Abstract
While the interfacial partitioning of charged or aromatic anchor residues may determine the preferred orientations of transmembrane peptide helices, the dependence of helix orientation on anchor residue position is not well understood. When anchor residue locations are changed systematically, some adaptations of the peptide-lipid interactions may be required to compensate the altered interfacial interactions. Recently we have developed a novel transmembrane peptide, termed GW5,19ALP23 (acetyl-GGALW5LALALALALALALW19LAGA-ethanolamide), which proves to be a well behaved sequence for an orderly investigation of protein-lipid interactions. Its roughly symmetric nature allows for shifting the anchoring Trp residues by one Leu-Ala pair inward (GW7,17ALP23) or outward (GW3,21ALP23), thus providing fine adjustments of the formal distance between the tryptophan residues. With no other obvious anchoring features present, we postulate that the inter-Trp distance may be crucial for aspects of the peptide-lipid interaction. Importantly, the amino acid composition is identical for each of the resulting related GWALP23 sequences, and the radial separation between the pairs of Trp residues on each side of the transmembrane α-helix remains similar. Here we address the adaptation of the aforementioned peptides to the varying Trp locations by means of solid-state 2H NMR experiments in varying lipid bilayer membrane environments. All of the GWx,yALP23 sequence isomers adopt transmembrane orientations in DOPC, DMPC and DLPC environments, even when the Trp residues are quite closely spaced, in GW7,17ALP23. Furthermore, the dynamics for each peptide isomer are less extensive than for peptides possessing additional interfacial Trp residues. The helical secondary structure is maintained more strongly within the Trp-flanked core region than outside of the Trp boundaries. Deuterium labeled tryptophan indole rings in the GWx,yALP23 peptides provide additional insights into the behavior of the Trp side chains. A Trp side chain near the C-terminus adopts a different orientation and undergoes somewhat faster dynamics than a corresponding Trp side chain located an equivalent distance from the N-terminus. In contrast, as the inter-Trp distance changes, the variations among the average orientations of the Trp indole rings at either terminus are systematic yet fairly small. We conclude that subtle adjustments to the peptide tilt, and to the N- and C-terminal Trp side-chain torsion angles, permit the GWx,yALP23 peptides to maintain preferred transmembrane orientations while adapting to lipid bilayers of differing hydrophobic thickness.
Keywords: peptide-lipid bilayer interactions, solid-state deuterium NMR, GWALP23 peptides, hydrophobic match
The influence of a lipid bilayer membrane on protein organization and function is well documented. For example, the orientation (tilt) angles of virus protein “u” (1) and of the GABAA receptor (2) have been shown to vary in response to the thickness of the lipid bilayer. The lipid acyl chain identities furthermore influence the assembly of M2 tetramer proton channels (3) and alter the functional equilibrium of rhodopsin (4).
Protein transmembrane domains and membrane protein function are governed in part by the interfacial or aqueous partitioning of aromatic or charged anchor residues that flank the transmembrane helical domains. These types of residues are enriched at the interfaces (5, 6), with the preferred position for lysines being about 3 Å farther from the bilayer center than the preferred positions for tryptophans (7). In order to better understand the influence of interfacial anchoring residues, systematic approaches are quite helpful. It is in this regard useful to consider combinations of synthetic lipids and model peptides, which make it feasible to adjust systematically the distances between anchor residues and the thickness of the lipid matrix (8–11). Such model peptide sequences typically have been based on a repeating sequence unit, wherein the addition or removal of an extra hydrophobic block will alter the length of the transmembrane domain with minimal disruption of other properties. For example in “WALP” family peptides (GWW(LA)nLWWA) (9, 12, 13), the effective hydrophobic length of the core helix between the Trp residues changes as “n” is varied. In these peptides, the core helix is bounded by the aromatic side chains of Trp. With regard to the modulation of lipid phase behavior, the effective length of a WALP peptide is governed by the distance between the innermost Trp residues that flank the core helix (14). Furthermore, because Trp and also Tyr are particularly enriched at the membrane-water interface in membrane proteins of known structure (5, 6, 15), changing the placements of aromatic residues could offer a means of modifying the orientation and behavior of the membrane-spanning domain. Conversely, the lipid thickness can be varied by altering the acyl chain lengths, a factor which may compensate when anchor residue positions are changed. When acyl chains longer than 14 carbons are utilized, nevertheless, one or more double bonds must be introduced (in some or all of the chains), to maintain appropriate lipid bilayer fluidity at physiological temperature.
Even such seemingly “simple” peptide-lipid systems may pose a number of issues. The lipid unsaturation will influence also the lipid lateral pressure profile (16). For the peptide, in addition to the length of the core helix between the anchor residues, the identities of the polar, amphiphilic and/or aromatic interfacial residues may alter the response of the system (17). Furthermore, the geometry of a helix dictates that adding or removing core helix residues inevitably will change also the radial positions of the anchoring side chains (18). A further potential issue concerns the overall hydrophobicity of the peptide system in cases where small “blocks” of sequence are added or removed. The sequences with smaller numbers of amino acids thereby may become too polar to insert into a lipid bilayer or too short to form stable helices (19), which could lead to oligomerization (20).
Recently we improved the design of “WALP” family peptides by replacing two of the tryptophans with glycines (21). Unexpectedly, the dependence of the WALP peptide apparent tilt angle on the lipid bilayer thickness is not straightforward. The relatively minor response of the original WALP peptides toward changes in the lipid thickness was later explained in terms of extensive dynamics (22, 23). The dynamics can be rationalized in terms of an excess of interfacial tryptophan residues, dispersed around a helical wheel (18), and potentially competing among themselves for favorable interactions with the head groups of the lipids (17, 24).
To circumvent some of these limitations, we developed GW5,19ALP23 (GGALW[LA]6LWLAGA), which proves to be a well behaved transmembrane peptide for the systematic investigation of protein-lipid interactions (17, 25). In this study we further exploit its orderly sequence, which allows for shifting the remaining individual Trp residues either inward or outward in pairwise fashion, thereby leading to sequences of the form GWx,yALP23, where the “x, y” pairs designate the Trp sequence positions, either “5, 19” (original GWALP23), “3, 21” (Trps moved outward) or “7, 17” (Trps moved inward). These sequences allow for fine-tuning the length of the core (Leu-Ala)e helix, while maintaining identical amino acid composition and therefore identical overall peptide hydrophobicity. Furthermore, because in each case one Trp of GWALP23 is moved radially by +200° and the other by −200°, they effectively “meet” on another face of the helix and thereby maintain a similar radial separation on one side of an α-helix in each of the three peptides (Table 1, Figure 1). The Trp residues on both ends of GWx,yALP23 have a substantial propensity to reside at the lipid-water interface (26, 27). To maintain effective interactions, it is conceivable that the single indole rings at either end may alter their side chain orientations to adapt in different lipid environments (28). For these reasons, we undertake a comprehensive solid-state NMR investigation of the generalized whole-peptide and specific indole-ring responses of GWx,yALP23 transmembrane peptides to varying lipid environments. We employ deuterated alanines to investigate the peptide helix average orientations and dynamics, and deuterated tryptophans to investigate the indole side chain adjustments. The intent is to provide enhanced understanding of how lipids influence the behavior of membrane-spanning peptides.
Table 1.
Sequences for GWx,yALP23 peptidesa
| Peptide | Sequence |
|---|---|
| GW3,21ALP23 | GGWLALALALALALALALALWGA |
| GW5,19ALP23 | GGALWLALALALALALALWLAGA |
| GW7,17ALP23 | GGALALWLALALALALWLALAGA |
N-terminal Gly residue is acylated. C-terminal Ala residue is blocked with ethanolamide.
[NB: please use a fixed-width font for the sequences and align them as shown.]
Figure 1.
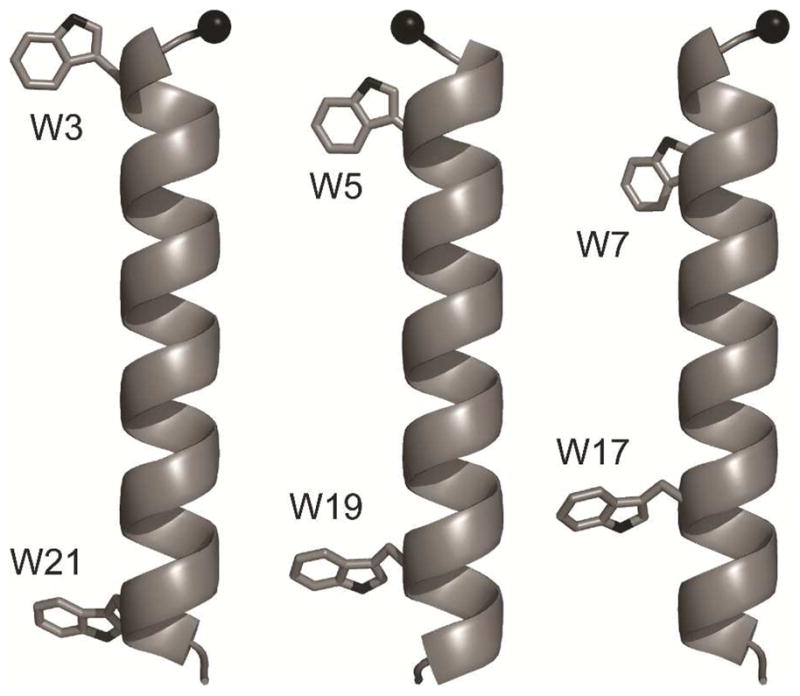
Molecular models of GW3,21ALP, GW5,19ALP23 and GW7,17ALP23 (left to right). Black sphere indicates the Cα carbon of Gly1. Note that the GW5,19ALP23 model is rotated by 180°.
Materials and Methods
All isotope enriched compounds were from Cambridge Isotope Laboratories (Andover, MA). Deuterium labeled alanine (Ala-d4) and tryptophan (Trp-d5; deuterons on the indole ring) were modified by manual synthesis to introduce fluorenylmethoxycarbonyl (Fmoc) group, using an identical protocol for both amino acids (29). Partial hydrogen/deuterium exchange on the Trp indole side chain was accomplished by incubating commercial Fmoc-Trp (NovaBiochem, San Diego, CA) with deuterated trifluoroacetic acid (TFA-d1) at 10 °C for 3 hr (30). This procedure facilitates deuterium incorporation at positions 2 and 5 of the indole ring, which was confirmed by 1H NMR spectroscopy in DMSO-d6, by means of the intensity reduction at position 2 and changes in the multiplet pattern at positions 4 and 6 (Figure S1 of the Supporting Information). The peptides synthesized with TFA-d1 treated Fmoc-Trp represent a mixture of GWx,yALP23-Trp-d0, GWx,yALP23-Trp-d1 and GWx,yALP23-Trp-d2. For brevity we further refer to them in the form of GWx,yALP23-Trp-d2.
