Abstract
CDC6 is conserved during evolution and is essential and limiting for the initiation of eukaryotic DNA replication. Human CDC6 activity is regulated by periodic transcription and CDK-regulated subcellular localization. Here, we show that, in addition to being absent from nonproliferating cells, CDC6 is targeted for ubiquitin-mediated proteolysis by the anaphase promoting complex (APC)/cyclosome in G1. A combination of point mutations in the destruction box and KEN-box motifs in CDC6 stabilizes the protein in G1 and in quiescent cells. Furthermore, APC, in association with CDH1, ubiquitinates CDC6 in vitro, and both APC and CDH1 are required and limiting for CDC6 proteolysis in vivo. Although a stable mutant of CDC6 is biologically active, overexpression of this mutant or wild-type CDC6 is not sufficient to induce multiple rounds of DNA replication in the same cell cycle. The APC–CDH1-dependent proteolysis of CDC6 in early G1 and in quiescent cells suggests that this process is part of a mechanism that ensures the timely licensing of replication origins during G1.
Keywords: APC, CDC6, cell cycle, DNA replication, proteolysis
Most of our knowledge concerning the molecular mechanisms responsible for the initiation of DNA replication in eukaryotes is based on genetic and biochemical data obtained in Saccharomyces cerevisiae. Genomic footprinting of S. cerevisiae autonomously replicating sequences (ARSs) has suggested a model for the occupation of origins of replication by protein complexes during the cell cycle (for review, see Diffley 1996; Stillman 1996; Dutta and Bell 1997). Prereplication complexes (preRCs) assemble at the ARS during G1 and render the DNA competent for replication. PreRCs persist throughout G1 and are not observed on origins in S phase. The firing of origins is most likely triggered by cyclin-dependent kinases (CDKs) and Cdc7p/Dbf4p kinases that lead to the conversion of preRCs to postRCs. The postRCs, which are present in S, G2, and M phase, are not competent for initiation of DNA replication. In vitro, the postRC features of the footprints can be reconstituted by a multisubunit complex termed the origin recognition complex (ORC; Bell and Stillman 1992).
ORC consists of six subunits (Orc1p–Orc6p) that associate with ARSs throughout the cell cycle (Aparicio et al. 1997; Liang and Stillman 1997). In addition to ORC, Cdc6p and Mcm proteins (Mcm2p–Mcm7p) are required for the formation of the preRC. These and their orthologs in Schizosaccharomyces pombe and Xenopus laevis are required for the initiation of DNA replication (for review, see Leatherwood 1998), suggesting a conserved mechanism of regulating DNA replication. Biochemical analyses of the ORC and the Mcms have shown that they contain ATPase motifs and that the Mcms have intrinsic DNA helicase activity (You et al. 1999).
CDC6 plays a key role in regulating the initiation of DNA replication. It is indispensable for the formation and maintenance of preRCs and for the association of Mcms with origins of replication (Leatherwood 1998). Cdc6p and its orthologs in S. pombe (Cdc18) and X. laevis (XlCDC6) are all essential for initiation of DNA replication (for review, see Diffley 1996; Dutta and Bell 1997). Cdc6p and Cdc18 are both unstable proteins and de novo synthesis of these two proteins in G1 is required for DNA replication (Diffley 1996). Like some Orc and Mcm proteins, Cdc6p and its orthologs contain an ATPase domain. Mutation of the ATPase domain indicates that binding and hydrolysis of ATP are necessary for the Cdc6 proteins to stimulate DNA replication (Perkins and Diffley 1998; Herbig et al. 1999; Weinreich et al. 1999).
Both Cdc6p and Cdc18 are CDK substrates, and phosphorylation at the G1/S boundary leads to ubiquitin-mediated proteolysis of the two proteins. The proteolysis is regulated by an SCF (Skp1, Cdc53/Cullin, F-box protein)–E3 ligase complex in both yeast species (Piatti et al. 1996; Drury et al. 1997; Jallepalli 1997; Kominami and Toda 1997). Interestingly, overexpression of Cdc18 is sufficient to induce re-replication (Kelly et al. 1993). The proteolysis of Cdc18 at the G1/S boundary appears to be an important mechanism for restricting origin usage to “once and only once” per cell cycle, as a stable mutant of Cdc18 with mutations in all putative CDK phosphorylation sites is more efficient than wild-type Cdc18 in inducing re-replication upon overexpression (Jallepalli et al. 1997). In contrast, wild-type Cdc6p or a stable Cdc6p mutant is not sufficient to induce re-replication in S. cerevisiae (Piatti et al. 1996; Drury et al. 1997). These data suggest that some aspects of regulation of DNA replication in the two yeast species differ significantly and that S. cerevisiae has developed other regulatory mechanisms that restrict origin usage. In agreement with this notion is the recent finding that overexpression of wild-type S. cerevisiae Cdc6p in S. pombe can induce re-replication (Sánchez et al. 1999).
Human CDC6 also plays a critical role in regulating DNA replication, as it is both limiting and essential for S phase entry (Hateboer et al. 1998; Stoeber et al. 1998; Yan et al. 1998). Because of its key role in regulating DNA synthesis, it is expected that several control mechanisms regulate the abundance and activity of CDC6. In agreement with this concept is the fact that CDC6 is absent from quiescent cells (Williams et al. 1997, 1998), and that its cell growth-dependent transcription is strictly controlled by the E2F transcription factors (Hateboer et al. 1998; Yan et al. 1998). Moreover, phosphorylation of CDC6 by cyclin A–CDK2 leads to a down-regulation of CDC6 activity during S phase by the relocalization of the protein from the nucleus to the cytosol (Saha et al. 1998; Jiang et al. 1999; Petersen et al. 1999). In contrast with its yeast orthologs, the level of CDC6 has been reported to be uniform during the cell cycle in proliferating cells (Saha et al. 1998; Jiang et al. 1999), and it is believed that the relocalization of the protein in S is sufficient to control CDC6 activity after origin firing. The recent findings that CDC6 is a very sensitive marker for proliferation and that CDC6 immunostaining of cervical smears can be used as an improved marker for premalignant cells (Williams et al. 1998) demonstrate that there is a very tight control on the abundance of CDC6 in the cell. These results may also indicate that mechanisms other than transcriptional control are involved in the regulation of CDC6 levels. Here, we show that CDC6 is targeted for proteolysis in early G1 and in quiescent cells by the anaphase promoting complex (APC) or cyclosome, (for review, see Zachariae and Nasmyth 1999) and suggest that the degradation of CDC6 is part of a mechanism that ensures correct temporal usage of origins of replication.
Results
Mammalian CDC6 is absent in early G1
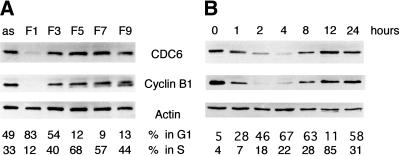
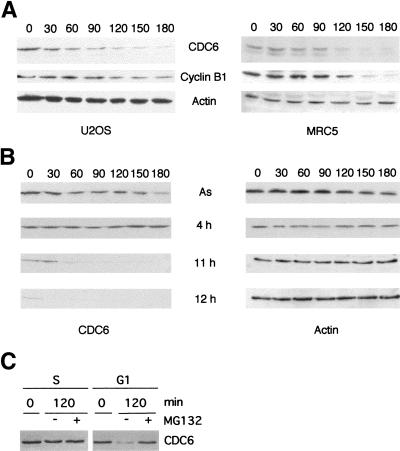
To determine whether CDC6 is present in all phases of the cell cycle, U2OS cells were synchronized by centrifugal elutriation, and protein extracts prepared from cells with different cell cycle profiles were analyzed by Western blotting. As shown in Figure 1A, very little CDC6 is present in the sample containing mainly early G1 cells (F1), whereas the extracts prepared from cells in other phases of the cell cycle contain an almost equal amount of CDC6. The degree of synchronization was confirmed by FACS analysis and by analysis of the expression of cyclin B1, which is targeted for proteolysis by APC and is absent in early G1 (Brandeis and Hunt 1996). The cell cycle–regulated expression of CDC6 also was observed in chemically synchronized cells. HeLa cells were synchronized with nocodazole. After release from the nocodazole block for various periods of time, samples were prepared for Western blotting and FACS analysis. In agreement with the data obtained by centrifugal elutriation, CDC6 was dramatically down-regulated in G1, whereas high levels of CDC6 were present in the remaining part of the cell cycle (Fig. 1B). These data show that the protein level of CDC6 fluctuates dramatically in every cell cycle in proliferating cells, not only when cells exit or enter the cell cycle.
