Abstract
A widely held viewpoint in the field of predictive biomarkers for disease holds that no single marker can provide high enough discrimination and that a panel of markers, combined in some type of algorithm, will be needed. Motivated by a recent study where 27 additional markers for ovarian cancer, many of which had good predictive value alone, failed to substantially increase the predictive ability of the primary marker of CA125, we explore the effect of additional markers on the area under the ROC curve (AUC). We develop a statistical model based on the multivariate normal distribution and linear algorithms and use it to explore how the magnitude and direction of statistical correlation among the markers (in diseased and in non-diseased) is critical in determining the added predictive value of additional markers. We show mathematically and empirically that if the additional marker(s) is negatively correlated with the primary marker, then it will always be able to provide increased AUC when combined with the primary marker (as compared to that obtained with the primary marker alone), even if it has little predictive ability on its own. In contrast, if the additional marker(s) is positively correlated with the primary marker, then it is unlikely to substantially increase the AUC when combined with the primary marker, even when it has good predictive ability on its own. Thus, univariate analyses alone may not be the best approach in choosing which markers to combine in a predictive panel of markers; patterns of statistical correlation should be considered in ranking top-performing biomarkers.
Keywords: correlation, ROC AUC, biomarkers, multivariate normal distribution, linear algorithm
Introduction
For various diseases, including many types of cancer, population screening using markers in the blood or urine is thought to be a promising strategy for reducing disease-associated morbidity and mortality. A widely held viewpoint of research on early biomarkers of disease is that no single marker can provide high enough discrimination between cases and non-cases for a screening test destined for clinical applications and that the use of multiple markers, combined in some type of algorithm, will be necessary in order to produce the requisite level of predictive ability.1–3
In a recent ovarian biomarker study, five such algorithms, each combining 6–8 biomarkers, and representing in total 28 distinct markers, were evaluated for their performance improvement over CA125 alone. The results were both surprising and intriguing: additional biomarkers, including those with near-comparable performance to CA125 by itself, added little to the predictive performance of CA125 alone when combined with CA125.4,5
In this report, we examine this phenomenon in more detail through statistical and mathematical analysis. We provide evidence, theoretical and confirmed with empirical data, that patterns of statistical correlation of a primary marker with other potential markers predict the extent to which those other markers will add value to a primary marker. Specifically, we show that a marker negatively correlated with a primary marker will tend to have good additive value and a marker positively correlated with a primary marker will tend to have poor additive value.
We first build a statistical framework for the problem and mathematically derive some basic results about the effects of correlations on added predictive value. We then use the ovarian cancer marker study referred to above to illustrate how our main theoretical findings play out with real-world data.
Statistical Framework for Analyzing the Effect of Marker Correlations on Added Predictive Value
The formal statistical framework that we develop here allows for a rigorous analysis of the effect of the pattern of correlations between markers on the added predictive ability of marker combinations as measured by the area under the ROC curve (AUC), a common metric of predictive performance. Added predictive ability is defined as the increase in AUC over the level obtained with the primary marker alone. In order to derive analytic results, we make two assumptions, one concerning the distributions of the marker levels in the populations of interest and the other concerning the nature of the algorithm for combining the multiple marker values. Later, we will show that in practice the basic findings of this analysis are still generally upheld even under deviations from these assumptions.
In biology, the term “correlation” is often used loosely to denote a relationship between two factors or effects. In statistics, correlation has a specific definition with respect to the values of a pair of quantitative variables (eg, concentrations of two markers). The level of correlation, as summarized in the correlation coefficient r, measures the extent or the tendency of one variable to increase (positive correlation) or decrease (negative correlation) as the other variable increases. The correlation coefficient r ranges from −1 to 1, with 1 indicating perfect correlation, 0 no correlation or independence, and −1 perfect negative correlation. On a two dimensional scatter plot, the more closely the points cluster around a positively (negatively) sloped regression line, the higher the magnitude of positive (negative) correlation.
The first assumption we make, about the distribution of marker levels, is that, within cases and within controls, each marker is lognormally distributed, ie, the log of the marker concentration is normally distributed. For many, but not all, markers, levels are approximately lognormal. Further, we assume that the multivariate normal (MVN) distribution describes the distribution (in cases and in controls) of log values of a set of markers of interest. The MVN specifies not only the parameters (mean and standard deviation) of the normal distribution of each (log) marker, but also the correlations between the markers.