Peptides were synthesized utilizing a model 433A peptide synthesizer (Applied Biosystems by Life Technologies, Foster City, CA) in a similar manner to GWALP23 (17), using Wang resin and Fmoc-protected amino acids (NovaBiochem). Deuterium-enriched alanines were introduced in pairs at different isotope abundance levels. Deuterium-enriched tryptophans were incorporated in separate peptides one at a time; namely, for each GWx,yALP23 sequence, four separate Trp-labeled peptides were synthesized (having full or partial deuteration of the N- or C-terminal Trp). Due to the mild conditions for peptide cleavage from the Wang resin (20% ethanolamine in dichloromethane), no side chain protecting groups were required (31). Peptides were purified by reversed-phase HPLC (C8), using the previously established conditions for GWALP23 (17). Confirming HPLC chromatograms and mass spectra are provided in Supporting Information (Figures S2–S3).
Circular dichroism (CD) spectra were obtained for peptides incorporated into small unilamellar vesicles (1/40, peptide/lipid) produced by ultrasonic treatment. Peptide concentrations were determined spectrophotometrically to be in the 100 μM range. CD spectra were collected using a 1 mm pathlength cell and a Jasco J710 spectropolarimeter (Easton, MD) operated at a 20 nm/min scan rate and 1.0 nm band width. Five spectra were averaged to enhance the signal intensities. Aliquots of the same samples were further diluted 50-fold for steady state fluorescence spectroscopy, using a Perkin Elmer LS-55 fluorescence spectrometer. The excitation wavelength was 284 nm, and emission was recorded between 300 and 500 nm at a rate of 200 nm/min. Slit widths were 7.5 nm, both for excitation and emission. An asymmetric cuvette was employed, having a 10 mm pathlength for excitation and a 1 mm pathlength for emission. Ten spectra were acquired and averaged.
Samples for solid-state 2H NMR were prepared by mechanical alignment, as described previously (12). A mixture of 2 μmol peptide and 80 μmol lipid (Avanti, Alabaster, AL) was deposited on glass slides (4.8 × 23 × 0.07 mm) from methanol/water (95/5), dried in vacuo (10−3 torr) and hydrated with 2H-depleted water to a 45% level of hydration (w/w). Glass slides were stacked and sealed in a glass cuvette (4.9 × 4.9 × 24 mm). Deuterium NMR spectra were recorded using two Bruker (Billerica, MA) Avance 300 spectrometers operating at a magnetic field of 7.0 T, utilizing probes with cylinder 5 mm coil and a quadrupolar echo pulse sequence with full phase cycling (32). The spectral width was 1,000,000 Hz, recycle delay 90 ms, and pulse durations 3.2 or 4.5 μs (depending on the spectrometer probe). Approximately 700,000 transients were collected for Ala-d4 peptides and twice that number for Trp-dx peptides. Spectra for Ala-labeled peptides were processed by zero filling the time domain to 5120 points, applying 100 Hz exponential apodization and Fourier transformation. The corresponding parameters for the spectra of Trp-labeled peptides were 2048 data points and 300 Hz. Spectra were recorded at two sample orientations, with the lipid bilayer normal either parallel (β=0°) or perpendicular (β=90°) to the applied magnetic field. An average value of the quadrupolar splitting magnitude was taken (after applying the scaling factor of two for the β=90° orientation), the standard deviation between the two sample orientations typically being within 0.5 kHz.
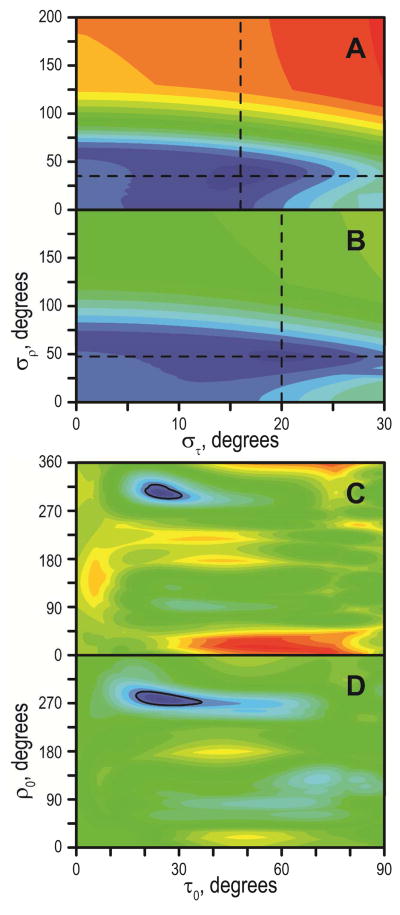
Geometric Analysis of Labeled Alanines (GALA; (12)) was performed by fitting a generalized order parameter Szz, and the apparent peptide tilt magnitude (τ) and direction (ρ), to a model of a tilted α-helical peptide, with an ε// angle between the alanine Cα-Cβ bond vector and peptide helix axis equal to 59.4° (12). Further, we refer to this Szz value as Spept to avoid confusion with the tryptophan side-chain order parameter (see below). Selected peptide/lipid systems were analyzed also by considering Gaussian distributions of tilt and rotation around the average values (τ0, ρ0) with standard deviations (στ, σρ) (22). In these cases, the order parameter was fixed at 0.88 and a multidimensional grid search was performed by varying σρ between 0° and 200°, στ between 0° and 30°, τ0 between 0° and 90°, and ρ0 between 0° and 359°, using 1° increments.
Deuterium NMR data from Trp-labeled peptides were analyzed by rotating the previously refined structure of 3-methyl-indole (33) by two angles, ρ1 and ρ2 (defined in Figure S4 of the Supporting Information), and considering the dynamics in the form of an order parameter, Szz (30, 34), which now would incorporate side-chain as well as backbone dynamics. Due to symmetry considerations, such analysis returns eight possible orientations of the indole ring; we report the values for one unique octet with 0°≤ρ1≤180° and 0°≤ρ2≤90° (Figure S4). While the fully deuterated Trp side chain has five deuterons, the C–D bond vectors at carbons 4 and 7 are nearly collinear (angle of 179.3°; (33)), and therefore are generally not resolved.
Indeed, it was assumed that deuterons at positions 4 and 7 are not distinguishable and produce identical signals, since none of the Trp-d5 spectra had five resolved peaks. Since the spectral assignments are not known, initially 4! = 24 possible assignment schemes were considered for each Trp. (The number was later reduced in systems where Trp-d2 data were available). For estimating the root mean squared deviation (RMSD), we treat positions 4 and 7 separately, under the assumption that the corresponding NMR signal represents a superposition of these deuterons. Assignment schemes were selected based on the Szz and RMSD values, as explained in Results.
For the conversion of backbone-independent (ρ1, ρ2) angles to Trp side chain (χ1, χ2) angles, models of GWx,yALP23 were constructed using Swiss-PdbViewer 4.0 (35) using (Φ, Ψ, Ω) of (−65°, −40°, 180°) and rotated by angles τ and ρ according to Table 3 (see Results) to yield the coordinates of the tilted peptides. The side chain of a tryptophan residue in question was rotated around the (χ1, χ2) angles to yield the indole orientation matching to the previously obtained (ρ1, ρ2) angles of 3-methyl-indole. Steric hindrance contours were generated by rotating the Trp side chains through the complete range of (χ1, χ2) angles. Steric clash was defined as the distance <2 Å between any of the non-hydrogen atoms of the indole ring and any non-hydrogen atoms of the peptide backbone.
Table 3.
GALA fit results for GWx,yALP23 peptides in lipid bilayer membranesa
| Peptide (Lipid) | ||||
|---|---|---|---|---|
| Spept | τ, deg | ρ, deg | RMSD, kHz | |
| (DLPC) | ||||
| GW7,17ALP23 | 0.79 | 6.7 | 223 | 0.1 |
| GW5,19ALP23a | 0.71 | 20.8 | 304 | 0.7 |
| GW3,21ALP23b | 0.63 | 18.0 | 281 | 2.0 |
| (DMPC) | ||||
| GW7,17ALP23 | 0.82 | 4.3 | 182 | 0.1 |
| GW5,19ALP23a | 0.88 | 9.1 | 310 | 1.1 |
| GW3,21ALP23 | 0.75 | 9.0 | 268 | 1.1 |
| (DOPC) | ||||
| GW7,17ALP23 | 0.83 | 4.0 | 186 | 0.1 |
| GW5,19ALP23a | 0.86 | 6.1 | 322 | 0.6 |
| GW3,21ALP23 | 0.80 | 4.3 | 257 | 0.8 |
Data for GW5,19ALP23 from ref. (17).
The RMSD for GW3,21ALP23 in DLPC reduces to 1.0 kHz if the A19 data point is omitted or to 1.5 kHz if the A5 data point is omitted. The corresponding (Spept, τ, ρ) values are (0.60, 19.7, 277) with A19 omitted or (0.68, 15.3, 283) with A5 omitted. Exclusion of both A5 and A19 leads to RMSD = 0.9 kHz, with the corresponding parameters of (0.63, 18.0, 279).
Results
Both leucine and alanine are considered to have high α-helix propensity, while tryptophan does not exhibit this property (36). A lipid bilayer typically offers a stabilizing environment for the transmembrane helices, because it is favorable to maximize the backbone hydrogen bonding in the nonpolar lipid environment (37). Nevertheless, in the case of a lipid with short acyl chains (DLPC), several residues at the peptide termini may protrude into the interfacial and/or aqueous phases, where deviations from the helical structure can occur more easily. To assess the secondary structure of the GWx,yALP23 peptides we have recorded CD spectra of the peptides in DLPC (Figure 2).
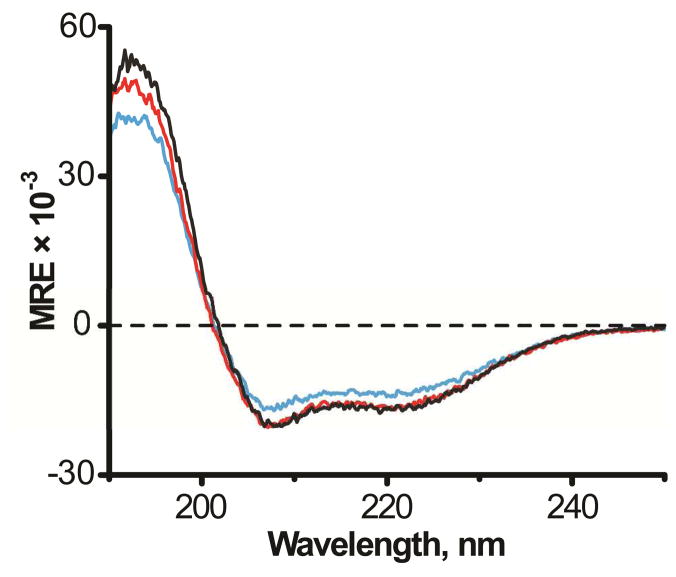
Figure 2.