Figure 1.
CDC6 is absent in early G1. (A) U2OS cells were fractionated by elutriation. The cell cycle profile of the different fractions was determined by FACS analysis (percentage of cells in G1 and S phase indicated below). Cell extracts (50 μg) from the indicated fractions were analyzed by Western blotting with antibodies specific to CDC6, Cyclin B1, and Actin (as a loading control). (as) Extracts from asynchronously proliferating cells. (B) HeLa cells were synchronized in metaphase by nocodazole treatment and released into the cell cycle by plating in nocodazole-free medium. Cells were harvested at the indicated number of hours after the release and analyzed as in A.
CDC6 mRNA is present in early G1
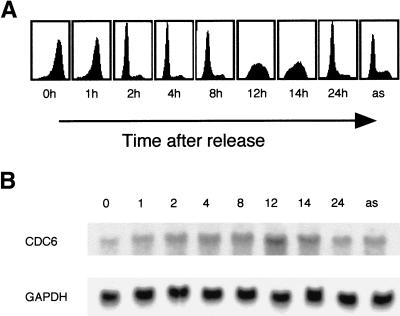
The absence of CDC6 in early G1 could be explained by a down-regulation of CDC6 transcription or a specific degradation of CDC6 in this phase of the cell cycle. To investigate whether CDC6 mRNA and protein levels correlate in early G1 of the cell cycle, HeLa cells were synchronized by nocodazole treatment. Samples were harvested for FACS analysis and Northern and Western blotting at different times after the release. As shown in Figure 2B, CDC6 mRNA is present throughout the cell cycle. A two- to threefold increase of CDC6 mRNA was observed in S phase cells when compared with asynchronously growing cells or cells in G1. Importantly, however, although CDC6 mRNA was present throughout G1, there was no correlation between CDC6 protein and mRNA levels in this phase of the cell cycle (Fig. 2B; data not shown). Moreover, these results show that CDC6 is detectable 4–6 hr prior to the initiation of S phase (Fig. 1 and data not shown). CDC6 appeared at the same time as cyclin E. However, complete expression of CDC6 was not observed before cyclin A was expressed at high levels. This pattern of CDC6 expression in proliferating cells is similar to the timing of expression in cells entering the cell cycle from quiescence and is in agreement with the requirement for CDC6 to initiate DNA replication (Stoeber et al. 1998; Yan et al. 1998; Petersen et al. 1999).
Figure 2.
CDC6 mRNA is present in early G1. HeLa cells were synchronized in metaphase as in Fig. 1. Cells were harvested for FACS analyses and isolation of total RNA at the indicated number of hours after the release. (A) Cell cycle profiles as determined by flow cytometry. (B) Total RNA was analyzed by Northern blotting. The blot was probed for expression of CDC6 and GAPDH.
CDC6 is degraded by the proteasome
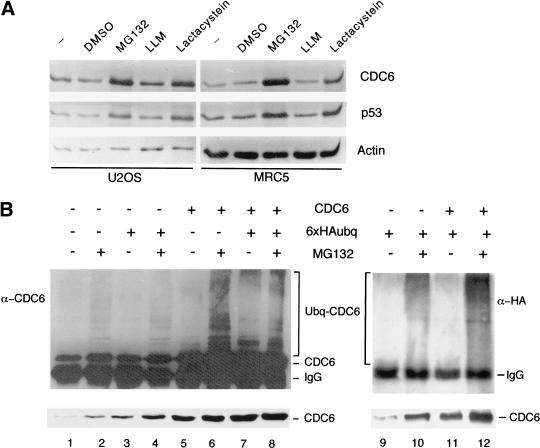
To test whether CDC6 can be stabilized by inhibition of the proteasome, U2OS and MRC5 cells were cultured in the presence of MG132, LLM, and lactacystein, potent inhibitors of either the proteasome (MG132 and lactacystein) or the calpain proteases (MG132 and LLM; Lee and Goldberg 1998). The treatment with MG132 and lactacystein led to a four- to fivefold increase in the level of CDC6, whereas the addition of DMSO and LLM had no effect (Fig. 3A). As a control for these experiments, the expression pattern of p53 was analyzed in the absence or presence of protease inhibitors. In agreement with previously published results (Maki et al. 1996), p53 was stabilized specifically by inhibitors of the proteasome. Thus, CDC6, like p53, is specifically targeted for degradation by the proteasome.
Figure 3.
Ubiquitin-mediated proteolysis of CDC6. (A) Stabilization of CDC6 by proteasome inhibitors. U2OS and MRC5 cells were treated for 8 hr with the indicated drugs. DMSO was used as a control. CDC6, p53, and actin were evaluated by Western blotting. (B) Identification of polyubiquitinated CDC6 in vivo. HeLa cells were transfected with pCMVCDC6 (CDC6) and pMT123 (6 × HA–Ubq) as indicated. Cell lysates were prepared with (+) or without (−) an 8-hr prior incubation with MG132. CDC6 was immunoprecipitated by use of polyclonal anti-CDC6 antibody (X27) and analyzed by Western blotting with a monoclonal antibody to CDC6 (DCS181; lanes 1–8) or an anti-HA antibody (12CA5; lanes 9–12). The presence of polyubiquitinated CDC6 is indicated. The relative amount of CDC6 expressed in each extract (lower part of B) was analyzed by Western blotting with DCS181 .
CDC6 is polyubiquitinated in vivo
To investigate whether CDC6 degradation is regulated by ubiquitination, we tested whether CDC6 is polyubiquitinated in vivo. HeLa cells were transfected with a CDC6 expression vector, a HA–ubiquitin expression vector, or with both. The transfections were performed in duplicate; one plate was treated with MG132 and the other with DMSO as a control. Cell extracts were prepared and used for immunoprecipitations of CDC6. The immunoprecipitations were analyzed by immunoblotting with an antibody to CDC6 (Fig. 3B, lanes 1–8). Endogenous CDC6 was found to be ubiquitinated in samples from cells treated with MG132. Ectopic expression of CDC6 further increased the amount of detected polyubiquitinated CDC6. Ubiquitinated CDC6 was observed as a ladder of immunoreactive proteins with higher molecular weights than CDC6 itself. The ligation of HA-tagged ubiquitin to CDC6 was evaluated by probing with an antibody to the HA tag, 12CA5 (Fig. 3B, lanes 9–12). In agreement with the results obtained with the anti-CDC6 antibody, the 12CA5 antibody recognized slower-migrating polyubiquitinated forms of CDC6. In summary, the stabilization of CDC6 by proteasome inhibitors and the identification of polyubiquitinated CDC6 in vivo strongly suggest that the proteolysis of mammalian CDC6 is controlled by ubiquitination.