The second assumption concerns the nature of the multi-marker algorithm. Specifically, we assume that the algorithm is linear, ie, that it is a weighted sum of (log) marker concentrations. With a linear algorithm, along with the MVN assumption about marker distributions, one can analytically compute the following: (1) the weights for the optimal algorithm involving all the markers in the set and (2) the AUC of the resulting optimal algorithm.6 Formulas for these are given in the Appendix.
We now demonstrate, and also mathematically prove (in Appendix), several important qualitative properties relating the pattern of marker correlations with the ability of a multi-marker linear algorithm to provide increased predictive ability (AUC). We initially concentrate on the case with only two markers, but later sketch out how the results can be extrapolated to three or more markers.
With two population groups, cases and controls, there is a separate MVN distribution for each group; thus for any pair of markers there are two correlation parameters, one in cases (say r1) and one in controls (say r0). Note that, in practice, these two correlation coefficients are often quite different. The pivotal quantity in determining the ability of the 2nd marker to add predictive value to a primary marker turns out to be a weighted average of r1 and r0, which we denote by C. Specifically, C = [σ11σ12 r1+ σ01σ02r0]/A, where σij is the standard deviation of the distribution of (log) marker j (j = 1,2) in group i (1 = cases, 0 = controls) and A = σ11σ12 + σ01σ02. Note that because the weights are positive, both correlations being negative assures that C is negative and both being positive assures that C is positive.
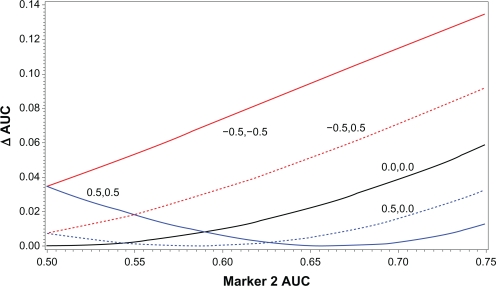
Figure 1 illustrates, for the situations of C > 0, C = 0, and C < 0, how the increase in AUC of the optimal two marker combination over that of the primary marker alone (denoted ΔAUC) is related to the AUC of the 2nd marker alone (denoted AUC-2). For C ≤ 0, ΔAUC is monotonically increasing as a function of AUC-2. Further, the greater the negative value of C, the higher the curves are throughout the entire range of AUC-2. In contrast, for C > 0, the curves have a quadratic shape. As AUC-2 increases from the null level (AUC = 0.5), ΔAUC decreases initially, eventually reaches the 0 mark, and finally begins increasing. Note that ΔAUC = 0 implies that the 2nd marker does not add at all to the AUC of the primary marker alone; in this case the optimal weight for the 2nd marker is 0. Unlike the C ≤ 0 situation, the curves for different C > 0 values intersect each other, with none being above another for the entire range of AUC-2.
Figure 1.
Relationship between correlation and ΔAUC for 2-marker combinations. ΔAUC for 2-marker combination is plotted against AUC of marker 2 alone; each curve represents different values of the correlation of marker 1 with marker 2. Solid black line is 0 correlation in both cases and controls. Two red lines are correlations of −0.5 in cases and controls (solid line) and correlation of −0.5 in cases and 0 in controls (dotted line). Two blue lines are correlations of 0.5 in cases and controls (solid line) and correlation of 0.5 in cases and 0 in controls (dotted line). Direction of regulation is assumed the same for both markers (ie, both up-regulated in cases or both down-regulated in cases).
From the figure, it is clear that the AUC of the 2nd marker alone does not by itself determine the level of increase in AUC with the combination. A marker with negative correlation (C < 0) may have lower AUC-2 than a marker with positive correlation but still have substantially greater ΔAUC. Even for the same level of positive correlation, a lower AUC-2 value may give rise to a greater value of ΔAUC.
To illustrate how, in the C > 0 situation, a second marker with predictive ability of its own may fail to add anything to the AUC of the primary marker alone, suppose that the second marker is simply the primary marker plus some additive noise (eg, measurement error). Then, although the second marker has predictive value, it clearly cannot add any predictive value to the noiseless version of itself. Note in this (added noise) situation the two markers will necessarily be positively, but not perfectly, correlated in cases and in controls.