Circular dichroism spectra of GW3,21ALP (black), GW5,19ALP23 (red) and GW7,17ALP23 (blue) in DLPC.
All three GWx,yALP23 peptides exhibit the spectral signature of an α-helix, with a distinct minimum at 208 nm and a broad shoulder around 222 nm. Nevertheless, the mean residue ellipticity values for GW7,17ALP23 are somewhat lower (about 15% reduced in magnitude) in comparison with the other peptides, suggesting reduced helical structure when the tryptophans are moved inward. It is widely accepted that due to their amphiphilic character the tryptophan residues prefer the membrane-water interface, which would position the terminal residues 1–6 and 18–23 of GW7,17ALP23 in more polar regions, where some helix unwinding may be expected (38). The spectral intensities of GW5,19ALP23 and GW3,21ALP23 overlap, indicative of similar helicity. Previously we have observed some fraying of the GW5,19ALP23 termini in DMPC (17); within this context the CD data suggest that the GW3,21ALP23 helix could possibly terminate prior to the Trp residues in DLPC.
Earlier studies with the peptides of the WALP family demonstrated a peptide length-dependent formation of non-bilayer phases of phosphatidylcholine membranes at high peptide/lipid ratio: in the case of negative hydrophobic mismatch, the lipid phase can undergo transitions from lamellar to isotropic to inverse hexagonal phase (9). To probe this possibility for GWx,yALP23 peptides, phosphorous NMR spectra of oriented samples were recorded. For all of the peptide-lipid combinations under investigation here, using a molar ratio of 1/40 (peptide/lipid), the 31P NMR spectra were characteristic of bilayer lipids, with chemical shift anisotropy of ~42 ppm (Figure S5).
To gain insight into the behavior of GWx,yALP23 peptides in lipid bilayer membranes of varying thickness, we introduced deuterium-labeled Ala residues into the core helical sequence between the tryptophans. Previously we have observed that GW5,19ALP23 readily incorporates into lipid bilayers composed of C12–C18 lipids, remains helical, and adopts well-defined average orientations that vary with the lipid thickness (17, 21). We find similar responses when the tryptophans are moved inward or outward to effectively decrease or increase the length of the Leu-Ala core sequence between the tryptophans (Figure 3). The Ala methyl 2H quadrupolar splittings (Δνq) of these peptides are dependent on the macroscopic sample orientation, indicating fast precession of each helix about the lipid bilayer normal (39). A full set of 2H NMR spectra is provided in Figures S6–S8, and the Δνq magnitudes are tabulated in Table 2. For the spectra in Figure 3 and Figures S6–S8, the peptide/lipid ratio is 1/40 (mol/mol); nevertheless, the Δνq magnitudes change very little when the ratio is reduced to 1/200 (Figure S9). The quadrupolar splittings of centrally positioned alanine residues change from 20.9 kHz to 23.2 kHz (A11) and from 3.8 kHz to 9.6 kHz (A13) as the peptide/lipid ratio is decreased five-fold, from 1/40 to 1/200. Since the magnitude of quadrupolar splittings is described by the equation 1, the Δνq values relate to changes in θ, the angle between a particular alanine Cα-Cβ bond vector and the applied magnetic field.
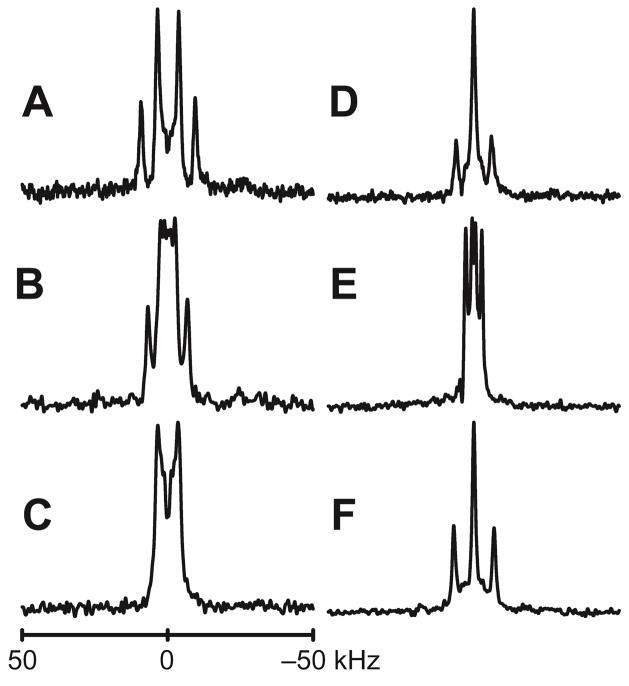
Figure 3.
Deuterium NMR spectra of peptides in DMPC. A–D: GW3,21ALP23; E–F: GW7,17ALP23. Labeled positions are: 5 and 7 (A), 9 and 11 (B, E), 13 and 15 (C, F), 17 and 19 (D). Sample orientation is β=0°. A complete set of deuterium NMR spectra is provided in Figures S6–S8.
Table 2.
Alanine CβD3 quadrupolar splittings for GWx,yALP23 peptides incorporated in lipid bilayersa
| Peptide | Lipid | Alanine position | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 7 | 9 | 11 | 13 | 15 | 17 | 19 | ||
| GW7,17ALP23 | DLPC | 8.0 | - | 6.1 | 6.1 | 12.5 | 0.8 | - | 7.4 |
| DMPC | 10.9 | - | 11.2 | 2.1 | 13.9 | 0.8 | - | 4.1 | |
| DOPC | 12.5 | - | 10.9 | 3.1 | 13.4 | 0.8 | - | 3.0 | |
| GW5,19ALP23b | DLPC | - | 26.4 | 25.5 | 26.9 | 14.6 | 20.7 | 3.4 | - |
| DMPC | - | 21.9 | 8.9 | 20.9 | 3.8 | 17.6 | 2.9 | - | |
| DOPC | - | 16.6 | 1.7 | 16.7 | 1.5 | 15.4 | 2.6 | - | |
| GW3,21ALP23 | DLPC | 19.6 | 23.8 | 15.7 | 18.7 | 0.9 | 9.6 | 11.4 | 0.8 |
| DMPC | 6.4 | 17.9 | 5.2 | 13.6 | 6.7 | 6.7 | 12.2 | 0.8 | |
| DOPC | 0.8 | 13.1 | 2.1 | 9.3 | 6.6 | 6.6 | 12.3 | 1.3 | |
Values in kHz. Entries left blank were not measured because Trp is present instead of Ala.
Data from (17).
| Equation 1 |
Substitution of QCC = 168 kHz, Spept = 0.88 (Table 3), β = 0 ° and γ = 109.5° (the tetrahedral angle to account for the methyl group rotation) leads to the angle θ (for the average peptide orientation) changing from 43.8° to 42.6° for A11, and from 52.7° to 49.6° for A13. From equation 1, it follows that the closer θ is to the magic angle (~54.7°), the more sensitive Δνq become toward even minor deviations of θ (note the steep slope of the helical wave in the vicinity of A13; Figure 4B). The small changes in the θ angles when the peptide/lipid ratio is diluted from 1/40 to 1/200 are in accord with the earlier finding of only a shallow dependence of Δνq or θ on the ratio of GW5,19ALP23/DMPC between 1/40 and 1/80 (17). These results furthermore illustrate that the 2H Δνq values are very sensitive to small changes in the Ala residue orientations.
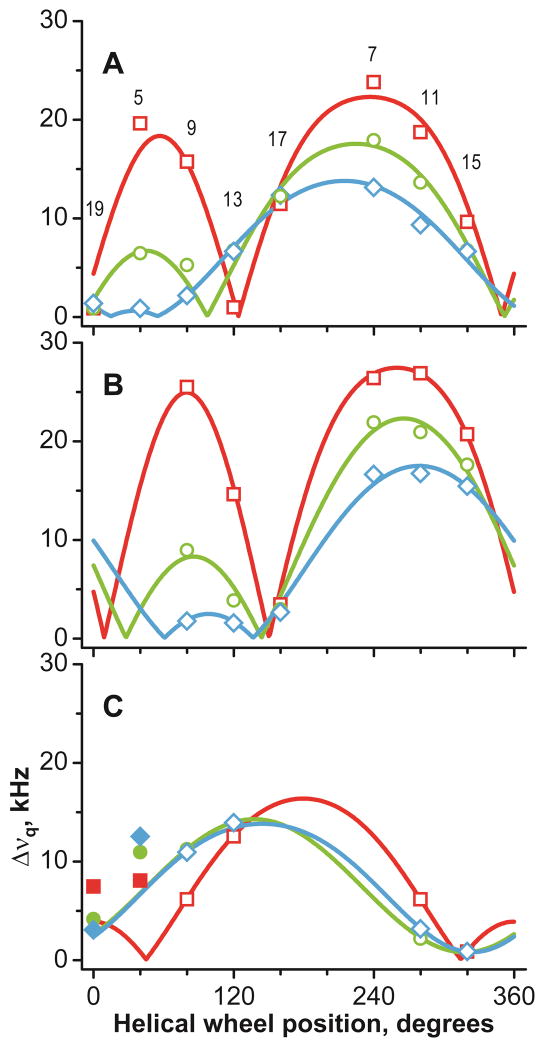
Figure 4.
GALA helical wave plots of peptides in DLPC (red squares), DMPC (green circles) and DOPC (blue diamonds). A: GW3,21ALP23; B: GW5,19ALP23; C: GW7,17ALP23. Deuterium labeled alanine positions are indicated in A. Filled symbols correspond to residues outside the inter-Trp core and not used for the analysis of helix orientation.
From the dependence of Δνq on the Ala residue position, it is apparent that each of the peptides is tilted to a preferred average orientation in the lipid bilayer membranes. It is notable that the peptide with only nine amino acids between the Trp residues (GW7,17ALP23) maintains a preferred, non-random, transmembrane orientation, not only in the thinner membranes but also in DOPC. While the membrane incorporation is not overly surprising, because of the overall hydrophobicity (40), the rather consistent helix orientation (on average) nevertheless contrasts with that of several peptides and peptaibols (which lack Trp residues), which switch from a transmembrane to an interfacially bound topology as a function of lipid bilayer thickness (41–43). Even for GW7,17ALP23 in DOPC, the tryptophans seem still to be important and significant for determining the average orientation and dynamics of the core helix.