Degradation of CDC6 is dependent on its amino terminus, but independent of CDK phosphorylation and subcellular localization
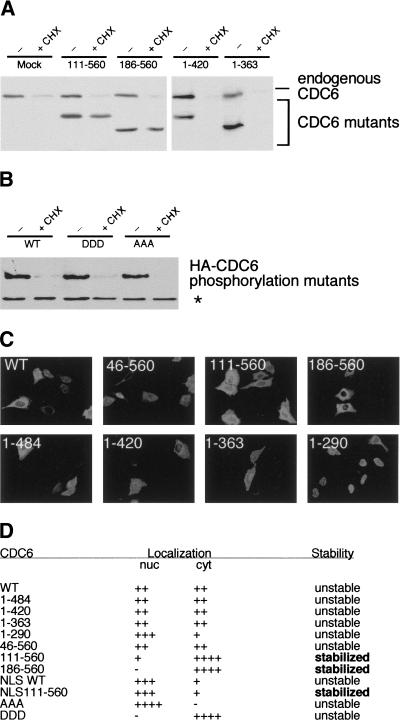
To understand which region of CDC6 targets it for degradation, different CDC6 deletion mutants were expressed in HeLa cells. The cells were treated with cycloheximide for 6 hr to block de novo protein synthesis and to allow most of the endogenous CDC6 to be degraded. As demonstrated in Figure 4A and summarized in Figure 4D, carboxy-terminal deletion mutants were as unstable as endogenous CDC6. In contrast, deletion of the first 110 amino-terminal residues led to stabilization of CDC6. This CDC6 mutant was expressed at almost the same level in cells treated with cycloheximide as in untreated cells. Similarly, a mutant lacking the 185 amino-terminal residues was also more stable than endogenous wild-type CDC6.
Figure 4.
Degradation of CDC6 is dependent on its amino terminus, but independent of CDK phosphorylation and subcellular localization. (A) Different CDC6 deletion mutants were expressed at low levels in HeLa cells and treated with cycloheximide (CHX) for 6 hr. One transfection mix was prepared per construct, and shared between three dishes to obtain equal transfection efficiency. The Western blots were probed with DCS 181. (B) The instability of CDC6 is independent on the CDK phosphorylation sites. The stability of the phosphorylation mutants was assayed as in A. The CDC6 mutants were identified with HA antibody to distinguish them from endogenous CDC6. (*) Cellular protein recognized by the HA antibody. (C) The indicated CDC6 deletion mutants were expressed in U20S cells, and the subcellular localization was analyzed by use of the HA antibody. (D) Summary of the localization and the stability of the CDC6 mutants. The stability of the mutants was determined as described above.
The amino terminus of CDC6 contains three CDK phosphorylation sites and a cyclin-binding motif (Petersen et al. 1999). The three amino-terminal phosphorylation sites appear to be the only functional CDK sites in CDC6, as mutation of these abolishes phosphorylation of CDC6 in vivo and in vitro (Jiang et al. 1999; Petersen et al. 1999). The amino-terminal regions of Cdc6p and Cdc18 have also been shown to regulate their stability (Drury et al. 1997; Kominami and Toda 1997). Phosphorylation of Cdc18 and Cdc6p by S and M phase CDKs target the proteins for SCF-dependent ubiquitination and subsequent degradation (Jallepalli 1997; Elsasser et al. 1999). Because this mechanism could also be implicated in the degradation of CDC6, we examined the stability of two mutants in which the three amino-terminal CDC6 phosphorylation sites were changed to alanine (CDC6-AAA) or aspartic acid residues (CDC6-DDD). If phosphorylation targets CDC6 for degradation, the CDC6-AAA mutant should have been stabilized, whereas the DDD mutants could have been destabilized. As shown in Figure 4B, the mutations in the phosphorylation sites had minimal effect on the stability of CDC6. Both mutants were degraded in the presence of cycloheximide, and, in addition, they were stabilized by inhibition of the proteasome (data not shown). These experiments showed that the degradation of CDC6 is independent of its phosphorylation status and that other amino-terminal sequences are involved in its degradation.
Recent experiments have demonstrated that the relocalization of p53 to the cytoplasm is part of the mechanism that regulates p53 degradation (Freedman and Levine 1998). Because CDC6 has been shown to relocalize to the cytoplasm during S phase (Saha et al. 1998; Stoeber et al. 1998; Petersen et al. 1999), we investigated whether the subcellular localization of CDC6 is important for its stability. As shown in Figure 4C and summarized in Figure 4D, the stable CDC6 111–560 and 186–560 deletion mutants are mainly cytoplasmic. To test whether the stabilization of these mutants could be due to their cytoplasmic localization, CDC6 111–560 fused to the nuclear localization signal from SV40 large T (Kalderon et al. 1984) was expressed. As expected, the NLSCDC6 111–560 mutant was shown to be nuclear, however, the nuclear localization did not confer instability to the protein (data not shown). Moreover, because the CDC6-DDD and CDC6-AAA mutants were shown previously to be exclusively cytoplasmic and nuclear, respectively (Jiang et al. 1999; Petersen et al. 1999), and both mutants were as unstable as wild-type CDC6 (Fig. 4B), it is unlikely that the subcellular localization of CDC6 is implicated in the regulation of its stability.
CDC6 degradation is dependent on amino-terminal destruction motifs
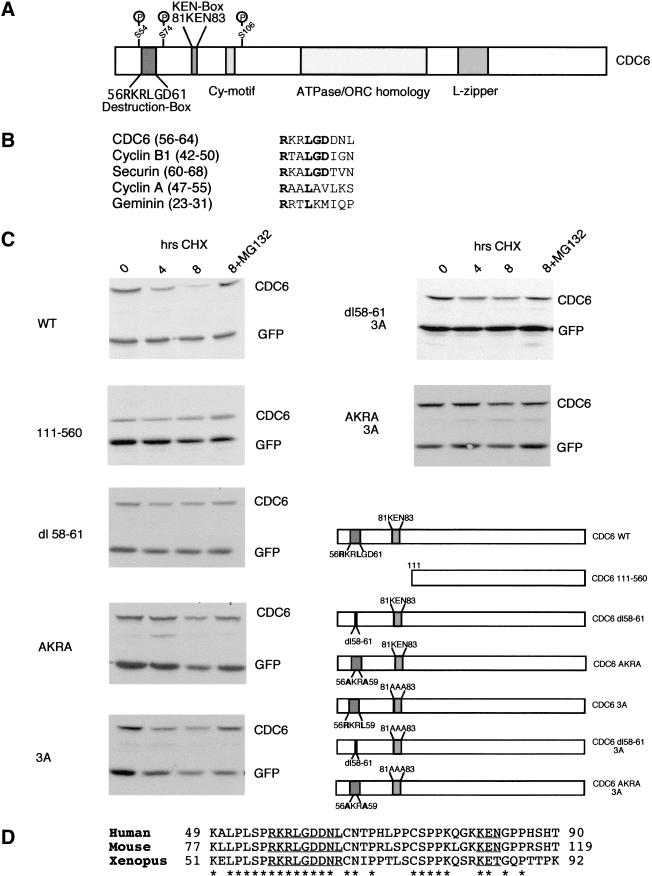
The fact that expression of CDC6 parallels cyclin B1 in synchronized U2OS and HeLa cells (Figs. 1 and 2) suggests that human CDC6, like cyclin B1, could be a target of the APC. Ubiquitination and degradation of cyclin B1 and other APC targets depend on a destruction box sequence frequently found in the amino terminus of the protein (Glotzer et al. 1991). A comparison of the primary sequence of CDC6 with destruction box (D box) sequences identified in cyclin B1 (Glotzer et al. 1991), cyclin A (Glotzer et al. 1991), securin (Zou et al. 1999), and geminin (McGarry and Kirschner 1998) revealed a putative D-box sequence at amino acids 56–64 (Fig. 5B), carboxy-terminal to the phosphorylated serine 54 (Petersen et al. 1999). This motif is highly conserved in human, mouse, and Xenopus CDC6 (Fig. 5D), whereas Cdc6p and Cdc18 are not similar to the CDC6 proteins from higher eukaryotes in this region. A deletion mutant removing 4 amino acids (58–61) of the putative D box was generated. As shown in Figure 5C, the putative D-box mutant (CDC6 dl 58–61) was stabilized, when compared with wild-type CDC6. Because we were concerned that the deletion of four amino acids in the destruction box would stabilize CDC6 because of changes in the overall structure of the protein, we also generated a CDC6 mutant in which two conserved residues in the destruction box were changed to alanines. This CDC6 mutant (AKRA) was also stabilized when compared with wild-type CDC6, however, the stabilization of the AKRA mutant was not as complete as that of the amino-terminal deletion mutant (111–560), suggesting that sequences outside the D box in the amino terminus could play a role in targeting CDC6 for degradation.
