Abstract
Bub3 is a conserved component of the mitotic spindle assembly complex. The protein is essential for early development in Bub3 gene-disrupted mice, evident from their failure to survive beyond day 6.5–7.5 postcoitus (pc). Bub3 null embryos appear normal up to day 3.5 pc but accumulate mitotic errors from days 4.5–6.5 pc in the form of micronuclei, chromatin bridging, lagging chromosomes, and irregular nuclear morphology. Null embryos treated with a spindle-depolymerising agent fail to arrest in metaphase and show an increase in mitotic disarray. The results confirm Bub3 as a component of the essential spindle checkpoint pathway that operates during early embryogenesis.
Keywords: Mitotic checkpoint, kinetochore, chromosome missegregation, mouse transgenic
The accurate attachment of chromosomes to the mitotic spindle via the kinetochore is vital for correct segregation of the genetic material into daughter cells. This process is overseen by a feedback-response mechanism, commonly known as the mitotic spindle checkpoint (for review, see Skibbens and Hieter 1998; Amon 1999). If defects are detected in the spindle, mitosis is halted to ensure that chromosomes achieve bipolar alignment before the cell proceeds through to anaphase.
Genetic screens in the budding yeast Saccharomyces cerevisiae have identified a series of genes (BUB1, BUB2, BUB3, MAD1, MAD2, MAD3, and MPS1) that fail to arrest in response to spindle damage (Hoyt et al. 1991; Li and Murray 1991; Weiss and Winey 1996). In the presence of microtubule-depolymerising drugs, the mutants accumulate severe mitotic errors because of their premature exit into anaphase. Higher eukaryotes contain several functional orthologs of these genes, including Bub1, Bub3, BubR1/Mad3, Mad1, and Mad2 (Chen et al. 1996; Li and Benezra 1996; Taylor and McKeon 1997; Basu et al. 1998; Chen et al. 1998; Taylor et al. 1998; Basu et al. 1999). Mutations and immunodepletion experiments on Bub1, BubR1/Mad3, Mad1, and Mad2 have shown that cells are unable to block at mitosis in response to microtubule-depolymerising agents, resulting in a premature exit into anaphase before the chromosomes have properly aligned (Chen et al. 1996; Li and Benezra 1996; Taylor and McKeon 1997; Cahill et al. 1998; Chen et al. 1998; Gorbsky et al. 1998; Waters et al. 1998; Basu et al. 1999; Chan et al. 1999). In humans, dominant-negative mutations of the Bub1 gene show a chromosomal instability phenotype in colorectal cancer cell lines (Cahill et al. 1998). These cell lines also fail to arrest at metaphase when treated with microtubule-depolymerising drugs.
Bub3 is found in most eukaryotes through evolution (Efimov and Morris 1998; Taylor et al. 1998; Martinez-Exposito et al. 1999). It contains four conserved WD40 repeats that are found in many proteins with diverse functions thought to be involved in protein–protein interactions (Neer et al. 1994). During mitosis, Bub3 appears on kinetochores during prophase, diminishing in quantity by metaphase. When kinetochores are unattached to the spindle, or lagging, the amount of kinetochore-associated Bub3 antigen increases (Martinez-Exposito et al. 1999). In higher eukaryotes, no mutation or depletion studies on Bub3 have been described. In this study, we have performed a genetic disruption of the Bub3 gene in mouse, using gene-targeting techniques. We describe a lethal phenotype for the gene-disrupted mice and show that Bub3 is an essential component of the mitotic spindle checkpoint pathway that is required for mitotic fidelity.
Results
Disruption of the Bub3 gene
To disrupt the mouse Bub3 gene, a promoterless targeting construct was used to obtain a high level of homologous recombination. The selection cassette, which contained the splice-acceptor/IRES/lacZ-neomycin resistance gene, was used to delete exons 2 and 3 (Fig. 1A). Only the first 65 amino acids of the 326–amino acid Bub3 protein would be correctly translated following gene disruption with the selection cassette. This disruption would result in the loss of three of the four WD40 repeat domains and the Bub1–interacting kinetochore domain and thus was expected to abolish any Bub3 activity at the kinetochore (Taylor et al. 1998). When theconstruct was transfected into two different ES cell lines,129/1 or W9.5, 20 of 107 (19%) neomycin-resistant cell lines were found to have the desired targeting event (Fig. 1B).