Note also from the figure that both in the C > 0 and the C < 0 situation (but not with C=0), a second marker with no predictive ability of its own (AUC-2 = 0.5) will necessarily add something to the AUC of the primary marker alone when optimally combined with it, a finding that may seem counter-intuitive. To help understand this phenomenon, consider predicting a person’s sex based on their own height (primary marker) and their father’s height (marker 2). Clearly, one’s father’s height is not at all predictive of sex, but is correlated with one’s own height. Consider an individual who is 5 foot 10 inches. With that information alone, one would predict that the person is male, since there are many times more men than women of that height. However, suppose we had additional information that the person’s father was 6 foot 6 inches. Then, such a man’s son would likely be much taller than 5′10″, and such a man’s daughter might likely be 5′10″, which could shift the prediction to female.
All of the above results can be demonstrated and proven mathematically; this is described in the Appendix.
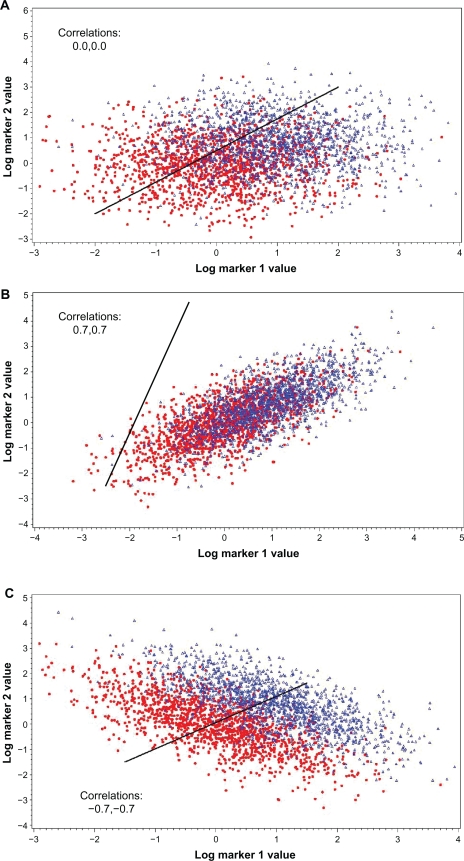
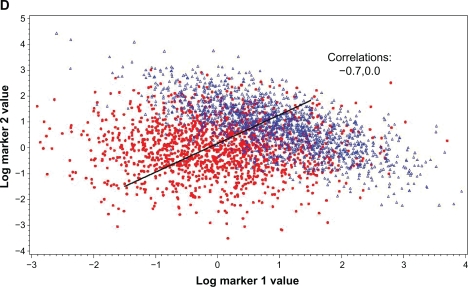
The effect of correlation on the ability of a second marker to add to the predictive ability of a primary marker can also be demonstrated with scatter plots. Figures 2A, B and C show the situation with C = 0, C > 0 and C < 0, respectively. Compared to the no correlation (C = 0) situation, the ability of the two markers to differentiate between cases and controls is substantially diminished when C < 0 and is substantially enhanced when C < 0 (note that the univariate AUCs of the markers are the same over all three plots). Also shown is Figure 2D, where the correlation in cases is the same as in Figure 2C but the correlation in controls is zero; the degree of differentiation between cases and controls is a bit less here than in 2C.
Figure 2.
Scatter plots of marker 1 by marker 2 values for cases (blue triangles) and controls (red dots). Individual AUCs of marker 1 and 2 are 0.760 and 0.714, respectively. Correlation (in both cases and controls) is 0.0 in (A), 0.7 in (B) and −0.7 in (C); in (D), correlation is 0 in controls and −0.7 in cases. AUCs for optimal linear combination are 0.817 in (A), 0.762 in (B), 0.950 in (C) and 0.869 in (D). Black line gives propensity score of optimal linear combination by perpendicular projection of points onto line.
Some of the mathematical results described above for two markers can be extended to the realm of three or more markers. For example, if markers B and C each do not add value when linearly combined (optimally) with marker A, then B and C together will also not add any value when linearly combined (optimally) with A.