Figure 5.
CDC6 degradation is dependent on amino-terminal destruction motifs. (A) Schematic drawing of CDC6 depicting features such as the previously identified CDK phosphorylation sites (S54, S74, and S106), the Cy-box, a leucine zipper, and the ATPase/ORC homology domain. Moreover, the presence of a putative destruction box and a KEN box is shown. (B) Alignment of CDC6 amino acids 56–64 with destruction box sequences identified in cyclin B1, securin, Pds1p, and geminin. Amino acid numbers from each protein are shown in parentheses. (C) Plasmids expressing Myc-tagged versions of CDC6 were transfected into asynchronously growing HeLa cells together with a plasmid expressing Myc-tagged EGFP as a control for transfection efficiency. Cells were treated for the indicated time with cycloheximide or cycloheximide and the proteasome inhibitor MG132. At the indicated number of hours, cells were harvested and the expression of CDC6 and GFP was evaluated by Western blotting with the Myc− antibody. (D) Clustal W alignment of destruction box sequences in human (Williams et al. 1997), mouse (Hateboer et al. 1998), and Xenopus (Coleman et al. 1996) CDC6. The putative destruction box and KEN-box motifs are underlined. (*) Conserved amino acids. Amino acid numbers from each protein are indicated.
Recent results have suggested that a KEN-box motif (composed of the amino acids K-E-N) could be an APC-targeting motif distinct from the D box (Pfleger and Kirschner 2000). CDC6 contains a KEN box in its amino terminus (amino acids 81–83), which is conserved in mouse CDC6, but not in Xenopus CDC6, where the asparagine is substituted for a threonine (Fig. 5D). To test whether the KEN box could be involved in targeting human CDC6 for proteolysis, we mutated the three amino acids to alanines. The mutation of the KEN box partially stabilized CDC6 in vivo, suggesting that the KEN box in CDC6 is also involved in CDC6 turnover (Fig. 5C). The stability of double mutants containing mutations in the KEN box (3A) and in the D box (either dl 58–61 or AKRA) was then tested (Fig. 5C). The double mutants were as stable as CDC6 (111–560) with a half-life of >8 hr. In contrast, the half-life of wild-type ectopically expressed CDC6 in asynchronously growing cells is ∼2–3 hr. In summary, our results suggest that the CDC6 is targeted for proteolysis through the coordinated action of at least two different motifs in its amino terminus.
APC-dependent degradation of CDC6 in quiescent cells
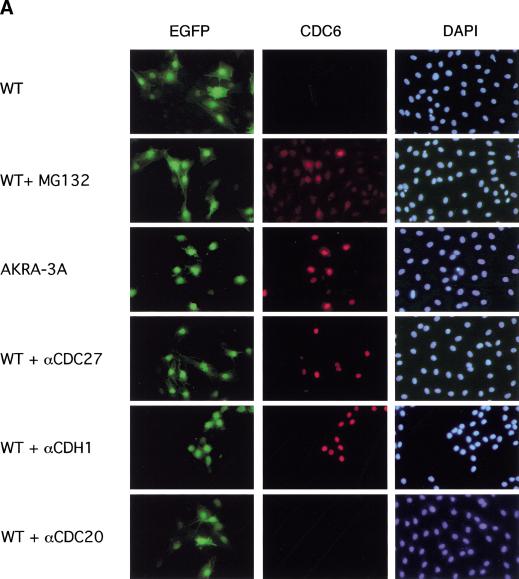
Previously, we have been unsuccessful in efficiently expressing CDC6 from a constitutive strong cytomegalovirus promoter in quiescent cells (Hateboer et al. 1998; Petersen et al. 1999), suggesting that CDC6 levels are controlled by not only transcription in nonproliferating cells. To test whether the inability to express CDC6 in quiescent cells is dependent on ubiquitin-mediated degradation and the putative destruction motifs in CDC6, expression vectors encoding wild-type and mutant CDC6 were injected into serum-starved Rat1 cells together with an enhanced version of the green fluorescent protein (EGFP), which served as a injection marker. Four hours after injection, CDC6 expression was evaluated by immunofluorescence (Fig. 6A). Wild-type CDC6 was not detected under these conditions. However, if the cells were cultured in the presence of MG132, expression of CDC6 was detected in 75% of the injected cells (Fig. 6). These results suggest that the inefficient expression of human CDC6 in quiescent cells is due to proteasome-mediated degradation. To test whether degradation of CDC6 in quiescent cells is dependent on the same regions of CDC6 as in proliferating cells, the stability of the different CDC6 mutants was tested by injection of expression plasmids in quiescent cells. As shown in Figure 6, the mutations of the destruction motifs allowed efficient expression of CDC6 (AKRA-3A) in quiescent cells in the absence of proteasome inhibitors. Similar data were obtained for the amino-terminal deletion mutant (CDC6 111–560), but not for a mutant lacking the Cy-motif (CDC6 dl 93–100; data not shown).
Figure 6.
APC–CDH1- and destruction motif-dependent degradation of CDC6 in quiescent cells. (A) Rat1 cells were synchronized by serum deprivation and injected with different CDC6 expression plasmids together with pCMVMYEGFP. Where indicated, wild-type (WT) CDC6 expression plasmids were coinjected with affinity-purified antibodies to CDC27, CDH1, or CDC20. Treatment with MG132 is indicated (+MG132). The cover slips were fixed 4 hr after injection. The antibody DCS181 was used for staining of CDC6, and positively injected cells were identified by the expression of GFP. Nuclei were stained with DAPI. (B) Quantification of CDC6-positive cells compared with the number of EGFP-positive cells. Results of three independent experiments are shown.
The role of the destruction motifs predicts that CDC6 is ubiquitinated by the APC. To obtain more direct evidence for a role of the APC in the regulation of CDC6 proteolysis, an antibody to the APC subunit CDC27 was injected into quiescent cells together with the CDC6 expression plasmid. As a control, purified rabbit immunoglobulin (data not shown) or an antibody to CDC20 was coinjected with the CDC6 expression plasmid. As shown in Figure 6, the injection of the CDC27 antibody, but not the antibody to CDC20 allowed efficient expression of CDC6 in quiescent cells, demonstrating that APC activity is required for proteolysis of CDC6 in quiescent cells.
CDH1 is required and limiting for CDC6 proteolysis
Two WD40-containing proteins termed CDC20 and CDH1/Hct1 associate with the APC and regulate its activity (for review, see Peters 1998; Zachariae and Nasmyth 1999). CDH1, but not CDC20, is associated with APC in G1 and in differentiated cells (Fang et al. 1998; Kramer et al. 1998; Gieffers et al. 1999), suggesting that it could be required for the degradation of CDC6. To test this hypothesis, an affinity-purified rabbit polyclonal antibody to CDH1, which neutralizes APC–CDH1 in vitro (E.R. Kramer and J.M. Peters, unpubl.), was injected together with the CDC6-expression plasmid. Consistent with the finding that CDH1 is the only known activator of APC in quiescent cells (and in G1), injection of the neutralizing CDH1 antibody led to efficient expression of CDC6 in the majority of the injected cells.