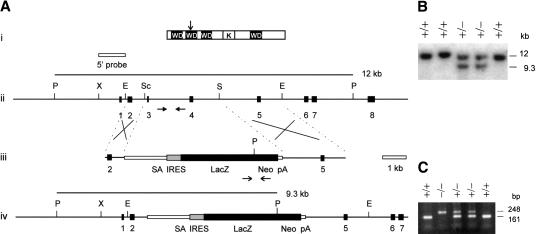
Figure 1.
Targeted disruption of the mouse Bub3 gene. (A) Gene disruption construct and restriction maps. (i) Mouse Bub3 protein showing positions of four WD40 motifs (WD) and the Bub1-kinetocore-interacting domain (K) (Taylor et al. 1998; Martinez-Exposito et al. 1999). The disruption site is indicated by the vertical arrow. Restriction-enzyme maps for (ii) the Bub3 gene covering exons 1 to 8, (iii) the neomycin-resistance gene targeting construct, and (iv) the Bub3 locus following targeted disruption. Black boxes represent exons. The selectable marker cassette contained in the targeting construct consists of a splice acceptor site (SA), a picornaviral internal ribosome-entry site (IRES), a lacZ–neomycin fusion gene (beta-geo) and a SV40 polyadenylation sequence (pA). A 1.2-kb XbaI–EagI fragment (designated 5′ probe) spanning exon 1 was used in the Southern-screening strategy and detected a 12-kb wild-type PstI fragment in the untargeted locus or a 9.3-kb PstI fragment in the targeted locus. The positions of the primers for nested PCR genotyping of cultured embryos up to day 3.5 + 4 are shown by the horizontal arrows in (ii) and (iii). Wild type primer pairs include BI3H-BI3I for the first round and BI3J-BI3K for the second round of amplification (closed horizontal arrows in ii), giving a final product of 161 bp for the untargeted allele. Beta-geo primer pairs include GF1–GR1 for first-round synthesis and GF2–GR2 for second-round synthesis (open horizontal arrows in iii), giving a final product of 248 bp for the targeted allele. Crosses denote expected sites of homologous recombination. Abbreviations for restriction enzymes are (E) EagI, (P) PstI, (Sc) SacI, (S) SalI, and (X) XbaI. (B) Southern blot analysis of wild-type and gene-targeted ES cell lines. The sizes of wild-type 12-kb and homologous recombinant 9.3-kb bands are shown. (C) Nested PCR analysis of cultured 3.5 + 4 day embryos from heterozygous crosses showing the expected bands for the targeted (248 bp) and untargeted (161 bp) alleles.
Production of gene-disrupted mice
For the generation of chimeric mice, heterozygous cell lines from the 129/1 and W9.5 embryonic stem (ES) background were injected into C57BL/6 blastocysts. This resulted in independent germ line–transmitting chimeric mice from the 129/1 and W9.5 substrains carrying the targeted allele. The chimeric mice were successfully bred to produce heterozygous animals.
Embryonic lethality of Bub3 gene-disrupted mice
Bub3+/− mice were healthy, fertile, and showed no apparent phenotype due to haplo-insufficiency. PCR genotype analysis of 91 live-born mice from Bub3+/− intercrosses showed 32 wild-type and 59 heterozygote animals with no homozygous mutants detected. The observed wild type:heterozygote:mutant homozygote ratio of ∼1:2:0, therefore, suggests an embryonic-lethal phenotype for the null mutant.
To further pinpoint the time of embryonic lethality, day 8.5 embryos from heterozygous crosses were removed from the mice and PCR genotyped. No Bub3 null embryos were observed out of a sample of 29 embryos, which consisted of eight wild types and 21 heterozygotes. This result suggested that embryonic death occurred prior to day 8.5 pc.