We note here a technical consideration. Heretofore, we have been assuming that each marker is up-regulated in cases, ie, that it tends to have higher levels in cases than in controls. Mathematically, it can be shown that, for the purposes of assessing the increase in predictive value, positive correlation for a marker that is up-regulated is equivalent to negative correlation for a marker that is down-regulated and vice-versa (note we are assuming that the primary marker is up-regulated). Therefore, all of the conclusions above about negative and positive correlation are reversed if the secondary marker(s) is down-regulated in cases (or more generally, has the opposite regulation of the primary marker). Thus, if a second marker is down-regulated in cases, positive correlation of it with a primary (up-regulated) marker will lead to greater added AUC and negative correlation to a lesser added AUC. Since most cancer biomarkers are up-regulated in cases, we stated in the abstract and introduction that negative correlations are conducive to added predictive value; the caveat should be added that this is assuming that both markers have the same direction of regulation.
Analysis of Ovarian Biomarker Data
We applied the above statistical framework to the biomarker data from the recent ovarian cancer biomarker study discussed earlier. In this study, five investigator groups assayed 28 different biomarkers, including CA125 (Table 1).4,5 A total of 118 ovarian cancer cases and 951 controls who were enrolled in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial were evaluated for five panels of biomarkers, using the blood sample most proximate (and prior) to the cancer diagnosis in cases and matched controls. The current analysis uses marker data on the 65 cases diagnosed within one year from the date of the serum sample and 439 general population controls for which results from all 28 assays were available.
Table 1.
Ovarian cancer biomarkers evaluated.
| Marker name (Gene symbol) | AUC | Marker # (Rank in AUC) |
|---|---|---|
| Apolipoprotein A-I (APOA1) | 0.527 | 21 |
| Beta-2-microglobulin (B2M) | 0.508 | 27 |
| B7-H4 (VTCN1 ) | 0.662 | 7 |
| CA125 (MUC16) | 0.896 | 1 |
| CA 15–3 (MUC1) | 0.668 | 5 |
| CA 19–9 | 0.502 | 28 |
| CA 72–4 | 0.753 | 3 |
| CTAPIII (PPBP) | 0.525 | 23 |
| EGFR (EGFR) | 0.529 | 20 |
| Eotaxin (CCL11) | 0.569 | 14 |
| HE4 (WFDC2) | 0.797 | 2 |
| Hepcidin-25 (HAMP) | 0.511 | 25 |
| IGFBPII (IGFBP2) | 0.571 | 13 |
| IGF-II (IGF2) | 0.584 | 12 |
| ITIH4 (ITIH4) | 0.522 | 24 |
| Kallikrein-6 (KLK6) | 0.685 | 4 |
| Leptin (LEP) | 0.509 | 26 |
| Mesothelin (MSLN) | 0.665 | 6 |
| MIF (MIF) | 0.600 | 9 |
| MMP-3 (MMP3) | 0.540 | 19 |
| MMP-7 (MMP7) | 0.604 | 8 |
| OPN (SPP1) | 0.527 | 22 |
| Prolactin (PRL) | 0.598 | 10 |
| SLPI | 0.558 | 16 |
| Spondin 2 (SPON2) | 0.597 | 11 |
| sVCAM-1 (VCAM1) | 0.559 | 15 |
| Transferrin (TF) | 0.548 | 17 |
| Transthyretin (TTR) | 0.543 | 18 |
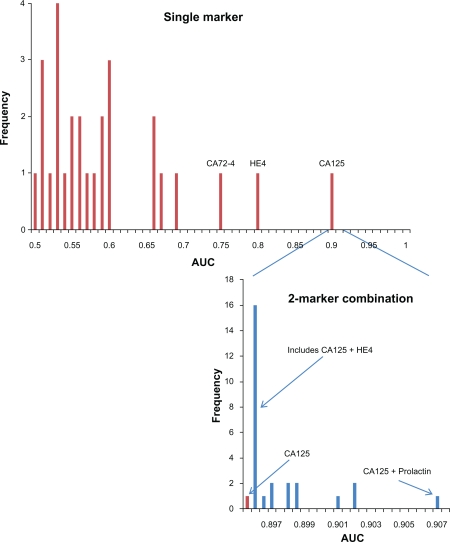
We first calculated the AUC for CA125 and each of the 27 other markers, and then the AUC for all 2-marker (CA125 plus one other marker) optimal linear combinations of log marker values. Figure 3 displays a AUC histogram of CA125 and the 27 other markers individually, as well as all 2-marker combinations (CA125 plus another marker). A number of the markers, aside from CA125, had relatively high AUCs, with 8 markers having AUCs between 0.6 and 0.8. However, the distribution of 2-marker AUCs clustered very closely at the AUC level of CA125 alone. The majority of the markers added essentially zero to the AUC of CA125 alone. Algorithms with 2–4 added markers also added relatively little to the AUC of CA125 (data not shown).