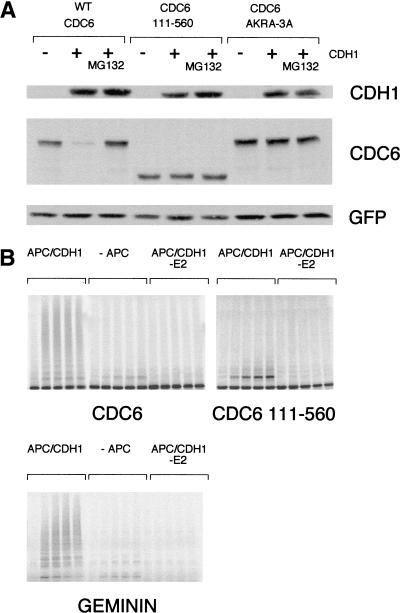
To test whether CDH1 is limiting for the degradation of CDC6, we transfected asynchronously growing HeLa cells with plasmids expressing wild-type CDC6, amino-terminally deleted CDC6 (111–560), or the destruction motif mutant (AKRA-3A) with or without an expression plasmid for human CDH1 (Fig. 7A). Overexpression of CDH1 resulted in the destruction of wild-type CDC6, but only marginally affected the expression of the two CDC6 mutants. The induced proteolysis of wild-type CDC6 by CDH1 was prevented by proteasome inhibitors, and because the expression of CDH1 in asynchronously growing HeLa cells does not alter cell cycle progression (data not shown; Kramer et al. 2000), our results show that CDH1 is limiting for the degradation of CDC6.
Figure 7.
CDC6 is targeted for degradation by CDH1 in vivo and is ubiquitinated by APC–CDH1 in vitro. (A) HeLa cells were transfected with plasmids expressing either wild-type CDC6 or stable mutants of CDC6 (AKRA-3A or 111–560) together with Myc-tagged EGFP as a control for transfection efficiency and with or without a plasmid encoding wild-type HA-tagged CDH1. Where indicated, cells were treated with MG132 for the last 8 hr before harvesting. Twenty-four hours after transfection, lysates were prepared from the transfected cells, and expression of CDC6, CDH1, and GFP (as a loading control) was evaluated by Western blotting. (B) APC–CDH1 ubiquitination of CDC6 in vitro. In vitro-translated wild-type CDC6, CDC6 111–560, and geminin were tested for APC–CDH1 ubiquitination in vitro. The reactions were performed in the presence of immunopurified APC and purified CDH1 (APC/CDH1), in the absence of APC (−APC) or in the absence of E2 (APC/CDH1 −E2). The absence of APC and E2 served as negative controls in the assays; geminin served as a positive control. Reactions performed for 0, 1, 2.5, 5, and 10 min were separated by SDS-PAGE.
CDC6 is ubiquitinated by APC–CDH1 in vitro
To test whether CDC6 can serve as a substrate for the APC, in vitro ubiquitination assays with immunopurified APC from Xenopus and recombinant CDH1 were performed. As shown in Figure 7B, CDC6 becomes polyubiquitinated in vitro dependent on the presence of APC and E2. Moreover, the presence of CDH1 in the reaction was also required for detecting ubiquitinated forms of CDC6 (data not shown). CDC6 (111–560) was not polyubiquitinated in these assays, showing that the amino-terminal region of CDC6 is required for APC–CDH1 ubiquitination. However, CDC6 (AKRA-3A), with specific mutations in the D box and the KEN box, was still ubiquitinated in vitro, although to a lesser extent than wild-type CDC6 (data not shown). Although the AKRA-3A mutant is significantly stabilized in vivo, these in vitro data indicate that there may be other regions in the amino terminus of CDC6 that targets it for degradation.
CDC6 has a short half-life in G1
So far, our results suggest that the destruction of CDC6 occurs through an APC-dependent mechanism in mitosis or during early G1. Because CDC6 mRNA is present throughout the cell cycle (Fig. 2), our data suggest that the degradation and, therefore, the half-life of CDC6 is cell cycle dependent. As shown in Figure 8A, CDC6 is a relatively short-lived protein with an estimated half-life of 2 hr in both transformed and primary human cell lines. To evaluate the half-life in specific phases of the cell cycle, HeLa cells were synchronized at the G1/S border by a double thymidine block and released for 4 hr into S phase, or for 11 and 12 hr into G1. Cycloheximide was added at the indicated times, and CDC6 levels were determined by Western blotting (Fig. 8B). No significant degradation of CDC6 was observed in S phase, whereas a very rapid turnover of CDC6 was evident in early G1. The half-life of CDC6 in early G1 is estimated to be ∼15–30 min. CDC6 was absent in cells released from the G1/S block for 12 hr. Moreover, the addition of proteasome inhibitors in early G1 stabilized the expression of CDC6 in this phase of the cell cycle, whereas no significant increase in CDC6 levels was observed with treatment of S phase cells (Fig. 8C).
Figure 8.
CDC6 has a short half-life in G1. (A) U2OS and MRC5 cells were treated with cycloheximide for the indicated times (in minutes). Cells were harvested and lysates prepared for Western blotting with antibodies specific to CDC6, cyclin B1 and Actin. (B) Asynchronously (As) growing HeLa cells, or HeLa cells presynchronized by a double-thymidine block and then plated in drug-free medium for 4 hr (4 h), 11 hr (11 h) or 12 hr (12 h) were treated with cycloheximide for the indicated time (in minutes). Cells were harvested, and the expression of CDC6 and actin was determined as described above. (C) Proteasome inhibitors stabilize CDC6 in G1, but not in S phase. To obtain an S phase population, HeLa cells were synchronized by a double thymidine block and released for 2 hr in the presence or absence of MG132. To obtain a G1 population, HeLa cells were synchronized by a nocodazole block and then released for 45 min (time point 0), and incubated further for 2 hr in the presence or absence of MG132. CDC6 expression was evaluated by Western blotting with a monoclonal antibody to CDC6.
CDC6 is degraded in early G1, not in mitosis
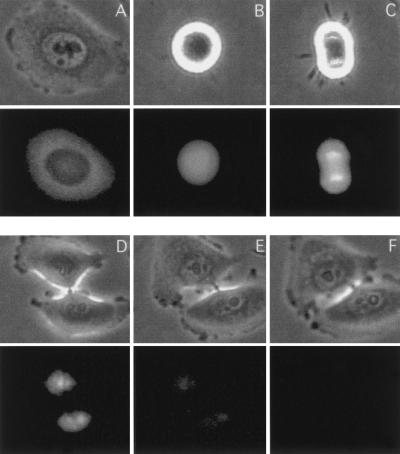
Because our data suggest that CDC6 is degraded slightly later than cyclin B1 (Fig. 1 and data not shown), which is degraded after prometaphase (Clute and Pines 1999), CDC6 is degraded in either late mitosis or early G1. A more precise determination of the timing of CDC6 degradation is essential for understanding its biological role. To resolve this issue, we used time-lapse microscopy. Accordingly, we constructed a plasmid expressing a wild-type EGFP–CDC6 fusion that, in HeLa cells, produced a protein of the expected size and that was stabilized in the presence of proteasome inhibitors (data not shown). Furthermore, as shown in Figure 9, the fusion protein was localized to the cytosol and the nucleus as has been described for wild-type CDC6 (Saha et al. 1998; Jiang et al. 1999; Petersen et al. 1999). To determine the timing of degradation of CDC6, HeLa cells were synchronized at the G1/S boundary and injected with the EGFP–CDC6 plasmid. Cells were released from the block and followed by time-lapse immunofluorescence microscopy for 12 hr. In agreement with previously published results, CDC6 is mainly cytoplasmic in S phase cells and nuclear in G1 (Fig. 9A,D). CDC6 was not degraded during mitosis (Fig. 9B,C), and it became an exclusively nuclear protein after cytokinesis (Fig. 9D). The degradation of CDC6 was initiated 2–3 hr after cytokinesis, and by 4 hr, CDC6 fluorescence was close to background levels (Fig. 9E,F).
Figure 9.
In vivo analysis of CDC6 proteolysis. Phase contrast microscopy (top), and GFP fluorescence (bottom) of HeLa cells injected with a plasmid expressing EGFP–CDC6 progressing from S phase into G1. HeLa cells were synchronized by a double-thymidine block, injected with 50 ng/μl of the expression plasmid, and released from the block. The protein expression was measured by time-lapse microscopy starting from 3 hr (A) until 12 hr after release (F). All fluorescence images were taken with the same exposure time with a cooled CCD camera during the following time periods: (B) in mitosis at 6 hr; (C) in telophase at 6 hr 45 min; (D) in G1 at 8 hr; (E) at 10 hr.