Morphological degeneration of early embryos
For morphological study and the further determination of the time of embryonic lethality, blastocysts were obtained from heterozygous crosses at day 3.5 and cultured in ES cell media. The embryos were monitored and photographed after 2, 3, and 4 days in culture (denoted as days 3.5 + 2, 3.5 + 3, and 3.5 + 4, respectively). At the end of day 3.5 + 4, the embryos were harvested and genotyped using the nested PCR strategy outlined in Figure 1. Of the 32 embryos studied, 10 +/+, 15 +/−, and 7 −/− were obtained (Fig. 1C). All 7 −/− embryos, but not any of the +/+ and +/− embryos, showed a rapidly degenerating phenotype (see below), suggesting a complete correlation between morphological deterioration and the −/− genotype. This result, combined with those for the day 8.5 embryos and the live-born pups, provided evidence that Bub3−/− embryos were viable at day 3.5 in utero and persisted in culture up to day 3.5 + 4 but were completely resorbed or proceeded to degenerate beyond experimental detection by the time they reached day 8.5 in utero.
By phase-contrast microscopy, no significant morphological difference was observed between the different genotypes at day 3.5 or 3.5 + 1 (not shown). In both the wild-type and heterozygous embryos, the transition from day 3.5 + 2 to 3.5 + 4 was characterized by an increase in the size of the inner cell mass and the trophectoderm (Fig. 2). This increase was due to the rapidly dividing inner cell mass population, whilst the cells of the outer trophectoderm simply increased in size without undergoing many divisions. In contrast, the inner cell mass of all the Bub3−/− embryos were significantly smaller in size and were obviously degenerated by day 3.5 + 3. The trophectoderm, however, appeared morphologically indistinguishable from those of the wild-type and heterozygous littermates throughout the culture period.
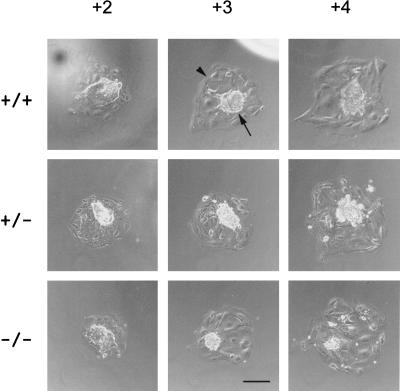
Figure 2.
Morphology of cultured blastocyst outgrowths from heterozygous crosses. Cultured wild-type, heterozygous and homozygous embryos were photographed using phase microscopy, from day 3.5 + 2 to +4. The tightly packed inner cell mass is found in the center of all the embryos (arrow), surrounded by the larger, flat cells of the trophectoderm layer (arrowhead). Scale bar represents 200 μm.
Severe mitotic phenotype in null embryos
To further examine the null phenotype, cultured embryos from heterozygous crosses were fixed and then stained with DAPI. Since the results of genotype analysis has indicated a complete concordance between embryo deterioration and the −/− genotype, and because of difficulties associated with PCR genotyping on fixed and stained cells, deteriorating or affected embryos were designated as presumed null mutants without further genotyping in the following studies. At day 3.5, no observable difference in nuclear morphology was detected between the embryos. After one day in culture, eight of 30 embryos exhibited a significantly increased number of micronuclei (Fig. 3B,C), compared to the normal littermate (Fig. 3A). An average of 7.4 micronuclei per affected embryo was scored, compared with a baseline level of 1.1 micronuclei for the unaffected embryos (Tab. 1). Other defects observed in the affected embryos included the occasional chromatin bridge and nuclear blebbing (not shown). No significant difference in the mitotic indices was seen between the normal and affected embryos (Tab. 1).