Figure 3.
Histogram of AUCs. Upper panel is histogram of AUCs of 28 individual ovarian markers; lower panel is histogram of AUCs of optimal combination of CA125 and an additional marker.
Next we explored whether the above observation could be explained by the patterns of correlation between the 2-marker pairs as analyzed in the prior section. Table 2 lists, for the top 15 markers, their individual AUCs, their correlations with CA125 and with the two other best performing markers (HE4 and CA72-4), and the increase in AUC (ΔAUC) over that of CA125, HE4 or CA72-4 alone when combined in a linear algorithm with that marker. Note all correlations are computed using the log-transformed marker values. The highest ranked markers in terms of univariate AUC each showed very small values of ΔAUC when combined with CA125, with the markers ranked 2–9 in univariate AUC (HE4 through MMP7; AUC range 0.598–0.797) each having ΔAUC ≤ 0.006. The largest ΔAUC value, of 0.011, was from prolactin, the 10th marker in AUC rank; this marker had a univariate AUC of only 0.598 but was slightly negatively correlated (r = −0.11) with CA125 in cases. In contrast, all but one of the markers ranked 2–9 had positive correlations with CA125 in cases of at least 0.43.
Table 2.
Marker correlations and AUC improvement with 2-marker panel.
| Secondary marker | AUC |
Primary marker |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
CA125 |
HE4 |
CA72-4 |
||||||||
| Correlations1 | ΔAUC2 | ΔAUCPr3 | Correlations1 | ΔAUC2 | ΔAUCPr3 | Correlations1 | ΔAUC2 | ΔAUCPr3 | ||
| CA-125 | 0.896 | – | – | – | – | – | – | |||
| HE4 | 0.797 | 0.72, 0.16 | 0.0009 | 0.002 | – | – | – | – | ||
| CA72-4 | 0.753 | 0.58, −0.05 | 0.001 | 0.0003 | 0.42, −0.01 | 0.031 | 0.017 | – | – | |
| Kallikrein-6 | 0.685 | 0.56, 0.12 | 0.0001 | 0.0004 | 0.57, 0.32 | 0.001 | 0.0003 | 0.43, −0.05 | 0.002 | 0.024 |
| CA15–3 | 0.668 | 0.61, 0.10 | 0.002 | 0.004 | 0.44, 0.15 | 0.0001 | 0.001 | 0.53, 0.0 | 0.0007 | 0.009 |
| Mesothelin | 0.665 | 0.43, −0.08 | 0.001 | 0.0005 | 0.61, 0.35 | 0.0002 | 0.0000 | 0.27, 0.10 | 0.017 | 0.027 |
| B7-H4 | 0.654 | 0.46, −0.01 | 0.006 | 0.0001 | 0.36, 0.07 | 0.014 | 0.006 | 0.41, −0.01 | 0.007 | 0.020 |
| MIF | 0.605 | 0.19, 0.05 | 0.0005 | 0.0002 | 0.30, 0.14 | 0.0004 | 0.0000 | 0.26, 0.04 | 0.0002 | 0.003 |
| MMP7 | 0.598 | 0.51, 0.07 | 0.0007 | 0.003 | 0.49, 0.44 | 0.0003 | 0.002 | 0.46, −0.10 | 0.0006 | 0.004 |
| Prolactin | 0.598 | −0.11, 0.02 | 0.011 | 0.007 | −0.25, −0.06 | 0.034 | 0.020 | −0.15, 0.02 | 0.017 | 0.018 |
| Spondin | 0.597 | 0.25, 0.04 | 0.0004 | 0.0001 | 0.18, 0.25 | 0.001 | 0.001 | 0.08, −0.01 | 0.003 | 0.011 |
| IGF-II | 0.584 | [−0.16, −0.07] | 0.0002 | 0.0001 | [−0.21, −0.20] | 0.0006 | 0.0001 | [−0.12, 0.04] | 0.003 | 0.006 |
| IGFBPII | 0.571 | 0.46, 0.05 | 0.0002 | 0.003 | 0.42, 0.33 | 0.0009 | 0.002 | 0.31, −0.09 | 0.0003 | 0.002 |
| Eotaxin | 0.569 | [0.04, 0.04] | 0.005 | 0.006 | [0.10, 0.04] | 0.026 | 0.012 | [−0.06, −0.04] | 0.004 | 0.008 |
| sVCAM-1 | 0.559 | [−0.16, 0.00] | 0.0004 | 0.0000 | [−0.05, 0.11] | 0.002 | 0.003 | [−0.08, −0.08] | 0.0002 | 0.002 |
Notes: Top 15 markers in terms of univariate AUC are listed.