For determination of the preferred molecular orientations, the deuterium NMR Δνq magnitudes for the GWx,yALP23 series peptides were subjected to GALA analysis, using implicit rigid-body dynamics in the form of a principal order parameter (Spept) and peptide average orientation, namely magnitude (τ0) and direction (ρ0) of the helix tilt as independent variables (12). The fit quality was assessed by means of RMSD between the observed and back-calculated Δνq values. A fit is typically considered good when the RMSD value is less than the 2H peak linewidth (usually on the order of 1 kHz). It can be seen in Table 3 that this condition is fulfilled for each peptide-lipid system, with an exception of GW3,21ALP23 in DLPC, where the RMSD approaches 2 kHz. This situation can be understood in terms of partial helix unwinding, as mentioned above. Even though the CD spectra were recorded from samples of sonicated vesicles, for which relatively high curvature is expected, whereas the NMR spectra were recorded from oriented bilayer samples; the deductions about the extent of helicity, or partial fraying, show substantial agreement between the distributions of the alanine Δνq values and the observed CD spectra. We note nevertheless that the amount of helix unraveling at the peptide termini may be different between the vesicles and the aligned bilayers, as the solid-state NMR observables do not allow for expressing the extent of fraying quantitatively. Furthermore, the isotope labels were incorporated two residues away from an anchoring Trp residue, leaving open the possibility that the helix unwinding could begin prior to the labeled alanines. Despite these caveats, the qualitatively similar results from the vesicle and oriented bilayer systems, suggest that helix formation does not couple strongly with the bilayer curvature. Indeed, the exclusion of the most N-terminal data point (A5) of GW3,21ALP23 in DLPC reduces the RMSD for the GALA fit to 1.5 kHz, while the exclusion of (only) the most C-terminal point (A19) leads to RMSD of 1.0 kHz (Table 3). Conversely, excluding any individual central data point from A7 to A17 does not improve the fit quality, as the RMSD remains high, in the 1.9–2.1 kHz range when the data for A5 and A19 are present. Interestingly, introducing Arg12 or Arg14 in the GW3,21ALP23 sequence leads to a large tilt and a good fit for the entire helix in DLPC (44). When the Δνq value for A19 is excluded, the average orientation and dynamics of GW3,21ALP23 in DLPC do not differ significantly from the ones obtained using all data points.
Earlier we established, using 2H labels for alanines 3 and 21, that the peptide helicity is not completely retained outside of the Trp-flanked core in GW5,19ALP23 (17). While helix fraying near the water exposed termini can be expected, the question as to where the helix distortion begins to occur remains to be answered. To explore this question, we introduced Ala-d4 at positions 5 and 19 in GW7,17ALP23. These amino acids are located outside the two Trp residues, yet still can be expected to be more buried in comparison with A3 and A21. Additionally, both A5 and A19 are capable of participating in a more extensive hydrogen bonding network due to the presence of a full complement of i ± 4 residues. The Δνq values for alanines 5 and 19 in GW7,17ALP23 are indicated as filled symbols in Figure 4C. The signals from A5 in each lipid differ by 5–10 kHz from theoretical values, calculated for a central (Leu-Ala)4.5 helix of GW7,17ALP23. On the other hand, the values for A19 appear quite close to the predicted values for the core helix in DMPC and DOPC, but not in DLPC. The results suggest that A5 is not helical in GW7,17ALP23, whereas it appears that A19 does remain helical in thicker lipids but not in the thinner DLPC. The N-terminal segment appears to be more sensitive to helix unwinding, perhaps in order to accommodate an interfacial location for W7 in each of the lipid bilayer environments.
As a way of visualizing the quality of the GALA analyses, theoretical quadrupolar splittings were calculated and plotted as helical wave plots along with the observed Δνq magnitudes. Several points of interest emerge from examination of the GALA fits (Table 3, Figure 4). The apparent tilt angles of the peptides fall within a relatively small range of about 4°–20°, in contrast to the rather large ranges of inter-tryptophan distances and lipid bilayers thicknesses used in the experiments (Table 4). We note for the respective peptide/lipid combinations that the difference between the inter-Trp distance and the bilayer hydrophobic thickness (not including the head groups) spans a range of ~20 Å (from −12 Å to +8 Å); see Table 4. The average orientations and their uncertainties can also be examined on RMSD contour plots, constructed as a function of the τ and ρ angles (Figure 5). The trend in the tilt angle magnitudes τ is not strictly linear; it appears instead that the tilt magnitudes reach limiting minimum and maximum values. Thus GW7,17ALP23 with the innermost Trps tilts by only ~4–6° in each of the lipids, probably so that the Trps can approach as closely as possible to the bilayer interface while still maintaining the favorable entropy of the peptide precession about the bilayer normal (45). Interestingly, both GW5,19ALP23 and GW3,21ALP23 have 18–21° apparent tilt values in DLPC. While dynamic averaging, beyond what can be treated in the analysis here using the available |Δνq| values, may increase the deduced apparent tilt by about 10° (22, 46, 47), it is nevertheless of interest that an apparent upper limit is reached with these neutral peptides. In terms of peptide dynamics, GW3,21ALP23 exhibits a tendency toward lower Spept values, suggestive of larger amplitude motions when the Trps flank the long (Leu-Ala)8.5 core sequence. As a general trend, the more widely space Trps in the environment of the shorter lipids tend to favor more extensive motions than vice versa.
Table 4.
Comparison of Trp residue spacing in GWx,yALP23 peptides with lipid bilayer membrane hydrophobic thicknessa
| Inter-Trp distance | ||||
|---|---|---|---|---|
| GW3,21ALP23 | GW5,19ALP23 | GW7,17ALP23 | ||
| 27.0 | 21.0 | 15.0 | ||
| Hydrophobic thickness | Difference | |||
| DLPC | 19.5 | +7.5 | +1.5 | −4.5 |
| DMPC | 23.0 | +4.0 | −2.0 | −8.0 |
| DOPC | 27.0 | 0.0 | −6.0 | −12.0 |
Distances are in Å. The inter-Trp distances are based on an increment of 1.5 Å per residue in a standard α-helix. The lipid bilayer hydrophobic thicknesses (from (59)) do not include the head-group regions.
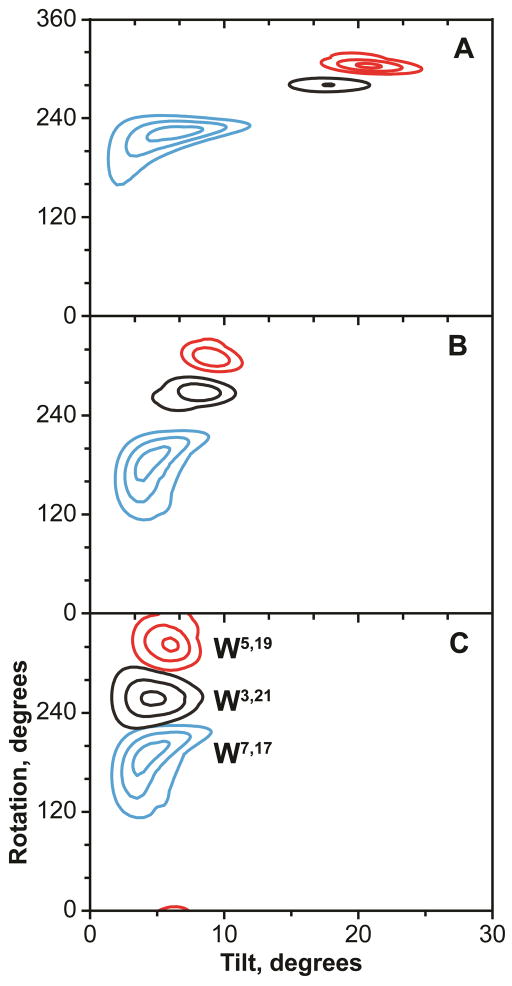
Figure 5.
RMSD contour plots of GW3,21ALP23 (black), GW5,19ALP23 (red) and GW7,17ALP23 (blue) in lipids. A: DLPC; B: DMPC; C: DOPC. Contour levels are plotted every 1 kHz; outer contour corresponds to 3 kHz.
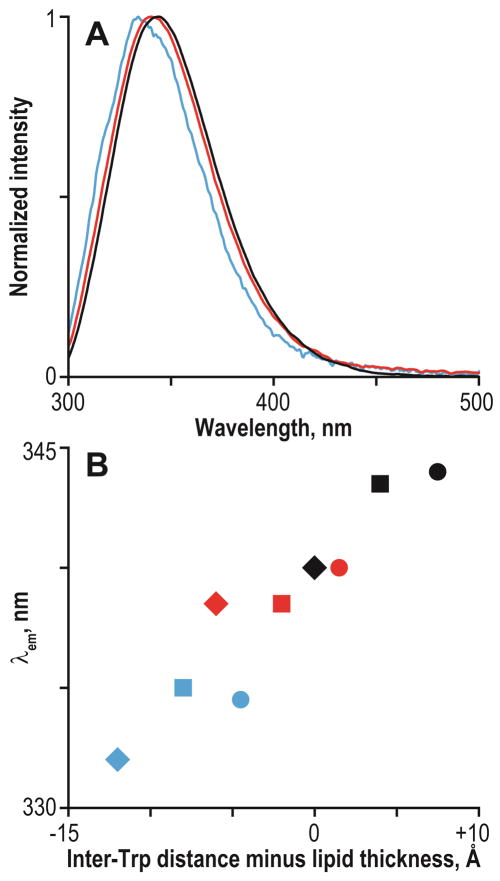
Tryptophans reside preferentially in the interfacial region (27, 48, 49). The region, nevertheless, does not have well-defined borders and spans quite a few angstroms (50). Tryptophan intrinsic fluorescence is a well-known metric of the polarity of the medium in the immediate vicinity of the Trp indole ring, for which more hydrophobic environments shift the emission maximum (λem) to lower wavelengths (blue shifts) (51). This property of Trp has been used extensively to probe the hydrophobicity of its immediate environment (11). The GWx,yALP23 sequences have one Trp residue on each side of the core α-helix; therefore steady-state fluorescence may report the average polarity at the peptide termini (subject to considerations of the quantum yield). Despite the potential limitation of averaging over two indoles, the λem values of GWx,yALP23 peptides in different lipids show good correlation with the difference between the lipid thickness and Trp spacing along the α-helix (Figure 6). The observed λem values span a range of 332–344 nm, indicative of an environment of graded polarity that is intermediate between those of the aqueous phase and the hydrophobic core of the lipid bilayer, as expected for the interfacial region.
Figure 6.
Steady-state fluorescence of GWx,yALP23 peptides. A: emission spectra of GW3,21ALP23 (black), GW5,19ALP23 (red) and GW7,17ALP23 (blue) in DLPC small unilamellar vesicles. B: Tryptophan emission maxima as a function of the difference between the inter-Trp distance and the lipid bilayer thickness (numerical values in Table 4). Peptides are GW3,21ALP23 (black), GW5,19ALP23 (red) and GW7,17ALP23 (blue); lipids are DLPC (circles), DMPC (squares) and DOPC (diamonds).