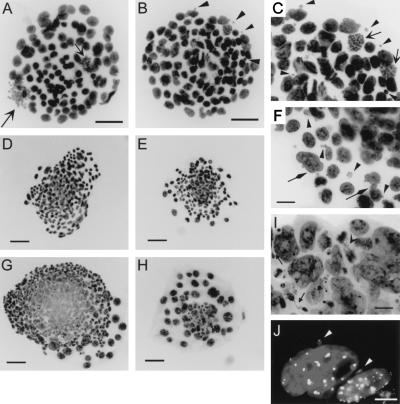
Figure 3.
Nuclear morphology of cultured embryos from heterozygous crosses. (A) DAPI-stained day 3.5 + 1 normal embryo, showing regular interphase nuclei and metaphase chromosomes (open arrow). (B,C) Two different magnifications of a DAPI-stained day 3.5 + 1 affected embryo, showing regular interphase nuclei, metaphase chromosomes (open arrow), and micronuclei (arrowheads). (D) A normal embryo at day 3.5 + 2. (E,F) Two different magnifications of an affected embryo at day 3.5 + 2, showing greatly reduced cell number, irregular nuclei with blebs (diamond arrows) and many micronuclei (some examples shown by arrowheads). (G) A normal day 3.5 + 3 embryo containing over 300 cells, with many metaphases (not shown). (H,I) Two different magnifications of a day 3.5 + 3 affected embryo, showing a dramatic reduction in the number of cells and a worsening mitotic phenotype, exhibited by micronuclei, chromatin bridging (closed arrows) and lagging chromosomes (open arrowhead). (J) Anticentromere CREST immunofluorescence of day 3.5 + 2 affected embryos, showing positively stained micronuclei (arrowheads). Scale bars, (A–B) 50 μm, (D,E,G,H) 100 μm, (C,F,I) 20 μm, and (J) 10 μm.
At day 3.5 + 2, 12 of 42 embryos were clearly affected, as characterized by an increase in the number of micronuclei, irregular nuclear morphology, and smaller inner cell mass (Fig. 3E,F). When immunofluorescence was performed using a previously described anti-CENP-A and CENP-B antisera (CREST#6) (du Sart et al. 1997) on these embryos, the micronuclei seen in the affected embryos were shown to contain one or more chromosomes, providing evidence that these micronuclei represented lagging chromosomes due to missegregation (Fig. 3J). The average cell number for an affected embryo was 130, which was significantly lower than 200 for a normal day 3.5 + 2 embryo (P = 0.0047) (Fig. 3D,E). Thus, while the normal embryos have undergone an approximate doubling in cell number compared to day 3.5 + 1, only a marginal increase of <10% in cell number was seen in the affected embryo over the same culture period (Table 1). The average mitotic index for an affected embryo at day 3.5 + 2 was also noticeably lower than that of a normal embryo at the same stage (3.2% vs. 7.0 %; P = 1.5 × 10−5). These results suggest a slowing down in development at 3.5 + 2 days in the affected group.
Table 1.
Analysis of mitotic phenotype in day 3.5 + 1 untreated or nocodazole-treated embryos from heterozygous crosses
|
|
+/− cross
|
+/+ cross
|
|||||
|---|---|---|---|---|---|---|---|
| Untreated
|
+ Nocodazole
|
+ Nocodazole
|
|||||
| Normal
|
Null
|
P value
|
Normal
|
Null
|
P value
|
Normal
|
|
| No. of embryos | 23 (74%) | 8 (26%) | 21 (78%) | 6 (22%) | 21 (100%) | ||
| No. of cellsa | 110 | 120 | 0.12 | 98 | 100 | 0.56 | 80 |
| Mitotic indexab | 2.1 | 1.6c | 0.55 | 19 | 3c | 6.1 × 10−6 | 17 |
| No. of micronucleia | 1.1 | 7.4d | 3.6 × 10−7 | 1.2 | 17d | 3.6 × 10−7 | 0.65 |
In addition to the normal litter mates from the heterozygous crosses, normal embryos from +/+ crosses were used as further controls to ascertain the effects of nocodazole.
Average value per embryo.
Expressed as a percentage of cells in mitosis over total cell number.
P = 0.071 for the mitotic indices of untreated versus nocodazole-treated null embryos.