Correlations (cases, controls) of primary marker with secondary marker. Brackets [,] denote that secondary marker is down-regulated in cases. Primary markers are all up-regulated in cases.
Observed increase in AUC for (optimal) combination of primary and secondary marker over that achieved with primary marker alone; AUCs computed non-parametrically. Bold indicates increase in AUC of 0.01 or more.
Predicted increase in AUC, based on assuming log MVN distribution, for (optimal) combination of primary and secondary marker over that achieved with primary marker alone.
We included the 2nd and 3rd highest markers in terms of AUC, HE4 and CA72-4, in the table for illustrative purposes, supposing that these each were in fact the primary marker. The results were similar to those obtained for CA125, with the markers with highest univariate AUC giving very low ΔAUC values and prolactin, which was negatively correlated (in cases) with both HE4 and CA72-4, giving the greatest ΔAUC value. Note that the largest ΔAUC value in the entire table, 0.034 for prolactin and HE4, corresponded to the largest negative correlation in the table (−0.25 in cases).
Also included in Table 2 are the predicted ΔAUC values (ΔAUCPr). These values were derived assuming that the log marker distributions were actually MVN, and using the formulas described in the statistical framework section and Appendix to calculate ΔAUC from the correlations of the markers (as well as the log means and variances). Note the ΔAUC values themselves (as opposed to the predicted ΔAUC values) were derived non-parametrically and did not assume an MVN, or any, distribution for the log marker values. Generally, ΔAUCPr agreed fairly closely with ΔAUC; the correlation coefficient of the two was r = 0.68 (P < 0.0001), and dichotomizing the ΔAUC values into whether they were above or below 0.005, ΔAUCPr agreed with ΔAUC 85% of the time.
We also examined combinations of greater than two markers. With CA125 fixed as the first marker, there were 351 possible combinations of 3 markers and 2925 combinations of 4 markers. Therefore, finding the optimal combinations for all of the panels in a training set environment could give rise to significant over-fitting. Nonetheless, the optimal 3 and 4 marker panels (including CA125) only gave ΔAUC values of 0.016 and 0.026. This is likely due to the fact that many of the markers are highly positively correlated with CA125.
Discussion
The findings here show that univariate analyses alone may not be that useful in choosing which markers can be productively combined with a given primary marker. An additional marker with substantial predictive ability by itself may add little or no predictive value to that achieved with the primary marker alone; conversely, additional markers with little or no predictive ability on their own may add substantial predictive value. To assess the potential for improvement in predictive ability, the correlations of potential secondary markers with a primary marker should be examined.
Biologically, the finding that the greatest improvement in predictive ability comes from combining markers with negative correlation (at least in cases) is intuitive. Essentially, this is the situation where the multiple markers are picking up different facets of the disease process. A simple, concrete example of this is where there are two sub-types (recognized or not) of the disease and two markers, with each marker differentially expressed in only a single (and different) sub-type. Over both disease sub-types combined, the two markers will then typically show a pattern of negative correlation. The fact that most cancers, including ovarian cancer, are quite heterogeneous, is what has, in part, led many to the conclusion that multiple markers will be needed to identify a high percentage of the cases. Unfortunately, it is often difficult to find such complementary markers.
For diseases with known sub-types (eg, cancers with different histologies), research studies should examine marker levels by sub-type. However, the differences among marker levels across sub-types need to be quite substantial to translate into negative correlations of any magnitude. For example, Minoo et al found significantly greater expression of CDX2 in distal colorectal (CRC) cancers (mean expression 87%) than in proximal CRC (mean expression 70%), whereas they found significantly greater expression of CD44s in proximal CRC (40%) than in distal CRC (25%).7 These differences alone, though, would translate only into a very slight negative correlation (less than 0.05) for these two markers. Mean marker levels need to be markedly different across subtypes to generate substantial negative correlations.