The fluorescence spectra indicate that the average polarity of the environment around the indole rings changes as a function of the difference between the Trp spacing and the bilayer thickness. This observation raises questions of how peptide tilting could affect the orientations of the Trp side chains. One may speculate that reorientation of the helix axis may alter the indole ring spatial orientations. Alternatively, the Trp side chains may have restricted sets of orientations such that preferential positioning of the Trp indole rings could restrict the peptide tilt. To probe these questions, we have synthesized GWx,yALP23 peptides with deuterium labels on the indole rings, and have recorded solid-state 2H NMR spectra in different lipid bilayer membranes. Deuterium NMR spectra of partially (d2) and fully (d5) labeled indole rings of Trp residues of GW5,19ALP23 are shown in Figure 7. A complete set of spectra for the fully labeled Trps in GW3,21ALP23 and GW7,17ALP23 are included in Figure S10 of the Supporting Information, and spectra for selected partially labeled Trps are shown in Figure S11. Similar to earlier observations for WALP peptides (28), larger quadrupolar splittings are observed for Trps that are near the N-terminus. Indeed, the largest Δνq value observed among the N-terminal Trps of the GWx,yALP23 peptides is 154 kHz, while the corresponding value for the set of C-terminal Trps is only 89 kHz. Alongside the signals from the 2H-labeled Trp residues, the large number of scans sometimes led to background signals from the lipids and residual HDO (25).
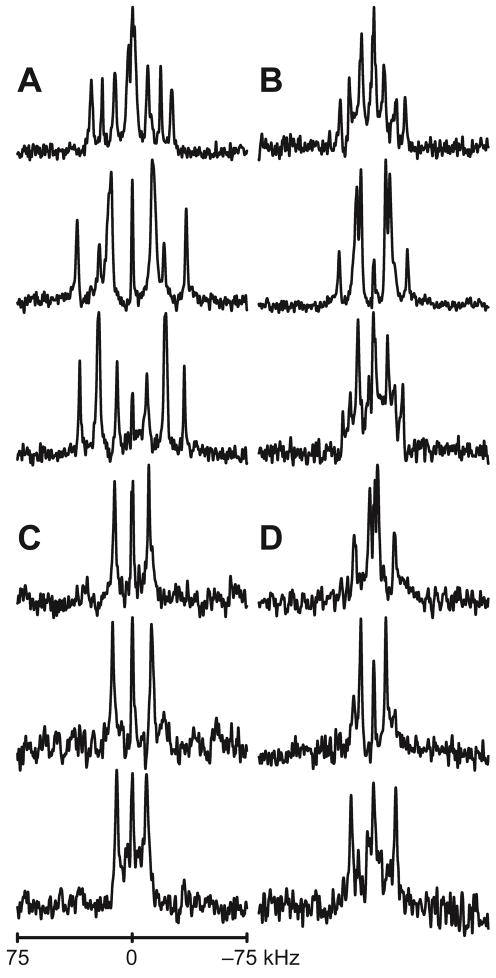
Figure 7.
Deuterium NMR spectra of GW5,19ALP23 labeled at Trp side chain in DLPC, DMPC and DOPC (top to bottom). A: N-terminal Trp5, full deuteration; B: C-terminal Trp19, full deuteration; C: N-terminal Trp5, partial deuteration; D: C-terminal Trp19, partial deuteration. Sample orientation β=90°.
Typically 3–4 resonances were observed for the d5 indole ring and 1–2 for the partially labeled d2 ring. Quadrupolar splitting magnitudes at β=0° and β=90° sample orientations were related by a factor of 1/2, as is expected when there is rapid whole-molecule rotational averaging about the bilayer normal. Nevertheless, large contingents of sometimes quite weak resonances made it hard to observe many of the signals at the β=0° orientation. For this reason, the reported data are derived from the spectra of the samples oriented at β=90°. The values observed at β=90° were then multiplied by a factor of two to simulate the expected values for β=0° (Table 5). Partially labeled samples often allowed the assignment of signals arising from the deuteron attached to indole carbon 2 and sometimes carbon 5. The assignments of these known signals were propagated to other samples where possible, using the least change principle. The remaining resonances were matched by fitting different assignment permutations to a model for the rotated indole ring, and eliminating assignment schemes that led to high values of RMSD or unrealistic order parameters. The order parameter reflects the overall motion experienced by a system; in the case of Trp, therefore, it is feasible to deconvolute Szz into terms for peptide Spept and side chain Ssc motion, such that Szz = Spept × Ssc and Si ∈ [1,0]. As reported above, the dynamics of GWx,yALP23 peptides encompass a range of 0.6–0.9 for Spept (Table 3). This range establishes upper limits for the indole ring Szz. Conversely, the side chain dynamics within the interfacial region are likely to be restricted due to steric hindrance. The value of Ssc is therefore likely to be quite high, which would place a lower limit on the overall Trp Szz value.
Table 5.
Tryptophan side chain CD quadrupolar splitting magnitudes for GWx,yALP23 peptides incorporated in lipid bilayer membranes.a
| Peptide | Lipid | Δνq, kHz, for (specific indole ring positions) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N-terminal Trp | C-terminal Trp | ||||||||
| (2) | (4/7) | (5) | (6) | (2) | (4/7) | (5) | (6) | ||
| GW7,17ALP23 | DLPC | 67 | 63 | 154 | 74 | 54 | 63d | 4 | 63d |
| DMPC | 54b | 81 | 154 | 85 | 49a | 67d | 6b | 67d | |
| DOPC | 42c | 77c | 55 | 67d | 8 | 67d | |||
| GW5,19ALP23 | DLPC | 43b | 8 | 105 | 76 | 61a | 30 | 6b | 85 |
| DMPC | 54b | 58 | 142 | 85 | 54a | 43 | 33b | 89 | |
| DOPC | 39b | 86 | 137b | 88 | 58b | 39 | 8 | 78 | |
| GW3,21ALP23 | DLPC | 64b | 120 | 51b | 15 | 53b | 36 | 4 | 58 |
| DMPC | 64c | 145c | 50bc | 42c | 40bc | 27c | 59c | ||
| DOPC | 62c | 141c | 52bc | 27bc | 19c | 7c | 51c | ||
Values were obtained from the β=90° sample orientation and were multiplied by two. Entries left blank were not observed.
Value is also observed in a partially deuterated sample.
Quadrupolar splittings have not been assigned to individual sites.
Signals not resolved.
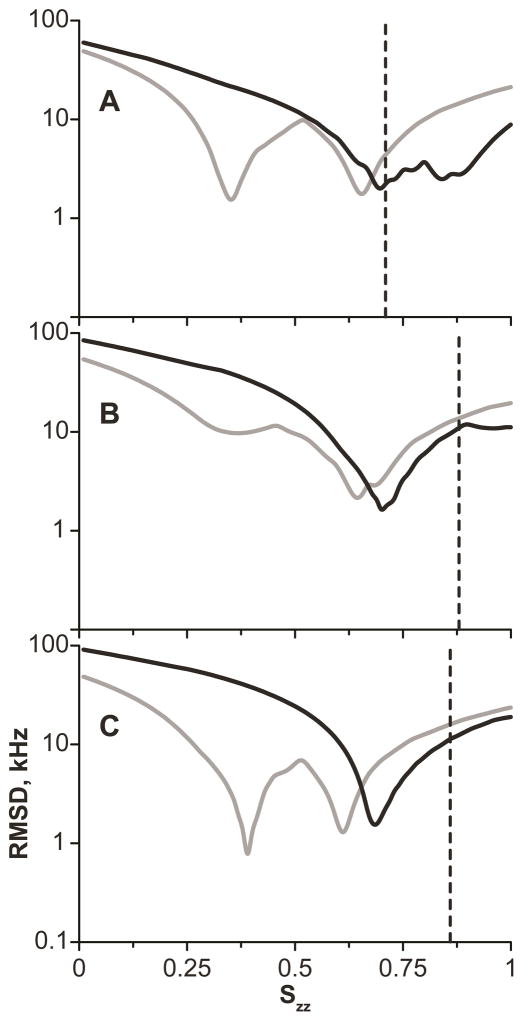
Figure 8 shows RMSD as a function of Szz for GW5,19ALP23. Typically, for each peptide, a unique minimum was observed in such a plot for the N-terminal Trp, largely due to the high magnitude of Δνq at carbon 5, which could not be fitted by lower values of Szz. The smaller range of quadrupolar splittings exhibited by the C-terminal tryptophans makes it possible to fit alternative ring orientations, manifest by several minima in the plots of RMSD versus Szz. Nevertheless, in some cases, such as W19 in DMPC (Figure 8B), or W17 in all three lipids, only one global minimum was observed, with an Szz value close to that observed also for the N-terminal Trp in the same peptide. Based on this finding, when a C-terminal Trp had multiple minima, we considered the one closest to the corresponding N-terminal Trp to be the global minimum, even if alternative fits might yield a slightly lower RMSD (Table 6). The uncertainties of the ρ1 and ρ2 angles at the Szz global minimum can be visualized in similar fashion to the peptide average orientation, using RMSD contour plots (Figure 9). It can be seen that the orientations of the N- and C-terminal Trp residues are distinct and differ primarily in the ρ2 angle.
Figure 8.
RMSD of fitting the N-terminal (black) and C-terminal (gray) tryptophans of GW5,19ALP23 to the rotated indole model (note the logarithmic scale). A: DLPC; B: DMPC; C: DOPC. Angles ρ1, ρ2 were optimized at each Szz value to achieve the lowest possible RMSD. Dashed lines indicate the order parameter of the peptide (Table 3) and correspond to the maximum possible value of the Trp Szz.
Table 6.
Tryptophan side chain free rotation fit values for GWx,yALP23 peptides incorporated in DLPC, DMPC or DOPCa
| Peptide | Lipid | Fit parametersb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N-terminal Trp | C-terminal Trp | ||||||||
| Szz | ρ1 | ρ2 | RMSD | Szz | ρ1 | ρ2 | RMSD | ||
| GW7,17ALP23 | DLPC | 0.68 | 137 | 0 | 2.8 | 0.54 | 165 | 36 | 0.3 |
| DMPC | 0.72 | 140 | 0 | 2.2 | 0.55 | 162 | 40 | 0.3 | |
| DOPC | 0.57 | 165 | 36 | 1.7 | |||||
| GW5,19ALP23 | DLPC | 0.70 | 133 | 30 | 0.9 | 0.65 | 158 | 44 | 1.6 |
| DMPC | 0.71 | 138 | 15 | 1.2 | 0.66 | 143 | 59 | 1.4 | |
| DOPC | 0.68 | 142 | 4 | 1.0 | 0.61 | 160 | 42 | 0.5 | |
| GW3,21ALP23 | DLPC | 0.50 | 11 | 10 | 1.3 | 0.50 | 162 | 41 | 3.5 |
| DMPC | |||||||||
| DOPC | |||||||||
Entries left blank were not fitted. The angles ρ1 and ρ2 are defined in Figure S4 of the Supporting Information; see also (30).