Biological effects of expressing stable CDC6 mutants
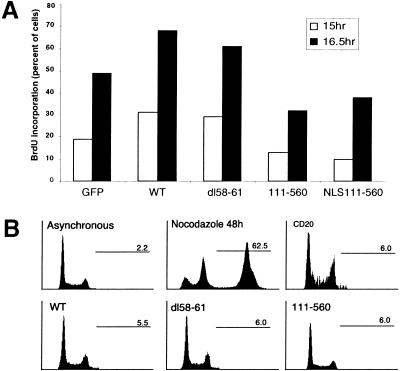
Ectopic expression of CDC6 has been shown previously to accelerate entry into S phase in mammalian cells (Hateboer et al. 1998; Stoeber et al. 1998). To investigate whether high levels of CDC6 in quiescent cells would lead to further acceleration of entry into S phase, serum-starved Rat1 cells were injected with plasmids expressing wild-type or stable mutants of CDC6. Cells were stimulated with serum in the presence of BrdU, and harvested 15 and 16.5 hr after serum addition (Fig. 10A). In agreement with previously published results, the expression of wild-type CDC6 led to premature entry into S phase. A similar acceleration of entry into S phase was observed by expression of the destruction box mutant of CDC6, demonstrating that the deletion of the four amino acids in this mutant does not interfere with its biological activity. However, these results also indicate that other events in addition to CDC6 are limiting for entry into S phase. For instance, both CDK activity and the MCM proteins are required for DNA replication, both of which are not present in quiescent cells and early G1. Interestingly, the expression of the two amino-terminally deleted mutants of CDC6 inhibited the initiation of DNA replication (Fig. 10A). This result is consistent with previous results showing that ectopic expression of nonphosphorylatable CDC6 leads to a delay in S phase entry (Jiang et al. 1999).
Figure 10.
Effects of CDC6 expression on DNA replication. (A) Wild-type and D-box mutant of CDC6 induce premature entry into S phase. Quiescent Rat1 cells were injected with plasmids expressing the indicated proteins. Three hours after injection, serum was added to the cells, and entry into S was monitored by BrdU incorporation 15 and 16.5 hr after addition of serum. A GFP-expression plasmid was used as an injection marker in combination with empty vector or expression plasmids encoding wild-type (WT) CDC6, a D-box mutant (dl58–61), or the two amino-terminal deleted mutants (111–560 or NLS111–560). (B) Overexpression of stable mutants or wild-type CDC6 does not lead to re-replication. Asynchronously growing HeLa cell were transfected with a CD20-expression plasmid as a transfection marker with or without plasmids expressing wild-type, a D-box mutant or an amino-terminal mutant of CDC6. Seventy-two hr after transfection, cells were harvested and DNA profiles of CD20-positive transfected cells were determined by FACS analysis. In parallel, the DNA profiles of non-transfected asynchronously growing cells and cells treated with nocodazole for 48 hr were determined. The numbers indicated in each profile show the percentage of cells with DNA content >4N.
Overexpression of Cdc18 leads to repeated rounds of DNA synthesis in the absence of mitosis whereas high levels Cdc6p do not induce re-replication. To test whether stable mutants of CDC6 are sufficient to induce re-replication, HeLa cells were transiently transfected with wild-type or stable CDC6 mutants. HeLa cells were used for these experiments as they do not contain functional p53, and, therefore, are prone to re-replication if incubated with nocodazole or colcemid (Cross et al. 1995; Lanni and Jacks 1998). Incubation of HeLa cells with nocodazole for 48 hr led to accumulation of DNA with >4N DNA content in 60% of the cells (Fig. 10B). In contrast, the overexpression of wild-type or stable CDC6 mutants for 24, 48, or 72 hr did not lead to any significant levels of re-replication (Fig. 10B, and data not shown). Because the overexpression of CDC6 does not lead to a block of mitosis, experiments were also performed in which wild-type and mutant CDC6 were overexpressed in the presence of nocodazole. In these experiments, we did not see any evidence for a cooperative role of CDC6 expression and loss of p53 in polyploidy (data not shown).
Discussion
Mechanism and regulation of CDC6 degradation
Our results demonstrate that CDC6 is absent from the mammalian cell cycle in early G1. This is not due to lack of CDC6 mRNA, but is a result of ubiquitin-mediated proteolysis of the protein. We have identified an amino acid stretch near the amino terminus of CDC6 resembling D box sequences found in cyclin B1, securin, cyclin A, and geminin (Glotzer et al. 1991; McGarry and Kirschner 1998; Zou et al. 1999) and a KEN box sequence found as in CDC20, Nek2, and B99 (Pfleger and Kirschner 2000). A CDC6 mutant in which the putative D box and KEN box are mutated is significantly stabilized compared with wild-type CDC6.
The yeast orthologs of CDC6 are targeted for proteolysis in S phase by an SCF complex. In contrast, our data show that degradation of mammalian CDC6 is regulated by the APC. First, CDC6 is very unstable in early G1 and in quiescent cells, and the APC has been reported to be active in these phases of the cell cycle (Peters 1998; Zachariae and Nasmyth 1999). Second, CDC6 contains functional amino-terminal D-box and KEN-box motifs found in other substrates of the APC. Third, the overexpression of CDH1 specifically leads to degradation of wild-type CDC6. Fourth, antibodies to the APC subunit CDC27 and to CDH1 prevent the degradation in quiescent cells. Fifth, CDC6 is ubiquitinated in vitro by APC–CDH1.
It was reported recently that APC, in association with CDH1, is expressed and presumably active in terminally differentiated cells (Gieffers et al. 1999). No targets for APC are known in differentiated cells. However, it is conceivable that APC activity is part of a regulatory program that maintains the differentiated state by degrading proteins that could otherwise activate entry into the cell cycle. For instance, deregulation of the pRB pathway leads to the expression of a series of E2F-regulated genes, whose products induce cell cycle progression and initiation of DNA replication (Dyson 1998). Deregulation of the pRB pathway would not be sufficient for the induction of S phase in differentiated cells if APC–CDH1 were actively preventing the accumulation of S phase factors, such as CDC6, that are required for the initiation of DNA replication. In agreement with such speculation is the finding that deregulated E2F activity is not sufficient to induce S phase in differentiated muscle cells (Pajalunga et al. 1999). Thus, the targeted proteolysis of CDC6 by APC in differentiated cells may be part of a mechanism designed to restrict cell proliferation.
Functional role of CDC6 degradation
The finding that mammalian CDC6 is degraded in early G1 is unexpected for two reasons. First, the yeast CDC6 orthologs are degraded as cells enter S phase, which is consistent with the role of CDC6 proteins in regulating initiation of DNA replication. Because the CDC6 proteins are required for the initiation of DNA replication, the degradation of the proteins as cells enter S restricts origin activation to once per cell cycle. However, mammalian CDC6 is present throughout S, G2, and M phase (Williams et al. 1997; Saha et al. 1998; Jiang et al. 1999; Petersen et al. 1999), and it is not degraded before entering of the next cell cycle. These results suggest that mammalian CDC6 may have additional functions in S, G2, or M phase. Because the degradation of several APC substrates is required for the exiting of distinct stages of mitosis (Glotzer et al. 1991; McGarry and Kirschner 1998), it is conceivable that high levels of CDC6 could prevent the exiting of mitosis. However, overexpression of wild-type or stable CDC6 mutants does not affect the progression through mitosis (M. Melixetian, B.O. Petersen, and K. Helin, unpubl.). These results are also in agreement with the observation that endogenous CDC6 is stable in mitosis. Alternatively, the presence of CDC6 during S, G2, and M phase may be implicated in regulating the timing of mitosis. In agreement with such a notion are results obtained in S. pombe and S. cerevisiae demonstrating that inactivation of cdc18 + and CDC6 results in mitosis in the absence of DNA replication (Kelly et al. 1993; Piatti et al. 1995). Future studies will be aimed at understanding whether CDC6 has a role in regulating mitosis in mammalian cells.
