesquet D, Capony JP, Cavadore JC, Le Bouffant F, Doree M. Dephosphorylation of cdc2 on threonine 161 is required for cdc2 kinase inactivation and normal anaphase. EMBO J. 1992;11:2381–2390. doi: 10.1002/j.1460-2075.1992.tb05302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey D, Yoo Y, Rechsteiner M. Ubiquitin metabolism in cycling Xenopus egg extracts. J Biol Chem. 1993;268:21205–21211. [PubMed] [Google Scholar]
- Maniatis T. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes & Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- ————— Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1:E47–53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Hayashi SI, Tanahashi N, Tanaka K. ATP-dependent inactivation and sequestration of ornithine decarboxylase by the 26S proteasome are prerequisites for degradation. Mol Cell Biol. 1999;19:7216–7227. doi: 10.1128/mcb.19.10.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell-cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Dominoes and clocks: The union of two views of the cell cycle. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: Key regulators of the eukaryotic cell cycle. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Okumura E, Sekiai T, Hisanaga S, Tachibana K, Kishimoto T. Initial triggering of M phase in starfish oocytes: A possible novel component of maturation-promoting factor besides cdc2 kinase. J Cell Biol. 1996;132:125–135. doi: 10.1083/jcb.132.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Prinz S, Amon A. Dual control of mitotic exit. Nature. 1999;402:133–135. doi: 10.1038/45949. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. The 26S proteasome. In: Peters JM, Harris JR, Finley D, editors. Ubiquitin and the biology of the cell. New York, NY: Plenum Press; 1998. pp. 147–190. [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- Sherwood SW, Kung AL, Roitelman J, Simoni RD, Schimke RT. In vivo inhibition of cyclin B degradation and induction of cell-cycle arrest in mammalian cells by the neutral cysteine protease inhibitor N-acetylleucylleucylnorleucinal. Proc Natl Acad Sci. 1993;90:3353–3357. doi: 10.1073/pnas.90.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi N, Suzuki M, Fujiwara T, Takahashi E, Shimbara N, Chung CH, Tanaka K. Chromosomal localization and immunological analysis of a family of human 26S proteasomal ATPases. Biochem Biophys Res Commun. 1998;243:229–232. doi: 10.1006/bbrc.1997.7892. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yoshimura T, Kumatori A, Ichihara A, Ikai A, Nishigai M, Kameyama K, Takagi T. Proteasomes (multi-protease complexes) as 20 S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988;263:16209–16217. [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes & Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Ruderman JV. Proteolytic ratchets that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]