Szz is a dimensionless entity, ρ angles are in degrees, and RMSD is in kHz.
Figure 9.
RMSD contour plots of the N-terminal (black) and C-terminal (gray) tryptophans of GW5,19ALP23 as a function of indole ring orientation. A: DLPC; B: DMPC; C: DOPC. Contour levels are plotted every 1 kHz, with the outer contour corresponding to 5 kHz. Insets show one possible orientation of Trp side chains.
In the case of W17 in GW7,17ALP23, only three pairs of resonances can be identified in the spectra (Figure S10). As the Δνq range for the C-terminal tryptophan residues is fairly small, it is conceivable that the missing signal is present, but not resolved due to spectral overlap. To account for this possibility, in each of the lipids we performed three separate fits of the W17 data by entering one of the quadrupolar splittings twice. Solutions were rejected on the previously described principles; in addition, the best fits among the different lipids were compared, as the C-terminal Trp in GW5,19ALP23 has shown little variation in different bilayer membranes. A possible assignment with the intermediate Δνq value involving overlap of deuterons 4/7 and 2 was discarded for reasons of very different ρ1 and ρ2 angles (~70° and ~30°), in comparison with W19. Conversely, the conditions were readily fulfilled if the outermost quadrupolar splitting resulted from an overlap between deuterons at positions 4/7 and 6, which led to similar orientation angles and Szz values for W17 and W19. Furthermore, in the 2H NMR spectra of W17, the outermost signals typically were strongest. While the intensity alone is problematic to interpret in deuterium NMR spectroscopy (due to a number of factors, including radio frequency power profile, contributions from powder pattern, etc.), the consideration of peak intensity nevertheless provides a valuable clue in combination with other factors. N-terminal W7 produced four signals in DLPC and DMPC, allowing the assignments, but only two resolved resonances in DOPC, which did not provide sufficient restraints for the analysis.
For GW3,21ALP23, fits for both tryptophans were possible only in DLPC. Broad overlapped peaks for W3 in DMPC and DOPC make it hard to extract or assign Δνq values. Likewise, uncertainties in peak positions and observation of fewer than four resonances complicate the analysis also for W21. While combinations of plausible Szz, ρ1, ρ2 that are similar to the N- or C-terminal Trps in other peptides can be obtained, it is not possible to exclude with confidence alternative assignments and consequently alternative ring orientations.
Discussion
We have investigated the responses of model peptides having identical amino acid composition and similar sequences, but different spacing between a pair of Trp residues that flank a central (Leu-Ala)n core helix, to lipid bilayer environments of differing lipid thickness. [We note in passing that when only two Trps are present in the sequence, the relatively polar Gly residues at positions 2 and 22 are of practical significance for the ease of synthesis, purification and handling of the GWALP23 family peptides.] All of the peptides were able to incorporate into the DLPC, DMPC and DOPC bilayer membranes. In every lipid tested, furthermore, each peptide retained a well-defined tilt angle, even in the potentially problematic case of a short spacing of the Trp residues, where tilting in a thicker lipid such as DOPC would be expected to drag Trp7 and Trp17 away from the interface in the direction of the bilayer center. A similar result has been observed in umbrella sampling simulations of WALP peptides, and was explained in terms of a favorable entropy contribution arising from peptide precession about the lipid bilayer normal (45, 52).
Trp residues show preference for the lipid-water interface in membrane proteins of known structure. A stabilization energy of about 2 kcal/mol has been estimated per interfacial Trp residue in the E. coli OmpA protein (15). In a “five-slab” membrane model, both Trp and Tyr partition to an 8-Å slab corresponding to the lipid head-group region (53). In 29 integral membrane proteins, the Trp and Tyr residues show saddle-like distributions with respect to the bilayer center, defining aromatic “belts” that are about 10 Å wide and whose midpoints are separated by some 20–30 Å (6). In comparison, since the helical repeat is 1.5 Å, the Trp residue separation in our peptides encompasses a similar range, from about 15 Å in GW7,17ALP23 to about 27 Å in GW3,21ALP23. From the low end to the high end of the range of aromatic residue separation, our results indicate notable differences in peptide properties.
Whole-peptide orientations and dynamics
The analysis of GW3,21ALP23 behavior in lipid bilayers suggests that the longer (Leu-Ala)8.5 stretch undergoes more extensive whole-body motion relative to its shorter (Leu-Ala)6.5 counterpart in GW5,19ALP23 (Table 3). To gain additional insights into the nature of such motion, we have performed an analysis of explicit dynamics for both peptides in DLPC, using the 2H NMR data. The semi-static analysis (Table 3) has suggested that the tilt angles of the two peptides are similar in DLPC, yet the Szz values vary. To facilitate direct comparison, quadrupolar splittings from the identical alanine positions (7, 9, 11, 13, 15 and 17) were fitted for the two systems. The overall shapes of the tilt and rotation distributions (στ and σρ respectively) are similar for the W3,21 and W5,19 peptides in DLPC, with only moderate oscillations around the average values. The solution area for GW5,19ALP23 is nevertheless more compact and shifted toward the lower στ range (Figure 10). The somewhat higher στ range for GW3,21ALP23 is consistent with the correspondingly lower value of Szz from the semi-static analysis (Table 3). The variations around the average ρ angle are close and fairly small for both the W3,21 and W5,19 peptides, indicating that neither peptide (each with only a single Trp residue near each terminus) undergoes extensive reorientation around the helix axis (in contrast to the extensive reorientation that is observed for WALP peptides when two Trps are present at each terminus). Furthermore we note the close correspondence between the sets of average orientations of GWx,yALP23 peptides that are deduced from the semi-static (variable order parameter) and explicit (Gaussian distributions of τ and ρ) treatments of the whole-body dynamics. Both methods should be used with caution, as the semi-static approach will tend to overly simplify the dynamics, while the Gaussian approach introduces additional variable(s) in the analysis procedure, leading to a requirement for additional data points. The requirement can sometimes be met by combining the 2H methyl quadrupolar splittings with 15N derived restraints (39, 54) or, in selected cases, with backbone deuteron signals (44). It is further of note that a more extensive treatment of GW5,19ALP23 in DLPC—using combined 2H and 15N data with Gaussian dynamics—has led to the same tilt angle as found in a semi-static analysis (17).
Figure 10.
Explicit dynamics analysis of GW5,19ALP23 (A, C) and GW3,21ALP23 in DLPC (B, D), showing the standard deviations of Gaussian distributions (A, B) and their centers (C, D). Six alanine residues were used for the analysis (see text). Dashed lines in A and B indicate the best fit στ and σρ, which were used for generating the plots in C and D, respectively. The color scale is identical between A and B (0 to 17 kHz, blue to red) and between C and D (0 to 22 kHz, blue to red). Color increments are 1 kHz; solid line in C, D is drawn at 3 kHz level. Best fits (τ0, στ, ρ0, σρ) are (25, 16, 303, 36) for GW5,19ALP23 and (25, 20, 278, 48) for GW3,21ALP23.
Earlier we established that among different anchoring residues at the bilayer–water interface, tryptophans are major determinants of the transmembrane peptide orientation (17). The design of GWx,yALP23 offers a way to investigate further details, due to the similar projections of the N- and C-terminal Trps from one side of the helical wheel. Additionally, the radial positions of the two Trps in GW7,17ALP23 resemble closely those in GW3,21ALP23, while the Trps in GW5,19ALP23 project from a different face of the helix (Figure 1). Indeed, we find that GW7,17ALP23 and GW5,19ALP23 have nearly opposite ρ angles (Table 3), thus matching the change in the Trp radial positions, with each peptide tilting approximately in the direction of the tryptophans. Due to the similar projections of the two tryptophans, it is not yet possible to say whether either the N- or the C-terminal Trp may have a dominant role in determining the helix orientation in a lipid bilayer, although the advantageous design of GWALP23 will allow future testing of this feature. GW3,21ALP23, on the other hand, does not seem to follow the same trend, perhaps because of the close proximity of W3 and W21 to the peptide termini. Indeed the fraying of the termini could impact the net radial projection of one or both Trp residues, and thereby alter the preferred orientation of the backbone helix.
Minimum and maximum helix tilt angles in lipid bilayers
Examination of the GWx,yALP23 tilt angles reveals some correlation, but not a strictly linear trend between the Trp separation and the tilt magnitude (Table 3). This is particularly noticeable when the Trp spacing is small in a relatively thick bilayer, and vice versa.. Thus a minimum tilt angle of about 4° is observed for GW7,17ALP23 in DOPC and DMPC, with only a slight increase in DLPC. On the other end of the scale are GW5,19ALP23 and GW3,21ALP23 in DLPC, both tilting by approximately 20°. We note that the tilt angle of a single-span transmembrane peptide can be as much as 30°, when a charged residue is present within the core helix (44), and possibly larger in helical bundles, where protein-protein interactions become vital. On the other hand, in cases such as GWx,yALP23 where the peptide tilt is governed largely by the lipid interactions of aromatic anchoring residues, it seems that minimum and maximum values of the tilt magnitude are observed.
We note moreover that the acyl chain unsaturation in DOPC will influence the lipid lateral pressure profile (16) at the same time that the longer acyl chains increase the DOPC bilayer hydrophobic thickness (55). Indeed, both bilayer thickness and lateral pressure appear to be important for the protein shape changes that accompany the gating of the MscL mechanosensitive channel (56). Nevertheless, the rather flat behavior for the tilt of e.g. GW7,17ALP23 among the three lipids tested here suggests that the impact of lateral pressure as well as bilayer thickness upon helix tilt may be dampened in specific cases, particularly for neutral peptides with only aromatic anchor residues. We note furthermore that the Δνq magnitudes, for GW5,19ALP23 in DMPC, show little variation between 1/40 and 1/200 peptide/lipid (Figure S9). In particular cases where the segment tilt depends only minimally on the packing density or the bilayer thickness, additional mechanisms, such as (partial) helix unwinding and/or reorientation of the side-chain (indole ring) anchors, seem to help the peptides adapt to the lipid environment. These further mechanisms also could be important for conformational changes that relate to membrane protein function.