Second, why degrade CDC6 in early G1 when cells should start preparing for DNA replication? One answer to this question could be that degradation of CDC6 is important for regulating the timing of initiation of DNA replication. However, our data suggest that ectopic expression of CDC6 only leads to a marginal shortening of G1 (around 1 hr, see Fig. 10A; data not shown), and despite the fact that CDC6 is expressed in a stable form very early in the cell cycle that it is not sufficient to induce DNA replication. Therefore, these data are consistent with the existence of a window of opportunity in which the expression of CDC6 can induce DNA replication, and that other factors, such as the ORCs, the MCMs and/or CDK activity, are limiting for the induction of DNA replication. Therefore, our data may be more consistent with a two-step model, in which the availability of CDC6 is essential for the licensing of an origin, and that CDKs and CDC7/DBF4 primarily regulate the timing of DNA replication. Although very few data are available on the regulation of preRCs in mammalian cells, it is compelling that the specification of an origin decision point (ODP) for the DHFR origin in CHOC400 cells ∼3–4 hr after mitosis (Wu and Gilbert 1996) coincides with the timing of degradation of CDC6. This observation raises the question: Why do cells degrade CDC6 and then resynthesize the protein later in G1? Two models can be envisioned. In one model, the origins are licensed in mitosis or very early G1, and the resynthesis of CDC6 later in G1 is not required for origin licensing. In this model, the resynthesis of CDC6 is required only for cells that have been out of the cell cycle for a prolonged period of time. In the second model, some origins are licensed for replication in mitosis or very early G1, and the resynthesis of CDC6 later in G1 is required for the licensing of other origins. In this model, early firing origins may be licensed in mitosis and late origins in late G1. In support of the second model is the demonstration that injection of antibodies to CDC6 or dominant-negative CDC6 proteins in late G1 delays entry into S phase (Hateboer et al. 1998; Herbig et al. 1999). These experiments suggest that CDC6 resynthesis is required for the entry into S phase. In contrast with the second model is that in vitro replication assays using CHOC400 cells and Xenopus egg extracts suggest that the spatial position and the replication timing of chromosomal domains are established in early G1. These experiments indicate that CDC6 resynthesis is not required for the proper initiation of DNA replication (Dimitrova and Gilbert 1999). At this point, it is clear that further experiments are required to distinguish which of the two models is correct, in particular, the identification of an origin of replication to which the ORCs, CDC6, and the MCMs bind.
Our data also show that the overexpression of a stable mutant or wild-type CDC6 is not sufficient to induce re-replication. These results are consistent with data obtained in S. cerevisiae, in which Cdc6p synthesis can only promote DNA replication in a restricted window of the cell cycle, between the destruction of the Clbs after anaphase and activation of Clb5p and Clb6p/Cdk1p in late G1 (Piatti et al. 1996). Cdc6p can accumulate within nuclei of G2 and M phase cells without induction of re-replication. Therefore, it seems that control of Cdc6p degradation is not the only mechanism that prevents refiring of origins. In fact, it is known that CDK activity is essential for preventing re-replication, since the loss of CDK activity in yeast, Drosophila, and human cells results in DNA re-replication in the absence of mitosis, (Hayles et al. 1994; Sauer et al. 1995; Itzhaki et al. 1997). Central for this discussion is that CDK activity is thought to restrict DNA replication by phosphorylating (among other targets) the MCMs and CDC6, leading to their functional inactivation (e.g., see Labib et al. 1999). At the same time, CDK activity is required for mitosis. Thus, cells without CDK activity are unable to divide. It has been suggested that loss of CDK1 activity is essential for allowing preRCs to form and, therefore, for the induction of re-replication. Because the overexpression of CDC6 does not lead to a block in mitosis or a down-regulation of CDK1 activity, we also performed experiments in which cells were blocked in mitosis by nocodazole treatment. Prolonged treatment of p53-negative cells has been shown previously to lead to re-replication and loss of CDK1 activity (e.g., see Lanni and Jacks 1998). However, consistent with the observation that CDC6 is stable in cells blocked in metaphase, we did not observe any cooperation between loss of functional p53 and CDC6 overexpression in inducing re-replication. On the basis of these data, we conclude that CDC6 is not limiting for re-replication. However, it is most likely essential for re-replication as has been shown recently in S. cerevisiae (Noton and Diffley 2000).
Regulation of CDC6 activity in mammalian cells
In yeast, Cdc6p is essential for the binding and subsequent loading of other proteins (like the Mcms) to origins of replication (Aparicio et al. 1997; Tanaka et al. 1997). The binding of Cdc6p to origins occurs during G1 and is dependent on its nucleotide-binding motif (Perkins and Diffley 1998; Weinreich et al. 1999). Because the proteins involved in regulation of DNA replication are conserved during evolution, it is assumed that regulation of initiation of DNA replication is conserved as well. Mammalian CDC6, like Cdc6p, is required for entry into S (Hateboer et al. 1998; Yan et al. 1998), and nucleotide-binding mutants of CDC6 block entry into S phase as do their yeast counterparts (Herbig et al. 1999). Despite this conservation, the mechanisms regulating CDC6 activity in mammalian cells and in yeast are substantially different. First, CDC6 is not degraded in S phase, but rather, in early G1. Second, phosphorylation does not target CDC6 for degradation. Third, the E3 ligase involved in CDC6 degradation is APC and not SCF. However, both the yeast and mammalian CDC6 proteins appear active at similar stages of the cell cycle. The expression of the CDC6 genes is cell cycle-dependent, and the proteins are present in the nucleus during mid-to-late G1, at which time they are required to form preRCs. Although mammalian CDC6 is not degraded in S phase, it is relocalizing to the cytosol, rendering it unable to induce DNA replication.
In summary, periodic transcription, CDK-regulated subcellular localization, and proteolysis in early G1 regulate CDC6 activity. These regulatory mechanisms confine CDC6 activity to the G1 phase of the cell cycle, which contributes to ensuring the timely licensing of origins of replication. Interestingly, however, our data also suggest that CDC6 could play a role during S, G2, and M phase that is not related to initiation of DNA replication. Future studies are required to investigate this possibility.
Materials and methods
Cell culture
HeLa, U2OS, Rat-1, and MRC-5 cells were grown in DMEM (Euroclone) supplemented with 10% BCS (Hyclone) and 2 mM L-glutamine (Euroclone). Cells were synchronized in early S phase by a double thymidine block or in metaphase by a thymidine–nocodazole block (Petersen et al. 1999). To obtain quiescent Rat-1 cells, the cells were cultured in DMEM without serum for 48 hr and induced to enter the cell cycle by addition of fresh medium containing 10% BCS. Synchronization of U2OS cells by elutriation was performed as described previously (Müller et al. 1997). Entry into S phase was monitored by cell cycle profiles and FACS analysis and/or incorporation of BrdU.
To inhibit protein synthesis, cells were cultured in the presence of 10 μg/ml cycloheximide. Inhibition of the proteasome and other proteases was performed by culturing cells for up to 8 hr in the presence of 5 nM MG132 (Biomol), 10 μM LLM (Biomol), or 10 μM lactacystein (Biomol). Stock solutions of MG132 and lactacystein were made in DMSO and used in a 1:1000 dilution. The 1000× stock solution of LLM was made in ethanol. For analysis of potential effects of CDC6 or mutants on DNA replication, cells were transfected with 10 μg of the CDC6-expression plasmids together with 1 μg of pCMVCD20 plasmid as a transfection marker (van den Heuvel and Harlow 1993). Seventy-two hr after transfection, the cells were harvested with PBS containing 5 mM EDTA and incubated with a FITC-coupled anti-CD20 antibody (Becton Dickinson) for 30 min on ice. Then cells were fixed and the cell cycle profile was examined by FACS analysis.