P = 0.039 for the number of micronuclei in untreated versus nocodazole-treated null embryos.
P values were derived using the student's t test.
At day 3.5 + 3, ten of 37 embryos were abnormal. The abnormalities were more pronounced than those seen in earlier stages and included a greatly reduced cell number, grossly irregular nuclear morphology, chromatin bridging, lagging chromosomes, and the presence of many micronuclei (Fig. 3H,I). At this stage, accurate scoring of the number of cells or mitoses became difficult because of the large highly compacted inner cell mass in the normal embryos and the grossly deteriorated nuclear morphology of the inner cell population of the affected embryos. The approximate cell number for an affected embryo was estimated to be between 50 and 100 compared with the 300–500 cells in an unaffected littermate, suggesting that while the normal embryos continued to actively divide in culture at day 3.5 + 3, cells in the affected embryos had stopped dividing and/or were disintegrating. In addition, while mitoses were clearly visible in the normal embryos, little or no metaphases were observed in the affected embryos by this stage.
Failure of nocodazole to arrest mitosis
To assess whether the observed phenotype was due to the breakdown of a mitotic checkpoint, day 3.5 + 1 embryos from heterozygous crosses were incubated with 2 μM nocodazole for ∼5 hrs. This drug depolymerizes and destroys the function of the microtubules. The results indicated that in the presence of nocodazole, mitosis in the normal embryos from heterozygous or control wild-type crosses was severely arrested, as evident from a great increase (by eight–ninefold) in mitotic index from an untreated value of 2.1% to treated values of 19% and 17%, respectively (Table 1; also cf. Fig. 4A with Fig. 3A). In stark contrast to this increase in mitotic index, nocodazole treatment did not result in a significant alteration in the mitotic index of the null embryos (3%) compared to the untreated null embryos (1.6%) (Table 1). However, following the drug treatment, a noticeable deterioration of the mitotic missegregation phenotype was evident from the greatly increased number of micronuclei in the null embryos (from 7.4 micronuclei to 17 micronuclei per embryo before and after treatment) (Table 1; also cf. Fig. 4B with Fig. 3B). Other defects observed were an increase in irregular nuclear morphology, nuclear bridging, and blebbing in the treated null embryos. Control experiments using nocodazole-treated day 3.5 + 1 embryos from wild-type crosses did not result in embryos with a phenotype corresponding to that found in the null embryos (Table 1). This suggests that the effects of this drug seen in the null embryos from the heterozygous crosses were specifically related to the Bub3−/− genotype.
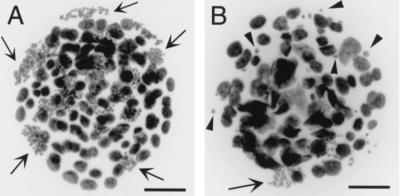
Figure 4.
Nocodazole-treated day 3.5+ 1 embryos from heterozygous crosses. (A) A normal embryo, showing regular-sized nuclei and many arrested metaphases (some examples indicated by arrows). (B) A similarly treated null embryo, showing irregularly sized nuclei, very few metaphases (arrow), and a large number of micronuclei (selected examples shown by arrowheads). Scale bars represent 50 μm.
Discussion
In order to understand the role of mitotic checkpoint control in mouse development, we have created Bub3 gene-disrupted mice. Bub3 is part of a protein complex that interacts with the kinetochore before all chromosomes have achieved bipolar attachment to the mitotic spindle. By deleting exons 2 and 3 of the Bub3 gene, we have interrupted the protein at amino acid No. 65, causing the loss of the Bub1 interaction domain and three of the four WD40 repeat regions (Taylor et al. 1998; Martinez-Exposito et al. 1999). Success in generating a relatively large number of heterozygous knockout ES cell lines indicates that the gene-targeting strategy is efficient and that the growth of heterozygous cells in culture has not been compromised.