Indole ring orientations
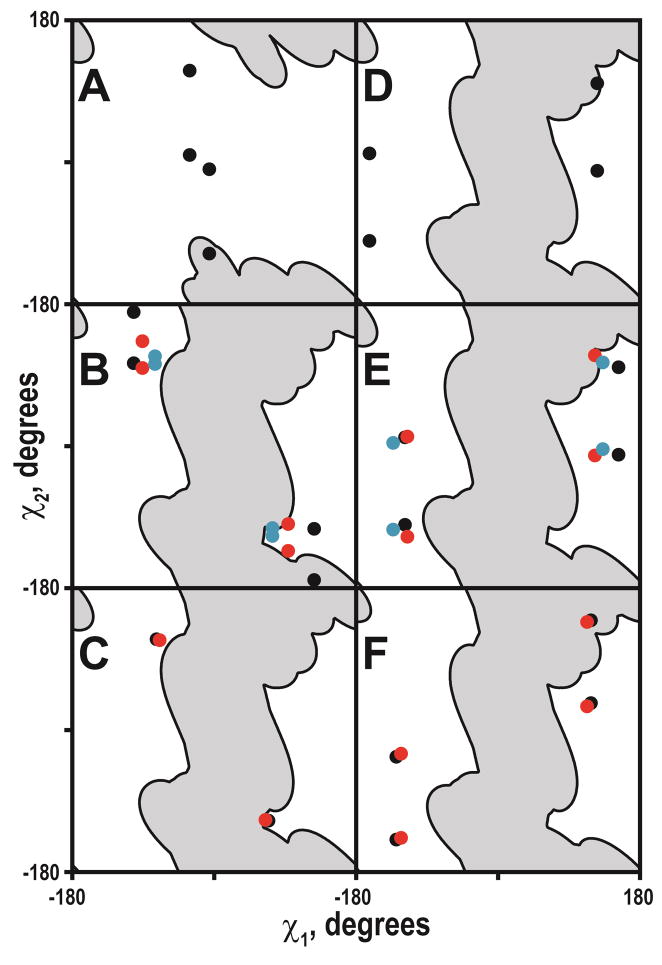
Deuterium labeling of the Trp residues allowed for defining the orientations of the indole ring moieties with respect to the membrane normal in several cases (Table 6). Both the N- and C-terminal Trps are tightly clustered, albeit in different regions of conformational space. To visualize the indole ring orientations with respect to the tilted peptides, the (ρ1, ρ2) backbone-independent angles of 3-methyl-indole were converted to the Trp side chain (χ1, χ2) torsion angles (Figure 11). Similar to the backbone-independent analysis, eight combinations of (χ1, χ2) lead to the identical orientation of the indole ring with respect to the applied magnetic field. However, in order to serve as a membrane anchor, the Trp side chain should be positioned so that the NεH bond vector points away from the bilayer center. This feature restricts the number of the ρ1,2 (or χ1,2) sets to four possible combinations (Figure S4), as the NεH bond vector should be directed along the positive Z-axis for the N-terminal Trps, but along the negative Z-axis for the C-terminal Trps (see also (28, 30, 57, 58)). Additionally, in cases where ρ2 is close to zero, the number of possible solutions is further reduced by half due to the symmetry collapse (Figure S4).
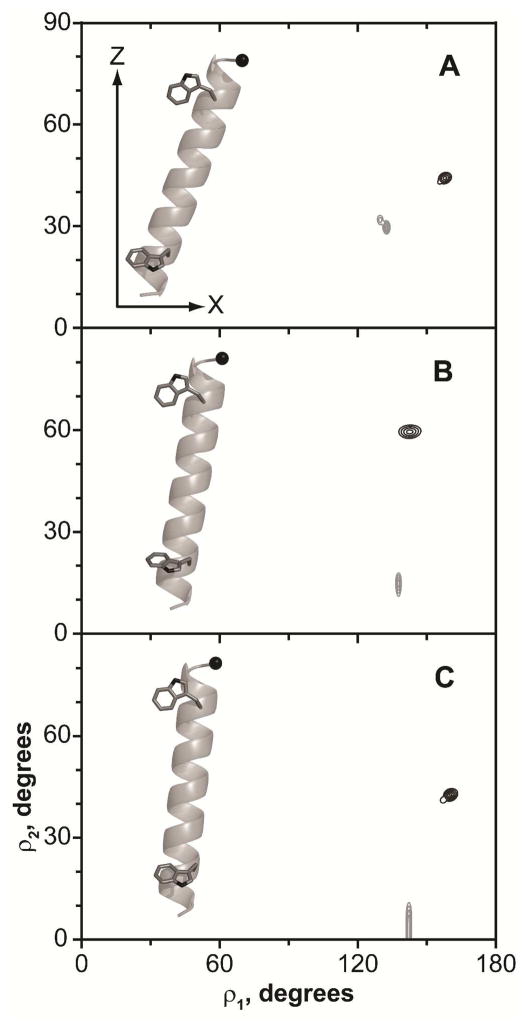
Figure 11.
Tryptophan side chain torsion angles. A: W3; B: W5; C: W7; D: W21; E: W19; F: W17. Gray regions indicate steric hindrance areas. Lipids are DLPC (black), DMPC (red) and DOPC (blue).
The possible side chain torsion angles of the N-terminal Trps fall into two major clusters (Figure 11, ABC), with the indole carbon-carbon “bridge” (the common bond between the 5- and 6-membered rings) essentially either co-aligned with the helix axis (negative χ1, positive χ2 cluster), or nearly perpendicular to it (positive χ1, negative χ2 cluster). Interestingly, both possible solutions are located close to the steric hindrance areas, suggesting that further changes of the Trp orientation are unfavorable, as they would include rearrangements of the backbone atoms. On the other hand, the orientations of the C-terminal Trps can be described by four possible combinations of (χ1, χ2) angles, in each case the “bridge” being virtually perpendicular to the helix axis (Figure 11, DEF). While it appears that the C-terminal Trps are located farther from steric hindrance regions, it should be noted that these areas are approximate and are likely to change upon deviation from α-helical geometry, which was noted for some of the GWx,yALP23 peptides (see Results). While deuterium NMR alone does not allow for distinguishing among the possible solutions, the choices of side-chain torsion angles could be further refined through distance measurements obtained by solution or magic angle spinning NMR.
The analysis of Trp side chain geometry has been previously reported for WALP peptides (having two sequential Trp residues in close proximity to the N- and C-termini) in DMPC and its ether analogue (28). Certain similarities can be seen for the Trps at the N- or C-termini of WALP and GWx,yALP23 peptides. The dynamic behavior of the N-terminal Trps is closely similar between the two systems, the order parameters being approximately 0.7. In the case of the C-terminal Trps, two minima have been observed for WALP peptides, with Szz values of 0.45 and 0.6. The results for the GWx,yALP23 peptides suggest that the minimum with the higher order parameter better reflects the state of the system. In terms of the average indole orientation with respect to the applied magnetic field, three of the Trps in WALP19 adopt ρ1 and ρ2 angles that resemble closely those found for corresponding Trps in GWx,yALP23 (Figure S12). Interestingly, only Trp18 in WALP19 seems not to fall within the cluster defined by the other C-terminal Trps that have been examined in WALP and GWALP peptides, mainly due to a difference in the ρ1 angle.
GW3,21ALP23 is similar to a WALP sequence in terms of close Trp proximity to the helix termini. It is notable that the Trp steric hindrance areas are smaller for this peptide. This result is particularly manifest for W3 due to the direction of the Cα-Cβ bond vector pointing toward the N-terminus, thereby effectively shifting all side chains marginally closer to the N-terminus. Interestingly, for GW3,21ALP23 it was possible to assign the indole quadrupolar splittings only in DLPC, as the resonances were broader and less well defined in other lipids, suggesting more complex dynamics. The steric hindrance patterns for WALP peptides are expected to be complicated due to the locations of two bulky Trp side chains next to each other, meaning that the orientation of each indole moiety could be influenced by the adjacent tryptophan. If such indole-indole restrictions are nevertheless ignored, the Trps at the C-terminus are expected to have less conformational freedom, as a χ1 angle near 0° would lead to the severe clashes with the backbone of the (i – 3) and (i – 4) residues. The steric hindrance area for the C-terminal Trps, therefore, changes only marginally as Trp approaches the peptide terminus (Figure 11, DEF).
Overall, the variations in the indole ring orientations were smaller than the variations in helix tilt angles for the different peptide-lipid combinations. The results imply that the Trp side chain undergoes changes in the (χ1, χ2) torsion angles to compensate in part for changes in the helix tilt. By this mechanism a particular Trp will be able to maintain a similar indole orientation and a similar lipid interaction, which presumably provides effective anchoring, as the peptide changes its tilt. A single Trp side chain near the N-terminal samples a larger range of conformational space in response to a change in the lipid thickness, although the primary adjustment would seem to involve only one of the χ angles (Figure 11). The response of a C-terminal Trp appears to be less systematic and to involve both χ angles to similar extents.
Concluding perspective
GWALP23 peptide isomers having different core helix lengths of 10, 14 or 18 residues between a single pair of anchoring Trp residues display systematic responses to lipid bilayer membranes of differing thickness and extent of acyl-chain unsaturation. The responses include adjustments to the peptide tilt between apparent limits of about 5° and just over 20° with respect to the bilayer normal. Further adjustments to the helix tilt seem to be dampened by potentially compensating adjustments involving the indole ring orientations and the whole-body dynamics, along with variable fraying of the ends of the helix. The peptide responses to differing lipid bilayer environments therefore involve combinations of multiple factors. Similar principles are likely to govern the behavior and function of the transmembrane domains of single-span membrane proteins.
Supplementary Material
Acknowledgments
We thank James Hinton and Denise Greathouse for helpful discussions.
Abbreviations
- CD
circular dichroism
- DLPC
1,2-dilauroyl-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- Fmoc
fluorenylmethoxycarbonyl
- GALA
Geometric Analysis of Labeled Alanines
- RMSD
root mean squared deviation
- TFA
trifluoroacetic acid
Footnotes
This work was supported in part by NSF grant MCB-0841227 and by the Arkansas Biosciences Institute. The peptide and NMR facilities were supported by NIH grants RR31154 and RR16460.