For the ovarian cancer marker data set analyzed here, about half of the cases were the same histology, serous cystadenocarcinoma, with the others being a grab-bag of various histologies. For CA-125, HE4 and the other top markers, no significant differences were observed in mean marker levels between the serous cystadenocarcinomas and the other histologies (taken as a whole). Thus, these markers do not appear to be complementary in terms of the histologies in which they are over-expressed.
Marker correlation in non-diseased populations is likely of small magnitude, as observed in the ovarian cancer biomarker data set. For all 378 pairs of markers, only 3% had correlations in non-diseased of magnitude 0.25 or more, with most of those being positive correlations. In contrast, among the diseased, 19% of pairs had positive correlations of at least 0.25, but only 6% had negative correlations of that magnitude or more. This is not surprising as the majority of the markers are over-expressed in cases, and may be up-regulated under the same pathogenic mechanism. Note for marker pairs with opposite directions of effect, the signs of the correlations were reversed for the above statistics.
Although the current analysis focuses on continuous-valued markers, the same principles with respect to correlation hold for binary markers. In the literature, correlations among binary markers are often not described directly but may be inferred. For example, Ries et al examined 12 MAGE-A antigens for oral squamous cell carcinoma (OSSC).8 They reported that singly, six of these markers were positive at rates between 40% and 55% for OSSC; each was negative in all control subjects. They further reported that with the best combination of six markers, the proportion of subjects with at least one positive marker was 69%. A relatively simple calculation shows that if the six markers were un-correlated, the expected proportion of subjects with at least one positive would be 98%. Strong positive correlations among the markers would be needed to reduce this probability to the observed 69%.
The theoretical findings described here were derived assuming the MVN distribution for the log marker values, and assuming a linear algorithm for combining log marker values. However, we have shown with experimental data that qualitatively, and to a large extent quantitatively as well, these findings hold with real-world marker distributions, not all of which are even approximately lognormal. It should also be noted that the findings assuming an MVN distribution for the log marker values will also hold if the non log-transformed marker values are MVN distributed, as long as the linear algorithm is also defined based on the non-transformed values (in this case, the relevant correlations are computed using the non log-transformed values as well). Of the 28 ovarian cancer markers considered here, 20 were closer to lognormally than normally distributed in both cases and controls, so it is likely that in general for these types of markers, lognormal distributions are more common than normal distributions.
We also explored, with these experimental data, the dependence of our theoretical findings on the assumption of a linear algorithm. Specifically, for the data in Table 2, we additionally fit the optimal quadratic algorithm (note this involves squares and products of log concentrations). We found that the overall results agreed quite well with those obtained using the linear algorithm. The rank correlation of the ΔAUCs (derived with the linear compared with the quadratic algorithm) was 0.76, indicating that those 2-marker combinations with greatest AUC increases with the linear algorithm also tended to yield the greatest increase with the quadratic algorithm. Further research is needed to analyze more generally the relation between marker correlations and AUC increase when non-linear algorithms are employed.
In conclusion, univariate analyses alone may not be the best approach in choosing which markers to combine in a predictive algorithm; patterns of statistical correlation should also be considered.
Appendix
MVN distribution and AUC increase
For a normally distributed variable in cases and controls, with mean u1 and u0 and variance σ12 and σ02, the area under the ROC curve (AUC) is equal to Φ(|u1–u0|/(σ02 + σ12)1/2) where Φ is the normal cumulative distribution function.6 If X1 … Xn are a group of markers that have a multivariate normal distribution (in both cases and controls), then the above formula applies to any linear combination of the markers, since any such linear combination, eg, a1X1 … + … anXn, or aX in vector form, where the ai are parameters (weights), is also normally distributed.