Heterozygous Bub3 mice show no apparent abnormalities in development or fertility, showing that one functional copy of the gene is sufficient for normal development. Genotyping of the progeny of heterozygous crosses indicates the absence of Bub3−/− pups and suggests an embryonic lethal phenotype. Morphological analysis of embryos grown in culture from the blastocyst stage (day 3.5) demonstrates a dramatic decrease in the size of the inner cell mass of the null embryos from +3 days to +4 days in culture, suggesting that rapid cell divisions have ceased in the mutant embryos by these stages. Closer examination of the embryos from the +/− crosses at the nuclear level reveals mitotic abnormalities in approximately 25% of day 3.5 + 1, 2, and 3 embryos, with no evidence of such abnormalities in any of the progeny of wild-type crosses. Determination of cell number indicates that mitotic division in the null embryos has decreased greatly at day 3.5 + 2 and ceased completely by day 3.5 + 3. By this latter stage, the embryos have accumulated such abundance of mitotic errors, in the form of micronuclei, nuclear bridging, and abnormal nuclear morphology, that cessation of embryo development becomes inevitable. These results show that Bub3 is essential for normal mitosis and for early embryonic development in the mouse. Such an essential role, therefore, contrasts the phenotype seen in previously reported null mutants of the BUB3 gene in S. cerevisiae and Aspergillus nidulans because these mutants are viable, albeit slower in growth relative to the wild type (Hoyt et al. 1991; Efimov and Morris 1998).
When compared to a number of other essential centromere proteins that have recently been disrupted in the mouse, such as Cenpc, Incenp and Cenpa, the Bub3−/− mice show a similar phenotype in that the accumulation of severe mitotic errors contribute directly to early lethality (Kalitsis et al. 1998; Cutts et al. 1999; Howman et al. 2000). Closer examination reveals that the Bub3−/− embryos appear to survive slightly longer, by two to three days in culture. This may be explained by the fact that a defect in the constitutive centromere proteins Cenpa and Cenpc is expected to immediately disrupt centromere function, leading to a rapid breakdown in kinetochore-microtubule binding and mitotic arrest. Similarly, perturbation of the Incenp protein results in a gross dysfunction in cytokinesis that is expected to have an immediate detrimental effect on mitotic progression. In comparison, a defect in a protein such as Bub3 that plays a checkpoint role (discussed below) may allow mitosis to proceed aberrantly for a slightly greater number of cycles before the accumulation of excessive errors eventually brings it to a halt.
In previous studies on the BUB and MAD genes in yeast and higher eukaryotes, spindle-depolymerizing drugs have been used to assess whether the mutants have a compromised spindle assembly checkpoint. These mutants characteristically fail to arrest in response to mitotic spindle damage, leading to chromosome missegregation. We have used a similar strategy to investigate whether Bub3 functions as a component of a mitotic spindle checkpoint in the developing mouse embryo. Examination of the effect of the spindle-disrupting drug nocodazole on normal embryos indicates that, in the presence of this drug, normal embryos are severely blocked in mitosis, as evident from a large increase in mitotic index, compared to untreated normal embryos. This result suggests that early mouse embryos utilize a mitotic spindle checkpoint mechanism that is sensitive to spindle-depolymerizing drugs, similar to those that have been described in other organisms and somatic cell lines.
To investigate the specific role of Bub3, we studied the effect nocodazole has on the Bub3 null embryos at day 3.5 + 1, when the total cell number and the extent of mitotic disarray are still sufficiently moderate to allow accurate ascertainment of mitotic indices and micronuclei numbers. At this time point, null embryos that have not been drug treated show clear signs of mitotic errors, as indicated by an approximately sevenfold increase in the number of micronuclei per embryo compared to an untreated normal embryo. However, the relatively unaltered mitotic index in the untreated null embryos compared to the normal embryos indicates that cell division appears to proceed at a normal rate. This mitotic index remains unaffected when the null embryos are challenged with nocodazole, which drastically contrasts with the situation in the normal embryos, in which treatment with the drug precipitates a large rise in mitotic index. This observation indicates that the Bub3 null embryos are able to escape the block imposed by the mitotic spindle checkpoint pathway that operates upon the normal embryos. A predicted consequence of an escape from such a checkpoint, especially when the mitotic process is seriously compromised by a potent spindle-disrupting drug, is an elevation in the level of mitotic errors. This prediction is supported by the observed significant increase in the number of micronuclei, from 7.4 per untreated null embryo to 17 per nocodazole-treated null embryo. These results are characteristic of mutants that are defective in mitotic spindle checkpoint mechanisms (Hoyt et al. 1991; Li and Murray 1991; Taylor and McKeon 1997; Cahill et al. 1998; Basu et al. 1999), thus lending strong support that mouse Bub3 participates directly in such a checkpoint system.