Supporting Information Available
Proton NMR spectra of Fmoc-Trp-d2. Definition of ρ angles. Physical data for GWx,yALP23 peptides (HPLC, mass spectrum). Deuterium NMR spectra of peptides in DLPC, DMPC, DOPC. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys J. 2006;91:3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kandasamy SK, Lee DK, Nanga RP, Xu J, Santos JS, Larson RG, Ramamoorthy A. Solid-state NMR and molecular dynamics simulations reveal the oligomeric ion-channels of TM2-GABAA stabilized by intermolecular hydrogen bonding. Biochim Biophys Acta. 2009;1788:686–695. doi: 10.1016/j.bbamem.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Schick S, Chen L, Li E, Lin J, Koper I, Hristova K. Assembly of the M2 tetramer is strongly modulated by lipid chain length. Biophys J. 2010;99:1810–1817. doi: 10.1016/j.bpj.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 5.Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RA. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 6.Ulmschneider MB, Sansom MSP. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 7.de Planque MR, Kruijtzer JA, Liskamp RM, Marsh D, Greathouse DV, Koeppe RE, 2nd, de Kruijff B, Killian JA. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane α-helical peptides. J Biol Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 8.Davis JH, Clare DM, Hodges RS, Bloom M. Interaction of a synthetic amphiphilic polypeptide and lipids in a bilayer structure. Biochemistry. 1983;22:5298–5305. [Google Scholar]
- 9.Killian JA, Salemink I, de Planque MR, Lindblom G, Koeppe RE, 2nd, Greathouse DV. Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane α-helical peptides: importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry. 1996;35:1037–1045. doi: 10.1021/bi9519258. [DOI] [PubMed] [Google Scholar]
- 10.Harzer U, Bechinger B. Alignment of lysine-anchored membrane peptides under conditions of hydrophobic mismatch: a CD, 15N and 31P solid-state NMR spectroscopy investigation. Biochemistry. 2000;39:13106–13114. doi: 10.1021/bi000770n. [DOI] [PubMed] [Google Scholar]
- 11.Krishnakumar SS, London E. Effect of sequence hydrophobicity and bilayer width upon the minimum length required for the formation of transmembrane helices in membranes. J Mol Biol. 2007;374:671–687. doi: 10.1016/j.jmb.2007.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Wel PC, Strandberg E, Killian JA, Koeppe RE., 2nd Geometry and intrinsic tilt of a tryptophan-anchored transmembrane α-helix determined by 2H NMR. Biophys J. 2002;83:1479–1488. doi: 10.1016/S0006-3495(02)73918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strandberg E, Ozdirekcan S, Rijkers DT, van der Wel PC, Koeppe RE, 2nd, Liskamp RM, Killian JA. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys J. 2004;86:3709–3721. doi: 10.1529/biophysj.103.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, 2nd, Separovic F, Watts A, Killian JA. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 15.Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 16.Binder H, Gawrisch K. Effect of unsaturated lipid chains on dimensions, molecular order and hydration of membranes. J Phys Chem B. 2001;105:12378–12390. [Google Scholar]
- 17.Vostrikov VV, Daily AE, Greathouse DV, Koeppe RE., 2nd Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J Biol Chem. 2010;285:31723–31730. doi: 10.1074/jbc.M110.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrache HI, Zuckerman DM, Sachs JN, Killian JA, Koeppe RE, II, Woolf TB. Hydrophobic matching mechanism investigated by molecular dynamics simulations. Langmuir. 2002;18:1340–1351. [Google Scholar]
- 19.Liu J, Wang D, Zheng Q, Lu M, Arora PS. Atomic structure of a short α-helix stabilized by a main chain hydrogen-bond surrogate. J Am Chem Soc. 2008;130:4334–4337. doi: 10.1021/ja077704u. [DOI] [PubMed] [Google Scholar]
- 20.Froyd-Rankenberg JM, Greathouse DV, Koeppe RE., 2nd Half- anchored WALP peptides: Effect of anchor position on peptide orientation. Biophys J. 2009;96:455a–456a. [Google Scholar]
- 21.Vostrikov VV, Grant CV, Daily AE, Opella SJ, Koeppe RE., 2nd Comparison of “Polarization Inversion with Spin Exchange at Magic Angle” and “Geometric Analysis of Labeled Alanines” methods for transmembrane helix alignment. J Am Chem Soc. 2008;130:12584–12585. doi: 10.1021/ja803734k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strandberg E, Esteban-Martin S, Salgado J, Ulrich AS. Orientation and dynamics of peptides in membranes calculated from 2H-NMR data. Biophys J. 2009;96:3223–3232. doi: 10.1016/j.bpj.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt A, Rougier L, Reat V, Jolibois F, Saurel O, Czaplicki J, Killian JA, Milon A. Order parameters of a transmembrane helix in a fluid bilayer: case study of a WALP peptide. Biophys J. 2010;98:1864–1872. doi: 10.1016/j.bpj.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozdirekcan S, Rijkers DT, Liskamp RM, Killian JA. Influence of flanking residues on tilt and rotation angles of transmembrane peptides in lipid bilayers. A solid-state 2H NMR study. Biochemistry. 2005;44:1004–1012. doi: 10.1021/bi0481242. [DOI] [PubMed] [Google Scholar]
- 25.Vostrikov VV, Hall BA, Greathouse DV, Koeppe RE, 2nd, Sansom MSP. Changes in transmembrane helix alignment by arginine residues revealed by solid-state NMR experiments and coarse-grained MD simulations. J Am Chem Soc. 2010;132:5803–5811. doi: 10.1021/ja100598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connell AM, Koeppe RE, 2nd, Andersen OS. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 1990;250:1256–1259. doi: 10.1126/science.1700867. [DOI] [PubMed] [Google Scholar]
- 27.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 28.van der Wel PC, Reed ND, Greathouse DV, Koeppe RE., 2nd Orientation and motion of tryptophan interfacial anchors in membrane-spanning peptides. Biochemistry. 2007;46:7514–7524. doi: 10.1021/bi700082v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ten Kortenaar PBW, Van Dijk BG, Peeters JM, Raaben BJ, Adams PJHM, Tesser GI. Rapid and efficient method for the preparation of Fmoc-amino acids starting from 9-fluorenylmethanol. Int J Pept Protein Res. 1986;27:398–400. [Google Scholar]
- 30.Koeppe RE, 2nd, Sun H, van der Wel PC, Scherer EM, Pulay P, Greathouse DV. Combined experimental/theoretical refinement of indole ring geometry using deuterium magnetic resonance and ab initio calculations. J Am Chem Soc. 2003;125:12268–12276. doi: 10.1021/ja035052d. [DOI] [PubMed] [Google Scholar]
- 31.Greathouse DV, Koeppe RE, 2nd, Providence LL, Shobana S, Andersen OS. Design and characterization of gramicidin channels. Methods Enzymol. 1999;294:525–550. doi: 10.1016/s0076-6879(99)94031-4. [DOI] [PubMed] [Google Scholar]
- 32.Davis JH, Jeffrey KR, Bloom M, Valic MI, Higgs TP. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem Phys Lett. 1976;42:390–394. [Google Scholar]
- 33.Pulay P, Scherer EM, van der Wel PC, Koeppe RE., II Importance of tensor asymmetry for the analysis of 2H NMR spectra from deuterated aromatic rings. J Am Chem Soc. 2005;127:17488–17493. doi: 10.1021/ja054935x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun H, Greathouse DV, Andersen OS, Koeppe RE., 2nd The preference of tryptophan for membrane interfaces: insights from N-methylation of tryptophans in gramicidin channels. J Biol Chem. 2008;283:22233–22243. doi: 10.1074/jbc.M802074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 36.Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. Relationship of sidechain hydrophobicity and α-helical propensity on the stability of the single-stranded amphipathic α-helix. J Pept Sci. 1995;1:319–329. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 37.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 38.Granseth E, von Heijne G, Elofsson A. A study of the membrane-water interface region of membrane proteins. J Mol Biol. 2005;346:377–385. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 39.Aisenbrey C, Bechinger B. Tilt and rotational pitch angle of membrane-inserted polypeptides from combined 15N and 2H solid-state NMR spectroscopy. Biochemistry. 2004;43:10502–10512. doi: 10.1021/bi049409h. [DOI] [PubMed] [Google Scholar]
- 40.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 41.Bechinger B, Skladnev DA, Ogrel A, Li X, Rogozhkina EV, Ovchinnikova TV, O’Neil JD, Raap J. 15N and 31P solid-state NMR investigations on the orientation of zervamicin II and alamethicin in phosphatidylcholine membranes. Biochemistry. 2001;40:9428–9437. doi: 10.1021/bi010162n. [DOI] [PubMed] [Google Scholar]
- 42.Salnikov ES, Friedrich H, Li X, Bertani P, Reissmann S, Hertweck C, O’Neil JD, Raap J, Bechinger B. Structure and alignment of the membrane-associated peptaibols ampullosporin A and alamethicin by oriented 15N and 31P solid-state NMR spectroscopy. Biophys J. 2009;96:86–100. doi: 10.1529/biophysj.108.136242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salnikov ES, Bechinger B. Lipid-controlled peptide topology and interactions in bilayers: structural insights into the synergistic enhancement of the antimicrobial activities of PGLa and magainin 2. Biophys J. 2011;100:1473–1480. doi: 10.1016/j.bpj.2011.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vostrikov VV, Hall BA, Sansom MSP, Koeppe RE., 2nd . “Rescue” of a central arginine in a transmembrane peptide by changing the placement of anchor residues. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, Im W. Transmembrane helix tilting: insights from calculating the potential of mean force. Phys Rev Lett. 2008;100:018103. doi: 10.1103/PhysRevLett.100.018103. [DOI] [PubMed] [Google Scholar]
- 46.Ozdirekcan S, Etchebest C, Killian JA, Fuchs PFJ. On the orientation of a designed transmembrane peptide: toward the right tilt angle? J Am Chem Soc. 2007;129:15174–15181. doi: 10.1021/ja073784q. [DOI] [PubMed] [Google Scholar]
- 47.Esteban-Martin S, Salgado J. The dynamic orientation of membrane-bound peptides: bridging simulations and experiments. Biophys J. 2007;93:4278–4288. doi: 10.1529/biophysj.107.113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacCallum JL, Bennett WF, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 50.Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; 2006. [Google Scholar]
- 52.Kim T, Im W. Revisiting hydrophobic mismatch with free energy simulation studies of transmembrane helix tilt and rotation. Biophys J. 2010;99:175–183. doi: 10.1016/j.bpj.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sengupta D, Smith JC, Ullmann GM. Partitioning of amino-acid analogues in a five-slab membrane model. Biochim Biophys Acta. 2008;1778:2234–2243. doi: 10.1016/j.bbamem.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Bechinger B, Resende JM, Aisenbrey C. The structural and topological analysis of membrane-associated polypeptides by oriented solid-state NMR spectroscopy: Established concepts and novel developments. Biophys Chem. 2011;153:115–125. doi: 10.1016/j.bpc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Marsh D. Energetics of hydrophobic matching in lipid-protein interactions. Biophys J. 2008;94:3996–4013. doi: 10.1529/biophysj.107.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samuli Ollila OH, Louhivuori M, Marrink SJ, Vattulainen I. Protein shape change has a major effect on the gating energy of a mechanosensitive channel. Biophys J. 2011;100:1651–1659. doi: 10.1016/j.bpj.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koeppe RE, 2nd, Killian JA, Greathouse DV. Orientations of the tryptophan 9 and 11 side chains of the gramicidin channel based on deuterium nuclear magnetic resonance spectroscopy. Biophys J. 1994;66:14–24. doi: 10.1016/S0006-3495(94)80748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu W, Lazo ND, Cross TA. Tryptophan dynamics and structural refinement in a lipid bilayer environment: solid state NMR of the gramicidin channel. Biochemistry. 1995;34:14138–14146. doi: 10.1021/bi00043a019. [DOI] [PubMed] [Google Scholar]
- 59.de Planque MR, Killian JA. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol Membr Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.