Antibodies and Western blotting
Polyclonal (X27) and monoclonal antisera (DCS 181) raised against CDC6 were described previously (Petersen et al. 1999). The M20 antiserum was raised against a carboxy-terminal CDC6 peptide (KIEEKEIEHAL). Cdc27 antibodies were raised against a peptide encoding the last 17 carboxy-terminal amino acids of human Cdc27. Immunsera were subsequently affinity purified by use of a peptide column and the specificity of the purified antibodies was tested by immunoprecipitation and Western blotting. Affinity-purified rabbit polyclonal antibodies against CDC20 (Sat107) and CDH1 (Sat105) were described previously (Gieffers et al. 1999; Kramer et al. 2000). The 12CA5 antibody was used for the HA tag, and the 9E10 antibody was used for the MYC tag. The monoclonal antibody HE12 to cyclin E and the polyclonal antibody H432 to cyclin A were obtained from Santa Cruz. The monoclonal antibody to cyclin B1 (GNS 5) was a kind gift of S. Shiff (Rockefeller University, NY). The anti-actin antibody AC-40 was obtained from Sigma. Western blotting was performed as described previously (Petersen et al. 1999).
Ubiquitination assays
Asynchronously growing HeLa cells were transfected with 10 μg of pCMVCDC6 and 2 μg of pMT123 (HA–ubiquitin; Treier et al. 1994). Cells were lysed in RIPA buffer containing 5 mM N-ethyl-maleimide (NEM; Sigma). CDC6 was immunoprecipitated with polyclonal anti-CDC6 antibody (X27) and analyzed by Western blotting using a monoclonal anti-CDC6 (DCS 181) or monoclonal anti-HA (12CA5) antibody.
In vitro ubiquitination assay
[35S]Methionine and [35S]cysteine-labeled human CDC6 and Xenopus geminin proteins were prepared by coupled transcription–translation reactions in rabbit reticulocyte lysate (Promega). To obtain pure CDH1-activated APC, APC was immunopurified from interphase Xenopus egg extracts and activated by baculovirus-expressed human CDH1. The in vitro ubiquitination reaction was performed essentially as described previously (Kramer et al. 2000).
Plasmids
pCMVHACDC6 dl 58–61 was generated by ligation of two PCR fragments into the BamHI site of pCMVHA introducing a ClaI site into CDC6. The resulting cDNA was also cloned into pCMVMY. pCMVMYCDC6 (AKRA), pCMVMYCDC6 (3A), pCMVMYCDC6 (AKRA-3A), and pCMVMYCDC6 (dl58–61–3A) were generated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene). pCMVNLSCDC6 and pCMVNLS 111–560 were constructed by cloning of full-length CDC6 and CDC6 111–560 as BamHI fragments from pCMVHACDC6 and pCMVHACDC6 111–560 into pCMVHANLS (K. Helin, unpubl.). pCMVMYEGFP-CDC6 was generated by insertion of a BamHI fragment containing the full open reading frame of CDC6 into pCMVMYEGFP (K. Helin and K. Holm, unpubl.). All other CDC6 constructs were described previously (Petersen et al. 1999). pCMVHACDH1 was constructed by PCR amplification of the full-length open reading frame of CDH1, and the PCR product was subsequently cloned into the BamHI site of pCMVHA. All PCR reactions were performed with PfuI proofreading polymerase (Stratagene), and sequences were verified by sequencing.
RNA isolation and Northern blot analysis
RNA extraction and Northern blot analysis were performed as described previously (Hateboer et al. 1998).
Microinjection, immunofluorescence, and time-lapse imaging
For microinjection, Rat1 cells were grown on glass coverslips in serum-free medium for 48 hr. For assessing the synthesis of CDC6 in serum-starved cells, the cells were injected with 20 ng/μl of the CDC6-expression plasmids together with 10 ng/μl of EGFP plasmid directly into cell nuclei using a Zeiss automatic injection system. Where indicated, affinity-purified antibodies at 1 μg/μl were coinjected. Three to four hours after injection, the cells were fixed and CDC6 expression was evaluated by use of a monoclonal antibody to CDC6, DCS181. For measuring entry into the cell cycle, the serum-starved Rat1 cells were injected with various amounts of CDC6-expression plasmid together with 10 ng/μl EGFP expression plasmid. For the experiments shown in Figure 10A, 100 ng/μl CDC6 expression plasmid was used. Cells were stimulated with serum 3–4 hr after injection, BrdU was added, and DNA synthesis was evaluated at the indicated number of hours after addition of serum.
Immunofluorescence was performed as described previously (Petersen et al. 1999). For time-lapse imaging, cells were plated and synchronized in 6-well plates. After injection, the plates were kept at 37°C in an incubator chamber with a CO2 supply mounted on an Olympus inverted microscope. Analysis of multiple wells was performed with an X–Y motorized stage and autofocus control (PRIOR). Images were captured with a coded CCD camera (Hamamatsu). GFP was detected with a FITC filter set (Chromo Technology).
Acknowledgments
We gratefully acknowledge Emanuela Frittoli for help with microinjection experiments and Simona Ronzoni for FACS analysis. We thank Tim Hunt and Gordon Peters for discussions, and Nick Heintz, Andrea Musacchio, and Simonetta Piatti for discussions and critical reading of the manuscript. B.O.P. was supported by a fellowship from the Danish Research Academy, and C.W. was supported by a fellowship from the European Commission. This work was supported by grants from the Austrian Industrial Research Promotion Fund to J.-M.P., and the Associazione Italiana per la Ricerca sul Cancro (AIRC) and the Fondazione Italiana per la Ricerca sul cancro (FIRC) to K.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL khelin@ieo.it; FAX 39-02-5748-9851.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.832500.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;90:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S-phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nature Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- Diffley JFX. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JFX. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters J-M. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hateboer G, Wobst A, Petersen BO, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woolard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2/mitotic cyclin B complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Herbig U, Marlar CA, Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol Biol Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki JE, Gilbert CS, Porter ACG. Construction by gene targeting in human cells of a “conditional” CDC2 mutant that rereplicates its DNA. Nature Genet. 1997;15:258–265. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiatior protein p65cdc18 by CDK phosphorylation. Genes & Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wells NJ, Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation. Proc Natl Acad Sci. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin–proteasome-mediated degradation of the CDK inhibitor rum1 and the S-phase initiator cdc18. Genes & Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Geiffers C, Holzl G, Hengstschläger M, Peters J-M. Activation of the human anaphase promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–1212. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters J-M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Diffley JFX, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nature Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes & Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki CG, Huibregste JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Müller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton E, Diffley JFX. CDK inactivation is the only essential function of APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Pajalunga D, Tognozzi D, Tiainen M, D'Angelo M, Ferrantelli F, Helin K, Sacchi A, Crescenzi M. E2F activates late-G1 events but cannot replace E1A in inducing S phase in terminally differentiated skeletal muscle cells. Oncogene. 1999;18:5054–5062. doi: 10.1038/sj.onc.1202897. [DOI] [PubMed] [Google Scholar]
- Perkins G, Diffley JFX. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Peters J-M. SCF and APC: The Yin and Yang of cell cycle regulated proteolysis. Curr Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sørensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by Cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger C, Kirschner M. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Böhm T, Cocker JH, Diffley JFX, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, Lawlis SJ, Hou Z-H, Hendricks M, Parvin JD, Dutta A. Human CDC6/Cdc18 associates with Orc1 and is selectively eliminated from the nucleus at the onset of S-phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M, Calzada A, Bueno A. Functionally homologous DNA replication genes in fission and budding yeast. J Cell Sci. 1999;112:2381–2390. doi: 10.1242/jcs.112.14.2381. [DOI] [PubMed] [Google Scholar]
- Sauer K, Knoblich JA, Richardson H, Lehner CF. Distinct modes of cyclin E/cdc2 kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes & Dev. 1995;9:1327–1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewksi LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading of Mcm proteins onto DNA. Proc Natl Acad Sci. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, Gilbert DM. A distinct G1 step required to specify the chinese hamster DHFR replication origin. Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci. 1998;95:3063–3068. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Komamura Y, Ishimi Y. Biochemical analysis of the intrinsic Mcm4–Mcm6–Mcm7 DNA helicase activity. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]