Genetic and biochemical evidence indicates that Bub3 interacts with other proteins, including Bub1, to form a checkpoint kinase complex (Hoyt et al. 1991; Roberts et al. 1994; Basu et al. 1998; Farr and Hoyt 1998; Taylor et al. 1998; Brady and Hardwick 2000). Since the presence of both Bub3 and Bub1 is required for localization to the kinetochores that have not completely attached to the mitotic spindle, mutations in these two proteins are expected to share a common phenotype. Consistent with this, genetic disruption of the Drosophila bub1 gene, which also leads to the depletion of Bub3 at the kinetochore (Basu et al. 1998, 1999), results in embryonic lethality at the larval/pupal transition stage. Cells from these null embryos fail to block in mitosis in response to microtubule-depolymerizing drugs and contain severe mitotic abnormalities (lower mitotic index, premature chromatid separation, lagging chromosomes, and chromatin bridging). Recently, Mad2, another essential component of the MAD/BUB checkpoint complex has been disrupted in the mouse (Dobles et al. 2000). The phenotype of the Mad2−/− and Bub3−/− embryos share many features. Both mutants show few cells in mitosis beyond day 6.5, contain chromosome segregation errors in the form of lagging chromosomes, and fail to arrest in the presence of nocodazole. Taken together, the results of the above studies in mouse and Drosophila clearly indicate that the MAD/BUB complex constitutes a crucial mitotic checkpoint for early embryonic development, and that the mitotic checkpoint pathway is not only conserved at a molecular level but also at the functional level within metazoans.
Materials and methods
Targeting construct
Three BAC clones—195c21, 177c15, and 24e04—were identified in a mouse ES 129 BAC library (Incyte Genomics). DNA pools were screened using PCR with primers, B1 (5′-AGAAACGTTGCTTAGGCGG-3′), and B2 (5′-GAACTCGGAGTACCTTAACC-3′), which spanned exon 1 and 2 of the mouse Bub3 gene, generating a PCR product size of 437 bp. BAC#177c15 was used for further subcloning. A 12-kb PstI genomic fragment containing exons 1 to 7 was cloned into the PstI site of pAlter (Promega; Fig. 1A). A 3.1-kb SacI/SalI fragment spanning exons 3 and 4 was removed and then replaced by a splice-acceptor/IRES/lacZ-neomycin cassette.
Transfections and chimera production
Mouse ES cell culture and manipulations were carried out using standard procedures (Kalitsis et al. 1998). For the creation of heterozygous cell lines, 50 μg of linearized DNA was electroporated at 0.8 kV, 3 μF, ∞ Ω into 5 × 107 mouse 129/1 or W9.5 ES cells and selected with 200 μg/ml of G418 (GIBCO BRL). Correctly targeted events were detected using Southern hybridization. A XbaI/EagI 1.2-kb fragment containing exon 1 was used as a 5′ external probe in the hybridization assay (Fig. 1A). Genomic DNA was digested with PstI, generating a 12-kb wild-type or 9.3-kb targeted band. Targeted ES cell lines were used in the generation of chimeric and heterozygous mice using standard methods (Kalitsis et al. 1998).
Genotyping of mice
Mouse tail DNA was extracted using standard techniques. For PCR genotyping, a duplex strategy was employed using the following primers: BI3G (5′-AGTGAATGACCAAACCTGGG-3′) and BI4F (5′-CAACAGCACACTCTCCAACC-3′) for the wild-type allele (407 bp), and GF2 (5′-CCATTACCAGTTGGTCTGGTG-3′) and GR2 (5′-CCTCGTCCTGCAGTTCATTC-3′) for the targeted allele (248 bp).