Denoting by Σ0, Σ1 the variance/covariance matrix of the vector X in controls and cases, and μ0, μ1 the mean vectors of X in controls and cases, respectively, the optimal linear combination vector A* is proportional to (Σ0 + Σ1)−1 (μ1–μ0) and the optimal AUC is given as Φ( [(μ1–μ0)T(Σ0 + Σ1)−1(μ1–μ0)]½).6
For two markers, denote (d1,d2) = (μ1–μ0)T; ie, these are the differences in means between cases and controls for markers 1 and 2. It is straightforward to show in the 2-marker case that the derivative of the square of the argument of the optimal AUC generating function Φ with respect to d2, denoted d2', is as follows:
d2' = [−2(Σ0(1,2) + Σ1(1,2))d1 + 2d2(Σ0(1,1) + Σ1(1,1))]/Det(Σ0+ Σ1), where the denominator is the determinant of the matrix Σ0 + Σ1.
Since Φ is monotone increasing, as is the square root function, then d2' < 0 (>0) indicates that the AUC must be decreasing (increasing). Assuming d1 > 0 (ie, marker 1 is up-regulated), then d2' > 0 for d2 ≥ 0 if Σ0(1,2) + Σ1(1,2) < 0. Setting σij = Σi(j,j)1/2, and noting that the correlations ri = Σi(1,2)/(Σi(1,1) Σi(2,2))1/2, it is seen that Σ0(1,2) + Σ1(1,2) = σ11σ12 r1 + σ01σ02r0. For Σ0(1,2) + Σ1(1,2) > 0, d2' will be negative for small d2; for d2 large enough, with d1 fixed, d2' > 0.
Again for two markers, and setting A1* = 1, it can be shown that A2* = (d2[Σ0(1,1) + Σ1(1,1)]–d1[Σ0(1,2) + Σ1(1,2)])/(d1[Σ0(2,2) + Σ1(2,2)] – d2[Σ0(1,2) + Σ1(1,2)]). Setting d2[Σ0(1,1) + Σ1(1,1)] – d1[Σ0(1,2) + Σ1(1,2)] = 0 and solving for d2 gives the level where A2* = 0, indicating no possible AUC improvement. Assuming d1 > 0, then for d2 > 0, such a d2 exists if and only if Σ0(1,2) + Σ1(1,2) > 0. If d2 = 0, the argument of the AUC generating function Φ can be shown to be equal to d1/[Σ0(1,1) + Σ1(1,1)–(Σ0(1,2) + Σ1(1,2))2/(Σ0(2,2) + Σ1(2,2))]1/2. For the AUC of marker 1 alone, the corresponding argument is d1/[Σ0(1,1) + Σ1(1,1)]1/2, which is smaller as long as Σ0(1,2) + Σ1(1,2) ≠ 0.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Baker S. Identifying combinations of cancer markers for further study as triggers of early intervention. Biometrics. 2000;56:1082–7. doi: 10.1111/j.0006-341x.2000.01082.x. [DOI] [PubMed] [Google Scholar]
- 2.Pepe MS, Thompson ML. Combining diagnostic tests results to increase accuracy. Biostatistics. 2000;1:123–40. doi: 10.1093/biostatistics/1.2.123. [DOI] [PubMed] [Google Scholar]
- 3.Pepe MS, Cai T, Longton G. Combining predictors for classification using the area under the receiver operating characteristic curve. Biometrics. 2006;62:221–9. doi: 10.1111/j.1541-0420.2005.00420.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhu CS, Pinsky PF, Cramer DW. A framework for evaluating biomarkers for early detection: validation of biomarker panels for ovarian cancer. Cancer Prevention Research. 2011;4:375–83. doi: 10.1158/1940-6207.CAPR-10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cramer DW, Bast RC, Berg CE, et al. Ovarian cancer biomarker performance in Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial specimens. Cancer Prevention Research. 2011;4:365–74. doi: 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su JQ, Liu JS. Linear combinations of multiple diagnostic markers. J American Stat Assoc. 1993;88:1350–5. [Google Scholar]
- 7.Minoo P, Zlobec I, Petersen M, Terracciano L, Lugli A. Characterization of rectal, proximal and distal colon cancers based on clinicopathological, molecular, and protein profiles. Int J Oncol. 2010;37:707–18. doi: 10.3892/ijo_00000720. [DOI] [PubMed] [Google Scholar]
- 8.Ries J, Mollaoglu N, Toyoshima T, et al. A novel multiple-marker method for the early diagnosis of oral squamous cell carcinoma. Disease Markers. 2009;27:75–84. doi: 10.3233/DMA-2009-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]