Culturing and genotyping of mouse embryos
Embryos were dissected out at day 3.5 and cultured in ES media at 37°C 5% CO2 for the required duration. After culturing, the DNA was extracted and resuspended in 10 μl of TE. Three μl of the embryo DNA was used in the first round of a nested PCR strategy. The first-round primers BI3H (5′-GATGCCTTTGCGTTCTTAGC-3′) and BI3I (5′-GATTCCAGGAGCAGCATCA-3′) (for the wild-type allele) and GF1 (5′-AGTATCGGCGGAATTCCAG-3′) and GR1 (5′-GATGTTTCGCTTGGTGGTC-3′) (for the targeted allele) were used in a duplex reaction, using the following conditions: first cycle at 95°C for 2 min, 55°C for 3 min, 72°C for 90 sec, and cycles 2–30 at 95°C for 60 sec, 55°C for 60 sec, 72°C for 90 sec, in a final reaction volume of 25 μl. For the second round, 1 μl of the first-round amplified product was used in separate wild-type BI3J (5′-TGTGGCAGGATTTGGAATG-3′) and BI3K (5′-TGTGCTTCTCAGTCCACTCG-3′) and targeted (GF2 and GR2) reactions, producing 161- and 248-bp bands, respectively. The conditions for the second-round amplification were first cycle at 95°C for 2 min, 57°C for 60 sec, 72°C for 90 sec, and cycles 2–30 at 95°C for 60 sec, 59°C for 60 sec, 72°C for 90 sec, in a final reaction volume of 20 μl.
Nuclear staining, immunofluorescence, and statistical analysis
Day 3.5 and 3.5 + 1 embryos were fixed and stained in DAPI, as previously described (Kalitsis et al. 1998). Embryos cultured on coverslips were washed three times in PBS and fixed in 4% paraformaldehyde/PBS for 15 min, followed by 0.1% Triton X-100/PBS for 10 min. The embryos were then rinsed twice in PBS and stained in DAPI and Vectashield (Vector Laboratories). For immunostaining of the kinetochore using the CREST#6 autoimmune serum, day 3.5 + 2 embryos were treated as described previously (Howman et al. 2000). Images were captured using an Axioplan2 microscope (Zeiss), with a Sensys-cooled CCD camera (Photometrics) and processed with IP Lab software (Scanalytics). Statistical analyses were performed using the Student's t-test.
Acknowledgments
We thank S. Gazeas, A. Sylvain, J. Ladhams, and P. Rasaratnam for excellent technical assistance; R. Saffery, J. Craig, and D. Magliano for helpful comments; and G. Kay for the 129/1 and J. Mann for the W9.5 ES cell lines. This work was supported by the National Health and Medical Research Council of Australia. K.H.A.C. is a Principal Research Fellow of the council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL choo@cryptic.rch.unimelb.edu.au; FAX 61-3-9348-1391.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.827500.
References
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Basu J, Logarinho E, Herrmann S, Bousbaa H, Li Z, Chan GK, Yen TJ, Sunkel CE, Goldberg ML. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, Sunkel CE, Goldberg ML. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts SM, Fowler KJ, Kile BT, Hii LL, O'Dowd RA, Hudson DF, Saffery R, Kalitsis P, Earle E, Choo KHA. Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum Mol Genet. 1999;8:1145–1155. doi: 10.1093/hmg/8.7.1145. [DOI] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao J, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KHA. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nature Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics. 1998;149:101–116. doi: 10.1093/genetics/149.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ, Chen RH, Murray AW. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KHA. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–17. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Earle E, Hill J, Choo KHA. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc Natl Acad Sci USA. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Martinez-Exposito MJ, Kaplan KB, Copeland J, Sorger PK. Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc Natl Acad Sci USA. 1999;96:8493–8498. doi: 10.1073/pnas.96.15.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]